Evaluation of FMR4, FMR5 and FMR6 Expression Levels as Non-Invasive Biomarkers for the Diagnosis of Fragile X-Associated Primary Ovarian Insufficiency (FXPOI)
Abstract
:1. Introduction
2. Material and Methods
2.1. Subjects
2.2. RNA Extraction and cDNA Synthesis
2.3. Digital Droplet Polymerase Chain Reaction (ddPCR)
2.4. FMR1 Molecular Parameters
2.5. Statistical Analysis
3. Results
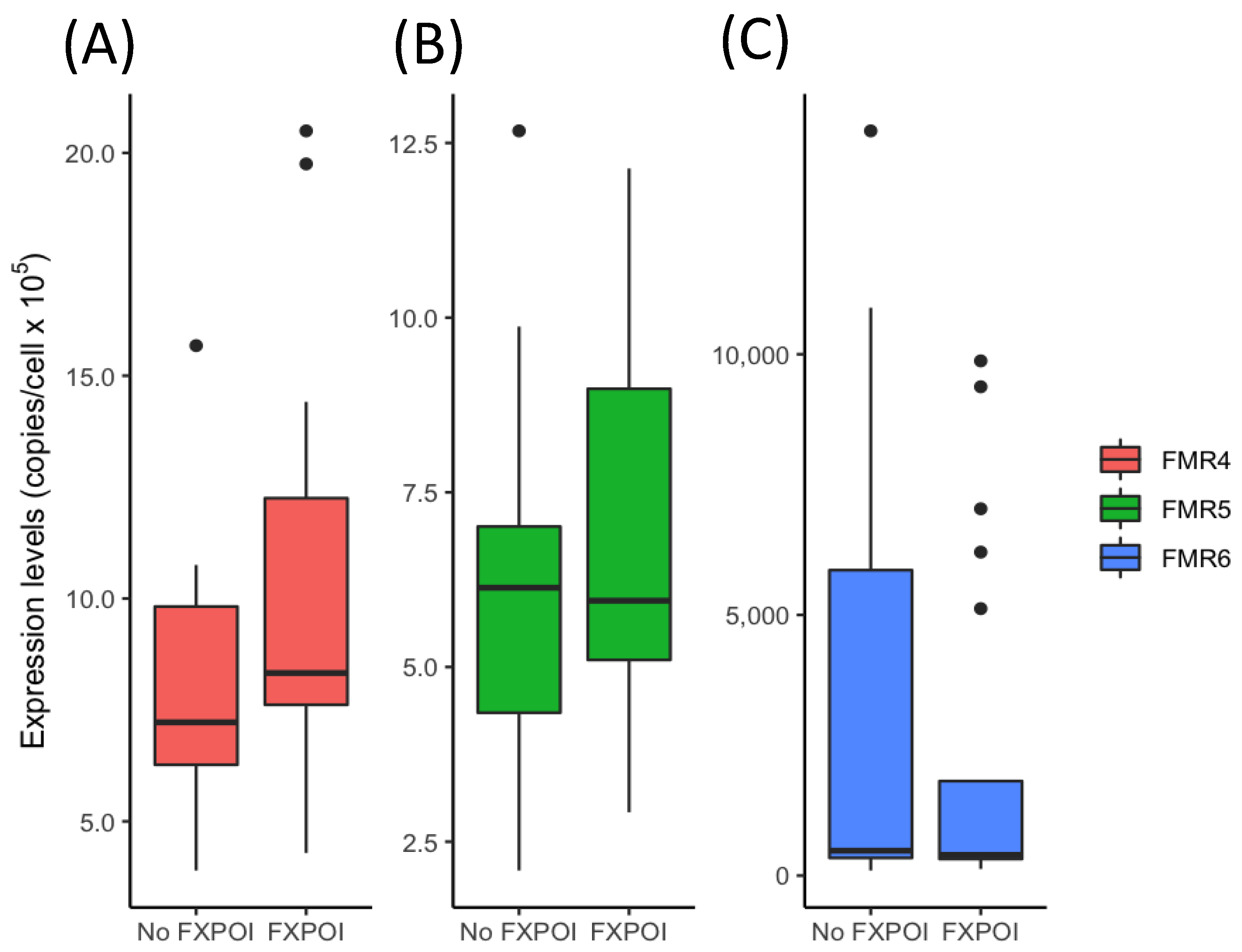
3.1. Expression Levels of FM4, FMR5 and FMR6 in FMR1 Premutation Carriers
3.2. Diagnostic Value of FMR4 for FXPOI
3.3. Association of lncRNAs Expression Levels with FMR1 CGG Repeat Size
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Verkerk, A.J.; Pieretti, M.; Sutcliffe, J.S.; Fu, Y.H.; Kuhl, D.P.; Pizzuti, A.; Reiner, O.; Richards, S.; Victoria, M.F.; Zhang, F.P.; et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 1991, 65, 905–914. [Google Scholar] [CrossRef]
- Oberle, I.; Rousseau, F.; Heitz, D.; Kretz, C.; Devys, D.; Hanauer, A.; Boue, J.; Bertheas, M.F.; Mandel, J.L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science 1991, 252, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Pritchard, M.; Kremer, E.; Lynch, M.; Nancarrow, J.; Baker, E.; Holman, K.; Mulley, J.C.; Warren, S.T.; Schlessinger, D.; et al. Fragile X genotype characterized by an unstable region of DNA. Science 1991, 252, 1179–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutcliffe, J.S.; Nelson, D.L.; Zhang, F.; Pieretti, M.; Caskey, C.T.; Saxe, D.; Warren, S.T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1992, 1, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Kenneson, A.; Zhang, F.; Hagedorn, C.H.; Warren, S.T. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 2001, 10, 1449–1454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tassone, F.; Iong, K.P.; Tong, T.H.; Lo, J.; Gane, L.W.; Berry-Kravis, E.; Nguyen, D.; Mu, L.Y.; Laffin, J.; Bailey, D.B.; et al. FMR1 CGG allele size and prevalence ascertained through newborn screening in the United States. Genome Med. 2012, 4, 100. [Google Scholar] [CrossRef] [Green Version]
- Sherman, S.L. Premature ovarian failure in the fragile X syndrome. Am. J. Med. Genet. 2000, 97, 189–194. [Google Scholar] [CrossRef]
- Hagerman, P.J.; Hagerman, R.J. The fragile-X premutation: A maturing perspective. Am. J. Hum. Genet. 2004, 74, 805–816. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, A.C.; Raspa, M.; Green, A.; Bishop, E.; Bann, C.; Edwards, A.; Bailey, D.B., Jr. Health and reproductive experiences of women with an FMR1 premutation with and without fragile X premature ovarian insufficiency. Front. Genet. 2014, 5, 300. [Google Scholar] [CrossRef] [Green Version]
- Rohr, J.; Allen, E.G.; Charen, K.; Giles, J.; He, W.; Dominguez, C.; Sherman, S.L. Anti-Mullerian hormone indicates early ovarian decline in fragile X mental retardation (FMR1) premutation carriers: A preliminary study. Hum. Reprod. 2008, 23, 1220–1225. [Google Scholar] [CrossRef] [Green Version]
- Bibi, G.; Malcov, M.; Yuval, Y.; Reches, A.; Ben-Yosef, D.; Almog, B.; Amit, A.; Azem, F. The effect of CGG repeat number on ovarian response among fragile X premutation carriers undergoing preimplantation genetic diagnosis. Fertil. Steril. 2010, 94, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Allingham-Hawkins, D.J.; Babul-Hirji, R.; Chitayat, D.; Holden, J.J.; Yang, K.T.; Lee, C.; Hudson, R.; Gorwill, H.; Nolin, S.L.; Glicksman, A.; et al. Fragile X premutation is a significant risk factor for premature ovarian failure: The International Collaborative POF in Fragile X study—Preliminary data. Am. J. Med. Genet. 1999, 83, 322–325. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes. Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [Green Version]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef]
- Huang, G.; Zhu, H.; Wu, S.; Cui, M.; Xu, T. Long Noncoding RNA Can Be a Probable Mechanism and a Novel Target for Diagnosis and Therapy in Fragile X Syndrome. Front. Genet. 2019, 10, 446. [Google Scholar] [CrossRef]
- Ladd, P.D.; Smith, L.E.; Rabaia, N.A.; Moore, J.M.; Georges, S.A.; Hansen, R.S.; Hagerman, R.J.; Tassone, F.; Tapscott, S.J.; Filippova, G.N. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 2007, 16, 3174–3187. [Google Scholar] [CrossRef] [Green Version]
- Khalil, A.M.; Faghihi, M.A.; Modarresi, F.; Brothers, S.P.; Wahlestedt, C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS ONE 2008, 3, e1486. [Google Scholar] [CrossRef] [PubMed]
- Pastori, C.; Peschansky, V.J.; Barbouth, D.; Mehta, A.; Silva, J.P.; Wahlestedt, C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum. Genet. 2014, 133, 59–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peschansky, V.J.; Pastori, C.; Zeier, Z.; Motti, D.; Wentzel, K.; Velmeshev, D.; Magistri, M.; Bixby, J.L.; Lemmon, V.P.; Silva, J.P.; et al. Changes in expression of the long non-coding RNA FMR4 associate with altered gene expression during differentiation of human neural precursor cells. Front. Genet. 2015, 6, 263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elizur, S.E.; Dratviman-Storobinsky, O.; Derech-Haim, S.; Lebovitz, O.; Dor, J.; Orvieto, R.; Cohen, Y. FMR6 may play a role in the pathogenesis of fragile X-associated premature ovarian insufficiency. Gynecol. Endocrinol. 2016, 32, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Zmienko, A.; Samelak-Czajka, A.; Goralski, M.; Sobieszczuk-Nowicka, E.; Kozlowski, P.; Figlerowicz, M. Selection of Reference Genes for qPCR- And ddPCR-Based Analyses of Gene Expression in Senescing Barley Leaves. PLoS ONE 2015, 10, e0118226. [Google Scholar] [CrossRef] [PubMed]
- Amos-Landgraf, J.M.; Cottle, A.; Plenge, R.M.; Friez, M.; Schwartz, C.E.; Longshore, J.; Willard, H.F. X chromosome-inactivation patterns of 1005 phenotypically unaffected females. Am. J. Hum. Genet. 2006, 79, 493–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tassone, F.; Hagerman, R.J.; Taylor, A.K.; Gane, L.W.; Godfrey, T.E.; Hagerman, P.J. Elevated levels of FMR1 mRNA in carrier males: A new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000, 66, 6–15. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Hu, Z.; Feng, Y.; Hu, X.; Yuan, J.; Zhao, S.D.; Zhang, Y.; Yang, L.; Shan, W.; He, Q.; et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015, 28, 529–540. [Google Scholar] [CrossRef] [Green Version]
- Haemmig, S.; Feinberg, M.W. Targeting LncRNAs in Cardiovascular Disease: Options and Expeditions. Circ. Res. 2017, 120, 620–623. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; He, P.; Bian, Z. Long Noncoding RNAs in Neurodegenerative Diseases: Pathogenesis and Potential Implications as Clinical Biomarkers. Front. Mol. Neurosci. 2021, 14, 685143. [Google Scholar] [CrossRef]
- Sullivan, A.K.; Marcus, M.; Epstein, M.P.; Allen, E.G.; Anido, A.E.; Paquin, J.J.; Yadav-Shah, M.; Sherman, S.L. Association of FMR1 repeat size with ovarian dysfunction. Hum. Reprod. 2005, 20, 402–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ennis, S.; Ward, D.; Murray, A. Nonlinear association between CGG repeat number and age of menopause in FMR1 premutation carriers. Eur. J. Hum. Genet. 2006, 14, 253–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, E.G.; Sullivan, A.K.; Marcus, M.; Small, C.; Dominguez, C.; Epstein, M.P.; Charen, K.; He, W.; Taylor, K.C.; Sherman, S.L. Examination of reproductive aging milestones among women who carry the FMR1 premutation. Hum. Reprod. 2007, 22, 2142–2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| FMR1 Premutation with FXPOI (n = 20) | FMR1 Premutation without FXPOI (n = 16) | p-Value | |
|---|---|---|---|
| Age (mean ± SD, years) | 41 ± 6.9 | 47 ± 8.3 | 0.03 * |
| CGG repeat (mean ± SD) | 100 ± 35 | 88 ± 26 | 0.3 |
| FMR1 mRNA (mean ± SD) | 1.5 ± 0.8 | 1.8 ± 1.3 | 0.3 |
| FMR4 expression level | p-Value | |||
| 1–7 | 7–12 | >12 | 0.039 * | |
| FXPOI (n = 20) | 2 (10%) | 12 (60%) | 6 (30%) | |
| No FXPOI (n = 16) | 7(44%) | 8 (50%) | 1 (6%) | |
| FMR5 expression level | 0.14 | |||
| 1–5 | 5–10 | >10 | ||
| FXPOI (n = 20) | 5 (25%) | 10 (50%) | 5 (25%) | |
| No FXPOI (n = 16) | 7 (44%) | 8 (50%) | 1 (6%) | |
| FMR6 expression level | 0.556 | |||
| <400 | 400–1000 | >1000 | ||
| FXPOI (n = 20) | 10 (50%) | 5 (25%) | 5 (25%) | |
| No FXPOI (n = 16) | 6 (38%) | 5 (31%) | 5 (31%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Mora, M.I.; Agusti, I.; Wijngaard, R.; Martinez-Barrios, E.; Barcos, T.; Borras, A.; Peralta, S.; Guimera, M.; Goday, A.; Manau, D.; et al. Evaluation of FMR4, FMR5 and FMR6 Expression Levels as Non-Invasive Biomarkers for the Diagnosis of Fragile X-Associated Primary Ovarian Insufficiency (FXPOI). J. Clin. Med. 2022, 11, 2186. https://doi.org/10.3390/jcm11082186
Alvarez-Mora MI, Agusti I, Wijngaard R, Martinez-Barrios E, Barcos T, Borras A, Peralta S, Guimera M, Goday A, Manau D, et al. Evaluation of FMR4, FMR5 and FMR6 Expression Levels as Non-Invasive Biomarkers for the Diagnosis of Fragile X-Associated Primary Ovarian Insufficiency (FXPOI). Journal of Clinical Medicine. 2022; 11(8):2186. https://doi.org/10.3390/jcm11082186
Chicago/Turabian StyleAlvarez-Mora, Maria Isabel, Ines Agusti, Robin Wijngaard, Estefania Martinez-Barrios, Tamara Barcos, Aina Borras, Sara Peralta, Marta Guimera, Ana Goday, Dolors Manau, and et al. 2022. "Evaluation of FMR4, FMR5 and FMR6 Expression Levels as Non-Invasive Biomarkers for the Diagnosis of Fragile X-Associated Primary Ovarian Insufficiency (FXPOI)" Journal of Clinical Medicine 11, no. 8: 2186. https://doi.org/10.3390/jcm11082186






