Latest Developments in Adapting Deep Learning for Assessing TAVR Procedures and Outcomes
Abstract
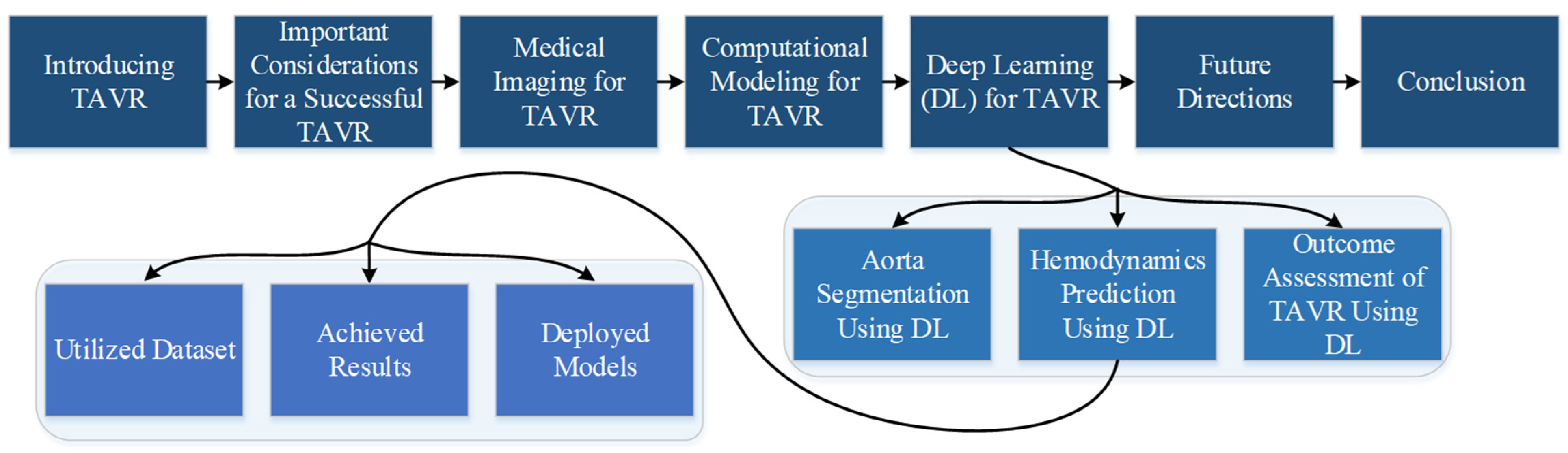
:1. Introduction
2. Important Considerations for a Successful TAVR
3. Medical Imaging for TAVR
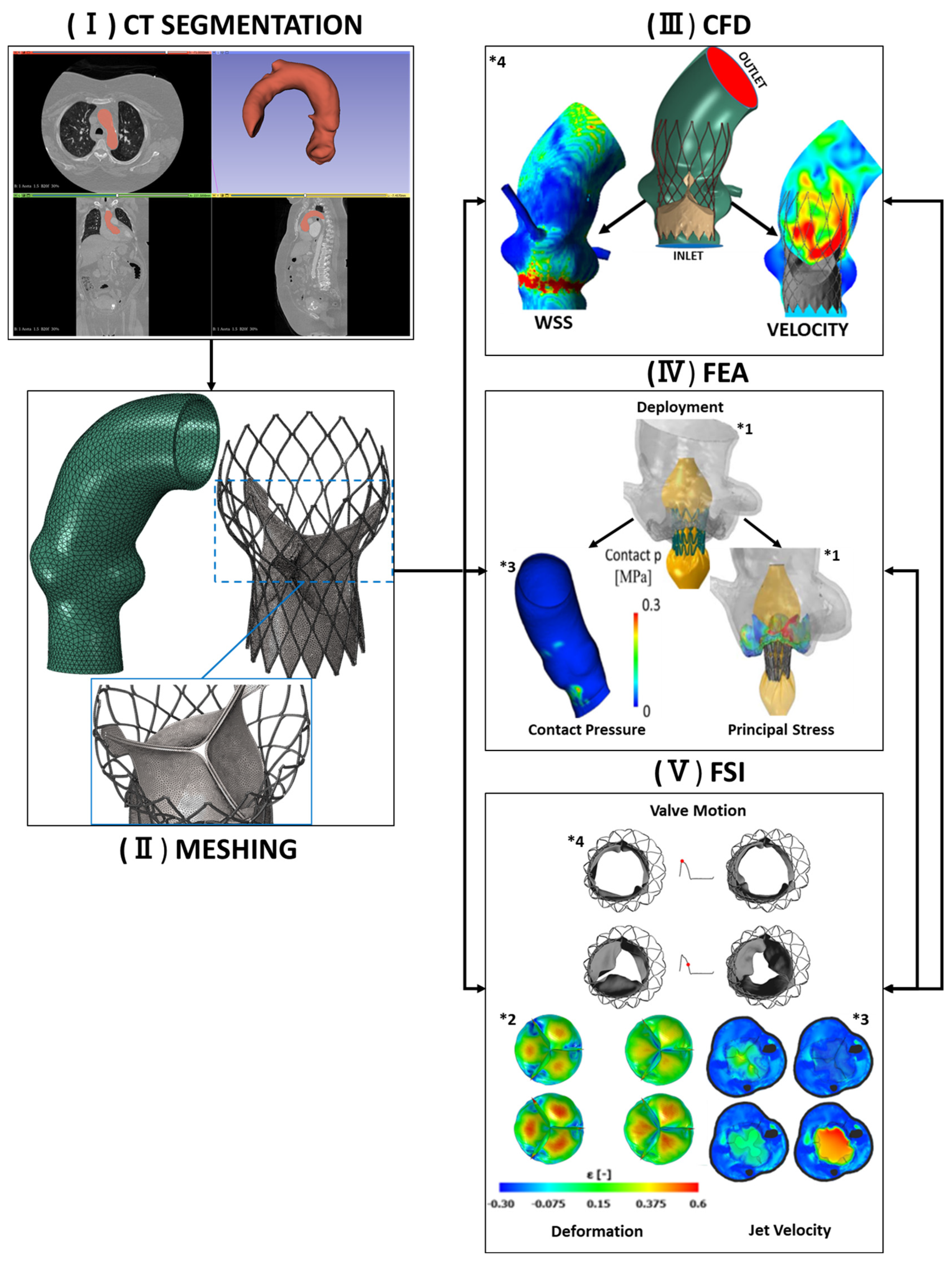
4. Computational Modeling for TAVR Procedures
5. Deep Learning for TAVR Procedures
5.1. Aorta Segmentation Using Deep Learning
5.2. Cardiovascular Hemodynamic Prediction Using Deep Learning
5.2.1. Utilized Dataset
5.2.2. Achieved Results
5.2.3. Deployed Models
5.3. Outcome Assessment of TAVR Procedures Using Machine/Deep Learning
6. Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TAVR | Transcatheter Aortic Valve Replacement |
| TAVI | Transcatheter Aortic Valve Implantation |
| AV | Aortic Valve |
| BHV | Bioprosthetic Heart Valve |
| CFD | Computational Fluid Dynamics |
| FEA | Finite Element Analysis |
| FSI | Fluid–Solid Interaction |
| DL | Deep Learning |
| ML | Machine Learning |
| GCN | Graph Convolutional Network |
| CNN | Convolutional Neural Network |
| E-D CNN | Encoder–Decoder CNN |
| CVD | Cardiovascular Disease |
| WHO | World Health Organization |
| MRI | Magnetic Resonance Imaging |
| CT | Computed Tomography |
| CCTA | Cardiac Computed Tomography Angiography |
| ECHO | Echocardiography |
| 3D TEE | 3D Transesophageal Echocardiography |
| 3D TTE | Transthoracic Echocardiography |
| PARTNER | Placement of Aortic Transcatheter Valve |
| AA | Ascending Aorta |
| AsAA | Ascending Aortic Aneurysm |
| CoA | Coarctation of the Aorta |
| LAD artery | Left Anterior Descending artery |
| LAA | Left Atrial Appendage |
| SSM | Statistical Shape Modeling |
| SDM | Statistical Distribution Modeling |
| PCA | Principal Component Analysis |
| MWSS, | Maximal Wall Shear Stress |
| TAWSS | Time-Averaged Wall Shaer Stress |
| WSS | Wall Shear Stress |
| SFD | Secondary Flow Degree |
| KE | Kinetic Energy |
| ECAP | Endothelial Cell Activation Pressure |
| AG-UCNet | Attention-Gate U-CliqueNet |
| UAD | Unsupervised Domain Adaptation |
| MLP | Multilayer Perceptron |
| MLR | Multilinear Regression |
| FCN | Fully Connected Neural Network |
| PLS | Partial Least Square |
| RPART | Recursive Partitioning and Regression Trees |
| LSTM | Long Short-Term Memory |
| Self-ONN | Self-Organized Operational Neural Network |
| GAN | Generative Adversarial Networks |
| GPU | Graphics Processing Units |
| DSC | Dice Similarity Coefficient |
| MAE | Mean Absolute Error |
| NMAE | Normalized Mean Absolute Error |
| RMSE | Root Mean Squared Error |
| MSE | Mean Squared Error |
| PVL | Paravalvular Leak |
| MLBCs | Major or Life-Threatening Bleeding Complications |
References
- McAloon, C.J.; Osman, F.; Glennon, P.; Lim, P.B.; Hayat, S.A. Chapter 4—Global Epidemiology and Incidence of Cardiovascular Disease. In Cardiovascular Diseases; Papageorgiou, N., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 57–96. [Google Scholar] [CrossRef]
- Faggiano, P.; Antonini-Canterin, F.; Baldessin, F.; Lorusso, R.; D’Aloia, A.; Cas, L.D. Epidemiology and cardiovascular risk factors of aortic stenosis. Cardiovasc. Ultrasound 2006, 4, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figulla, H.R.; Franz, M.; Lauten, A. The history of Transcatheter Aortic Valve Implantation (TAVI)—A personal view over 25 years of development. Cardiovasc. Revascularization Med. 2019, 21, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.R.; Leon, M.B.; Mack, M.J.; Miller, D.C.; Moses, J.W.; Svensson, L.G.; Tuzcu, E.M.; Webb, J.G.; Fontana, G.P.; Makkar, R.R.; et al. Transcatheter versus surgical aortic-valve replacement in high-risk patients. N. Engl. J. Med. 2011, 364, 2187–2198. [Google Scholar] [CrossRef] [Green Version]
- Reardon, M.J.; Van Mieghem, N.M.; Popma, J.J.; Kleiman, N.S.; Sondergaard, L.; Mumtaz, M.; Adams, D.H.; Deeb, G.M.; Maini, B.; Gada, H.; et al. Surgical or Transcatheter Aortic-Valve Replacement in Intermediate-Risk Patients. N. Engl. J. Med. 2017, 376, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- Popma, J.J.; Deeb, G.M.; Yakubov, S.J.; Mumtaz, M.; Gada, H.; O’Hair, D.; Bajwa, T.; Heiser, J.C.; Merhi, W.; Kleiman, N.S.; et al. Transcatheter Aortic-Valve Replacement with a Self-Expanding Valve in Low-Risk Patients. N. Engl. J. Med. 2019, 380, 1706–1715. [Google Scholar] [CrossRef]
- Mack, M.J.; Leon, M.B.; Thourani, V.H.; Makkar, R.; Kodali, S.K.; Russo, M.; Kapadia, S.R.; Malaisrie, S.C.; Cohen, D.J.; Pibarot, P.; et al. Transcatheter Aortic-Valve Replacement with a Balloon-Expandable Valve in Low-Risk Patients. N. Engl. J. Med. 2019, 380, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.G.; Ferreira, S.M.; Fonseca, P.; Dias, T.; Guerreiro, C.; Barbosa, A.R.; Teixeira, P.; Carvalho, M.; Ferreira, W.; Ferreira, N.D.; et al. Association between implantation depth assessed by computed tomography and new-onset conduction disturbances after transcatheter aortic valve implantation. J. Cardiovasc. Comput. Tomogr. 2017, 11, 332–337. [Google Scholar] [CrossRef]
- van der Boon, R.M.; Houthuizen, P.; Urena, M.; Poels, T.T.; van Mieghem, N.M.; Brueren, G.R.; Altintas, S.; Nuis, R.J.; Serruys, P.W.; van Garsse, L.A.; et al. Trends in the occurrence of new conduction abnormalities after transcatheter aortic valve implantation. Catheter. Cardiovasc. Interv. Off. J. Soc. Card. Angiogr. Interv. 2015, 85, E144–E152. [Google Scholar] [CrossRef]
- Bianchi, M.; Marom, G.; Ghosh, R.P.; Rotman, O.M.; Parikh, P.; Gruberg, L.; Bluestein, D. Patient-specific simulation of transcatheter aortic valve replacement: Impact of deployment options on paravalvular leakage. Biomech. Model. Mechanobiol. 2019, 18, 435–451. [Google Scholar] [CrossRef]
- Kadem, M.; Garber, L.; Abdelkhalek, M.; Al-Khazraji, B.K.; Keshavarz-Motamed, Z. Hemodynamic modeling, medical imaging, and machine learning and their applications to cardiovascular interventions. IEEE Rev. Biomed. Eng. 2022, 16, 403–423. [Google Scholar] [CrossRef]
- Enriquez, A.; Saenz, L.C.; Rosso, R.; Silvestry, F.E.; Callans, D.; Marchlinski, F.E.; Garcia, F.J.C. Use of intracardiac echocardiography in interventional cardiology: Working with the anatomy rather than fighting it. Circulation 2018, 137, 2278–2294. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, R.; Khodaei, S.; Ganame, J.; Keshavarz-Motamed, Z. Towards non-invasive computational-mechanics and imaging-based diagnostic framework for personalized cardiology for coarctation. Sci. Rep. 2020, 10, 9048. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, M.A.; Caso, P. Echocardiography: Basic Principles. In The ESC Textbook of Cardiovascular Imaging; Springer: Cham, Switzerland, 2010; pp. 1–38. [Google Scholar]
- Bushari, L.I.; Reeder, G.S.; Eleid, M.F.; Chandrasekaran, K.; Eriquez-Sarano, M.; Rihal, C.S.; Maalouf, J.F. Percutaneous transcatheter edge-to-edge MitraClip technique: A practical “step-by-step” 3-dimensional transesophageal echocardiography guide. Mayo Clin. Proc. 2019, 94, 89–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labrousse, L.; Dijos, M.; Leroux, L.; Oses, P.; Seguy, B.; Markof, M.; Lafitte, S. Guidance of the MitraClip® procedure by 2D and 3D imaging. Arch. Cardiovasc. Dis. 2018, 111, 432–440. [Google Scholar] [CrossRef]
- Howard-Quijano, K.; Methangkool, E.; Scovotti, J.C.; Mazor, E.; Grogan, T.R.; Kratzert, W.B.; Mahajan, A. Regional left ventricular myocardial dysfunction after cardiac surgery characterized by 3-dimensional strain. Obstet. Anesth. Dig. 2019, 128, 854–864. [Google Scholar] [CrossRef]
- Mollura, D.; Lungren, M.P. Radiology in Global Health; Springer: Cham, Switzerland, 2014; Volume 1. [Google Scholar]
- Pontone, G.; Cademartiri, F. Cardiac CT—Basic principles. In The ESC Textbook of Cardiovascular Imaging; Oxford University Press: Oxford, UK, 2021; p. 57. [Google Scholar]
- Blanke, P.; Weir-McCall, J.R.; Achenbach, S.; Delgado, V.; Hausleiter, J.; Jilaihawi, H.; Marwan, M.; Nørgaard, B.L.; Piazza, N.; Schoenhagen, P.; et al. Computed tomography imaging in the context of transcatheter aortic valve implantation (TAVI)/transcatheter aortic valve replacement (TAVR) an expert consensus document of the Society of Cardiovascular Computed Tomography. J. Am. Coll. Cardiol. Imaging 2019, 12, 1–24. [Google Scholar] [CrossRef]
- Ballocca, F.; Ruggeri, G.M.; Roscoe, A.; Thampinathan, B.; David, T.E.; Lang, R.M.; Meineri, M.; Tsang, W.J.E. Aortic root changes before and after surgery for chronic aortic dilatation: A 3D echocardiographic study. Echocardiography 2019, 36, 376–385. [Google Scholar] [CrossRef]
- Chen, S.A.; Ong, C.S.; Malguria, N.; Vricella, L.A.; Garcia, J.R.; Hibino, N.; Surgery, C.H. Digital design and 3D printing of aortic arch reconstruction in HLHS for surgical simulation and training. World J. Pediatr. Congenit. Hear. Surg. 2018, 9, 454–458. [Google Scholar] [CrossRef]
- Gatti, M.; Cosentino, A.; Cura Stura, E.; Bergamasco, L.; Garabello, D.; Pennisi, G.; Puppo, M.; Salizzoni, S.; Veglia, S.; Davini, O. Accuracy of cardiac magnetic resonance generated 3D models of the aortic annulus compared to cardiovascular computed tomography generated 3D models. Int. J. Cardiovasc. Imaging 2020, 36, 2007–2015. [Google Scholar] [CrossRef]
- Shinbane, J.S.; Saxon, L. Virtual medicine: Utilization of the advanced cardiac imaging patient avatar for procedural planning and facilitation. J. Cardiovasc. Comput. Tomogr. 2018, 12, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Bogaert, J.; Symons, R.; Wright, J. CMR—Basic principles. In The ESC Textbook of Cardiovascular Imaging; Oxford University Press: Oxford, UK, 2021; p. 67. [Google Scholar]
- Calamante, F.; Ittermann, B.; Kanal, E.; The Inter-Society Working Group on MR Safety; Norris, D. Recommended responsibilities for management of MR safety. J. Magn. Reson. Imaging 2016, 44, 1067–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, A.M.; Costa, F.; Tralhão, A.; Marques, H.; Cardim, N.; Adragão, P. MRI-conditional pacemakers: Current perspectives. Med. Devices Evid. Res. 2014, 7, 115–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaturvedi, A.; Hobbs, S.K.; Ling, F.S.; Chaturvedi, A.; Knight, P. MRI evaluation prior to Transcatheter Aortic Valve Implantation (TAVI): When to acquire and how to interpret. Insights Imaging 2016, 7, 245–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakerzadeh, R.; Hsu, M.-C.; Sacks, M.S. Computational methods for the aortic heart valve and its replacements. Expert Rev. Med Devices 2017, 14, 849–866. [Google Scholar] [CrossRef]
- Wu, W.; Pott, D.; Mazza, B.; Sironi, T.; Dordoni, E.; Chiastra, C.; Petrini, L.; Pennati, G.; Dubini, G.; Steinseifer, U.; et al. Fluid–structure interaction model of a percutaneous aortic valve: Comparison with an in vitro test and feasibility study in a patient-specific case. Ann. Biomed. Eng. 2016, 44, 590–603. [Google Scholar] [CrossRef]
- Bianchi, M.; Marom, G.; Ghosh, R.P.; Fernandez, H.A.; Taylor, J.R., Jr.; Slepian, M.J.; Bluestein, D. Effect of Balloon-Expandable Transcatheter Aortic Valve Replacement Positioning: A Patient-Specific Numerical Model. Artif. Organs 2016, 40, E292–E304. [Google Scholar] [CrossRef] [Green Version]
- Pasta, S.; Cannata, S.; Gentile, G.; Agnese, V.; Raffa, G.M.; Pilato, M.; Gandolfo, C. Transcatheter Heart Valve Implantation in Bicuspid Patients with Self-Expanding Device. Bioengineering 2021, 8, 91. [Google Scholar] [CrossRef]
- Sturla, F.; Ronzoni, M.; Vitali, M.; Dimasi, A.; Vismara, R.; Preston-Maher, G.; Burriesci, G.; Votta, E.; Redaelli, A. Impact of different aortic valve calcification patterns on the outcome of transcatheter aortic valve implantation: A finite element study. J. Biomech. 2016, 49, 2520–2530. [Google Scholar] [CrossRef] [Green Version]
- Kandail, H.S.; Trivedi, S.D.; Shaikh, A.C.; Bajwa, T.K.; Daniel, P.; Jahangir, A.; LaDisa, J.F., Jr. Impact of annular and supra-annular CoreValve deployment locations on aortic and coronary artery hemodynamics. J. Mech. Behav. Biomed. Mater. 2018, 86, 131–142. [Google Scholar] [CrossRef]
- Lavon, K.; Marom, G.; Bianchi, M.; Halevi, R.; Hamdan, A.; Morany, A.; Raanani, E.; Bluestein, D.; Haj-Ali, R. Biomechanical modeling of transcatheter aortic valve replacement in a stenotic bicuspid aortic valve: Deployments and paravalvular leakage. Med. Biol. Eng. Comput. 2019, 57, 2129–2143. [Google Scholar] [CrossRef]
- Wu, M.C.; Muchowski, H.M.; Johnson, E.L.; Rajanna, M.R.; Hsu, M.-C. Immersogeometric fluid–structure interaction modeling and simulation of transcatheter aortic valve replacement. Comput. Methods Appl. Mech. Eng. 2019, 357, 112556. [Google Scholar] [CrossRef] [PubMed]
- Yaakobovich, H.; Plitman Mayo, R.; Zaretsky, U.; Finkelstein, A.; Marom, G. Numerical models of valve-in-valve implantation: Effect of intentional leaflet laceration on the anchorage. Biomech. Model. Mechanobiol. 2020, 19, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Luraghi, G.; Migliavacca, F.; García-González, A.; Chiastra, C.; Rossi, A.; Cao, D.; Stefanini, G.; Rodriguez Matas, J.F. On the modeling of patient-specific transcatheter aortic valve replacement: A fluid–structure interaction approach. Cardiovasc. Eng. Technol. 2019, 10, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.P.; Marom, G.; Bianchi, M.; D’souza, K.; Zietak, W.; Bluestein, D. Numerical evaluation of transcatheter aortic valve performance during heart beating and its post-deployment fluid–structure interaction analysis. Biomech. Model. Mechanobiol. 2020, 19, 1725–1740. [Google Scholar] [CrossRef]
- Rocatello, G.; De Santis, G.; De Bock, S.; De Beule, M.; Segers, P.; Mortier, P. Optimization of a transcatheter heart valve frame using patient-specific computer simulation. Cardiovasc. Eng. Technol. 2019, 10, 456–468. [Google Scholar] [CrossRef]
- Pasta, S.; Gandolfo, C. Computational Analysis of Self-Expanding and Balloon-Expandable Transcatheter Heart Valves. Biomechanics 2021, 1, 43–52. [Google Scholar] [CrossRef]
- McGee, O.M.; Gunning, P.S.; McNamara, A.; McNamara, L.M. The impact of implantation depth of the Lotus™ valve on mechanical stress in close proximity to the bundle of His. Biomech. Model. Mechanobiol. 2019, 18, 79–88. [Google Scholar] [CrossRef]
- Rocatello, G.; El Faquir, N.; De Santis, G.; Iannaccone, F.; Bosmans, J.; De Backer, O.; Sondergaard, L.; Segers, P.; De Beule, M.; de Jaegere, P.; et al. Patient-specific computer simulation to elucidate the role of contact pressure in the development of new conduction abnormalities after catheter-based implantation of a self-expanding aortic valve. Circ. Cardiovasc. Interv. 2018, 11, e005344. [Google Scholar] [CrossRef] [Green Version]
- Pasta, S.; Cannata, S.; Gentile, G.; Di Giuseppe, M.; Cosentino, F.; Pasta, F.; Agnese, V.; Bellavia, D.; Raffa, G.M.; Pilato, M.; et al. Simulation study of transcatheter heart valve implantation in patients with stenotic bicuspid aortic valve. Med. Biol. Eng. Comput. 2020, 58, 815–829. [Google Scholar] [CrossRef]
- Morganti, S.; Conti, M.; Aiello, M.; Valentini, A.; Mazzola, A.; Reali, A.; Auricchio, F. Simulation of transcatheter aortic valve implantation through patient-specific finite element analysis: Two clinical cases. J. Biomech. 2014, 47, 2547–2555. [Google Scholar] [CrossRef]
- Morganti, S.; Brambilla, N.; Petronio, A.; Reali, A.; Bedogni, F.; Auricchio, F. Prediction of patient-specific post-operative outcomes of TAVI procedure: The impact of the positioning strategy on valve performance. J. Biomech. 2016, 49, 2513–2519. [Google Scholar] [CrossRef] [PubMed]
- Luraghi, G.; Matas, J.F.R.; Beretta, M.; Chiozzi, N.; Iannetti, L.; Migliavacca, F. The impact of calcification patterns in transcatheter aortic valve performance: A fluid-structure interaction analysis. Comput. Methods Biomech. Biomed. Eng. 2020, 24, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Jafar, R.; Labrosse, M.R.; Weaver, J.D.; Retta, S.M.; Wu, C.; Duraiswamy, N. A computational study on deformed bioprosthetic valve geometries: Clinically relevant valve performance metrics. J. Biomech. Eng. 2020, 142, 011003. [Google Scholar] [CrossRef]
- Mao, W.; Wang, Q.; Kodali, S.; Sun, W. Numerical parametric study of paravalvular leak following a transcatheter aortic valve deployment into a patient-specific aortic root. J. Biomech. Eng. 2018, 140, 101007. [Google Scholar] [CrossRef]
- Anam, S.B.; Kovarovic, B.J.; Ghosh, R.P.; Bianchi, M.; Hamdan, A.; Haj-Ali, R.; Bluestein, D. Assessment of Paravalvular Leak Severity and Thrombogenic Potential in Transcatheter Bicuspid Aortic Valve Replacements Using Patient-Specific Computational Modeling. J. Cardiovasc. Transl. Res. 2021, 15, 834–844. [Google Scholar] [CrossRef]
- Oks, D.; Samaniego, C.; Houzeaux, G.; Butakoff, C.; Vázquez, M. Fluid-structure interaction analysis of eccentricity and leaflet rigidity on thrombosis biomarkers in bioprosthetic aortic valve replacements. Int. J. Numer. Methods Biomed. Eng. 2022, 38, e3649. [Google Scholar] [CrossRef]
- Mao, W.; Li, K.; Sun, W. Fluid–structure interaction study of transcatheter aortic valve dynamics using smoothed particle hydrodynamics. Cardiovasc. Eng. Technol. 2016, 7, 374–388. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, R.P.; Marom, G.; Rotman, O.M.; Slepian, M.J.; Prabhakar, S.; Horner, M.; Bluestein, D. Comparative Fluid–Structure Interaction Analysis of Polymeric Transcatheter and Surgical Aortic Valves’ Hemodynamics and Structural Mechanics. J. Biomech. Eng. 2018, 140, 121002. [Google Scholar] [CrossRef]
- Dong, H.; Yang, G.; Liu, F.; Mo, Y.; Guo, Y. Automatic brain tumor detection and segmentation using U-Net based fully convolutional networks. In Proceedings of the Annual Conference on Medical Image Understanding and Analysis, Edinburgh, UK, 11–13 July 2017; pp. 506–517. [Google Scholar]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Shen, L.; Margolies, L.R.; Rothstein, J.H.; Fluder, E.; McBride, R.; Sieh, W. Deep learning to improve breast cancer detection on screening mammography. Sci. Rep. 2019, 9, 12495. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Halm-Lutterodt, N.V.; Tang, H.; Mecum, A.; Mesregah, M.K.; Ma, Y.; Li, H.; Zhang, F.; Wu, Z.; Yao, E.; et al. Automated MRI-based deep learning model for detection of Alzheimer’s disease process. Int. J. Neural Syst. 2020, 30, 2050032. [Google Scholar] [CrossRef] [PubMed]
- Tahir, A.M.; Chowdhury, M.E.H.; Khandakar, A.; Rahman, T.; Qiblawey, Y.; Khurshid, U.; Kiranyaz, S.; Ibtehaz, N.; Rahman, M.S.; Al-Maadeed, S.; et al. COVID-19 infection localization and severity grading from chest X-ray images. Comput. Biol. Med. 2021, 139, 105002. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.L.; Trout, A.T.; Somasundaram, E.; Anton, C.G.; Li, Y.; Dillman, J.R. Improving image quality and reducing radiation dose for pediatric CT by using deep learning reconstruction. Radiology 2021, 298, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Choi, S.-Y.; Hwang, J.A.; Lim, S.; Lee, M.H.; Yi, B.H.; Cha, J.G. The potential for reduced radiation dose from deep learning-based CT image reconstruction: A comparison with filtered back projection and hybrid iterative reconstruction using a phantom. Medicine 2021, 100, e25814. [Google Scholar] [CrossRef]
- Masutani, E.M.; Bahrami, N.; Hsiao, A. Deep learning single-frame and multiframe super-resolution for cardiac MRI. Radiology 2020, 295, 552–561. [Google Scholar] [CrossRef]
- Wang, W.; Xia, Q.; Hu, Z.; Yan, Z.; Li, Z.; Wu, Y.; Huang, N.; Gao, Y.; Metaxas, D.; Zhang, S. Few-Shot Learning by a Cascaded Framework With Shape-Constrained Pseudo Label Assessment for Whole Heart Segmentation. IEEE Trans. Med. Imaging 2021, 40, 2629–2641. [Google Scholar] [CrossRef]
- Zheng, Y.; Barbu, A.; Georgescu, B.; Scheuering, M.; Comaniciu, D. Four-Chamber Heart Modeling and Automatic Segmentation for 3-D Cardiac CT Volumes Using Marginal Space Learning and Steerable Features. IEEE Trans. Med. Imaging 2008, 27, 1668–1681. [Google Scholar] [CrossRef]
- Bratt, A.; Kim, J.; Pollie, M.; Beecy, A.N.; Tehrani, N.H.; Codella, N.; Perez-Johnston, R.; Palumbo, M.C.; Alakbarli, J.; Colizza, W.; et al. Machine learning derived segmentation of phase velocity encoded cardiovascular magnetic resonance for fully automated aortic flow quantification. J. Cardiovasc. Magn. Reson. 2019, 21, 1. [Google Scholar] [CrossRef] [Green Version]
- Degerli, A.; Zabihi, M.; Kiranyaz, S.; Hamid, T.; Mazhar, R.; Hamila, R.; Gabbouj, M. Early Detection of Myocardial Infarction in Low-Quality Echocardiography. IEEE Access 2021, 9, 34442–34453. [Google Scholar] [CrossRef]
- Kang, D.; Dey, D.; Slomka, P.J.; Arsanjani, R.; Nakazato, R.; Ko, H.; Berman, D.S.; Li, D.; Kuo, C.C.J. Structured learning algorithm for detection of nonobstructive and obstructive coronary plaque lesions from computed tomography angiography. J. Med. Imaging 2015, 2, 014003. [Google Scholar] [CrossRef]
- van Velzen, S.G.; Lessmann, N.; Velthuis, B.K.; Bank, I.E.; van den Bongard, D.H.; Leiner, T.; de Jong, P.A.; Veldhuis, W.B.; Correa, A.; Terry, J.G.; et al. Deep learning for automatic calcium scoring in CT: Validation using multiple cardiac CT and chest CT protocols. Radiology 2020, 295, 66–79. [Google Scholar] [CrossRef]
- Lessmann, N.; Ginneken, B.v.; Zreik, M.; Jong, P.A.d.; Vos, B.D.d.; Viergever, M.A.; Išgum, I. Automatic Calcium Scoring in Low-Dose Chest CT Using Deep Neural Networks With Dilated Convolutions. IEEE Trans. Med. Imaging 2018, 37, 615–625. [Google Scholar] [CrossRef] [Green Version]
- Zreik, M.; Hamersvelt, R.W.v.; Wolterink, J.M.; Leiner, T.; Viergever, M.A.; Išgum, I. A Recurrent CNN for Automatic Detection and Classification of Coronary Artery Plaque and Stenosis in Coronary CT Angiography. IEEE Trans. Med. Imaging 2019, 38, 1588–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jalali, A.; Lonsdale, H.; Do, N.; Peck, J.; Gupta, M.; Kutty, S.; Ghazarian, S.R.; Jacobs, J.P.; Rehman, M.; Ahumada, L.M. Deep Learning for Improved Risk Prediction in Surgical Outcomes. Sci. Rep. 2020, 10, 9289. [Google Scholar] [CrossRef]
- Chen, W.; Lu, Z.; You, L.; Zhou, L.; Xu, J.; Chen, K. Artificial intelligence–based multimodal risk assessment model for surgical site infection (AMRAMS): Development and validation study. JMIR Public Health Surveill. 2020, 8, e18186. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, S.M.; Nair, B.; Vavilala, M.S.; Horibe, M.; Eisses, M.J.; Adams, T.; Liston, D.E.; Low, D.K.-W.; Newman, S.-F.; Kim, J.; et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2018, 2, 749–760. [Google Scholar] [CrossRef]
- Scheeren, T.W.; Ramsay, M.A. New developments in hemodynamic monitoring. J. Cardiothorac. Vasc. Anesth. 2019, 33, S67–S72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Cannière, H.; Corradi, F.; Smeets, C.J.; Schoutteten, M.; Varon, C.; Van Hoof, C.; Van Huffel, S.; Groenendaal, W.; Vandervoort, P. Wearable monitoring and interpretable machine learning can objectively track progression in patients during cardiac rehabilitation. Sensors 2020, 20, 3601. [Google Scholar] [CrossRef]
- Rogers, M.A.; Aikawa, E. Cardiovascular calcification: Artificial intelligence and big data accelerate mechanistic discovery. Nat. Rev. Cardiol. 2019, 16, 261–274. [Google Scholar] [CrossRef]
- Chang, S.; Kim, H.; Suh, Y.J.; Choi, D.M.; Kim, H.; Kim, D.K.; Kim, J.Y.; Yoo, J.Y.; Choi, B.W. Development of a deep learning-based algorithm for the automatic detection and quantification of aortic valve calcium. Eur. J. Radiol. 2021, 137, 109582. [Google Scholar] [CrossRef]
- Li, G.; Wang, H.; Zhang, M.; Tupin, S.; Qiao, A.; Liu, Y.; Ohta, M.; Anzai, H. Prediction of 3D Cardiovascular hemodynamics before and after coronary artery bypass surgery via deep learning. Commun. Biol. 2021, 4, 99. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.K.; Bell, R.; Nair, A.; Menezes, L.J.; Patel, R.; Wan, S.; Chou, K.; Chen, J.; Torii, R.; Davies, R.H.; et al. A computationally efficient approach to segmentation of the aorta and coronary arteries using deep learning. IEEE Access 2021, 9, 108873–108888. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Fang, Z.; Gao, Y.; Xiong, N.; Zhong, C.; Tang, X. Coronary arteries segmentation based on 3D FCN with attention gate and level set function. IEEE Access 2019, 7, 42826–42835. [Google Scholar] [CrossRef]
- MM-WHS 2017. Available online: https://paperswithcode.com/dataset/mm-whs-2017 (accessed on 1 January 2023).
- Liu, T.; Tian, Y.; Zhao, S.; Huang, X.; Wang, Q. Automatic whole heart segmentation using a two-stage u-net framework and an adaptive threshold window. IEEE Access 2019, 7, 83628–83636. [Google Scholar] [CrossRef]
- Ye, C.; Wang, W.; Zhang, S.; Wang, K. Multi-depth fusion network for whole-heart CT image segmentation. IEEE Access 2019, 7, 23421–23429. [Google Scholar] [CrossRef]
- Wang, W.; Ye, C.; Zhang, S.; Xu, Y.; Wang, K. Improving whole-heart CT image segmentation by attention mechanism. IEEE Access 2019, 8, 14579–14587. [Google Scholar] [CrossRef]
- Vesal, S.; Gu, M.; Kosti, R.; Maier, A.; Ravikumar, N. Adapt Everywhere: Unsupervised Adaptation of Point-Clouds and Entropy Minimization for Multi-Modal Cardiac Image Segmentation. IEEE Trans. Med. Imaging 2021, 40, 1838–1851. [Google Scholar] [CrossRef]
- Liang, L.; Liu, M.; Martin, C.; Sun, W. A deep learning approach to estimate stress distribution: A fast and accurate surrogate of finite-element analysis. J. R. Soc. Interface 2018, 15, 20170844. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.; Mao, W.; Sun, W. A feasibility study of deep learning for predicting hemodynamics of human thoracic aorta. J. Biomech. 2020, 99, 109544. [Google Scholar] [CrossRef]
- Morales, X.; Mill, J.; Juhl, K.A.; Olivares, A.; Jimenez-Perez, G.; Paulsen, R.R.; Camara, O. Deep learning surrogate of computational fluid dynamics for thrombus formation risk in the left atrial appendage. In Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, Shenzhen, China, 13 October 2019; pp. 157–166. [Google Scholar]
- Acebes, C.; Morales, X.; Camara, O. A Cartesian Grid Representation of Left Atrial Appendages for a Deep Learning Estimation of Thrombogenic Risk Predictors; Springer: Cham, Switzerland, 2020; pp. 35–43. [Google Scholar]
- Yevtushenko, P.; Goubergrits, L.; Gundelwein, L.; Setio, A.; Heimann, T.; Ramm, H.; Lamecker, H.; Kuehne, T.; Meyer, A.; Schafstedde, M. Deep Learning Based Centerline-Aggregated Aortic Hemodynamics: An Efficient Alternative to Numerical Modelling of Hemodynamics. IEEE J. Biomed. Health Inform. 2021, 26, 1815–1825. [Google Scholar] [CrossRef]
- Farajtabar, M.; Larimi, M.M.; Biglarian, M.; Miansari, M. Machine-Learning Identification of Hemodynamics in Coronary Arteries in the Presence of Stenosis. arXiv 2021, arXiv:2111.01950. [Google Scholar]
- Morales Ferez, X.; Mill, J.; Juhl, K.A.; Acebes, C.; Iriart, X.; Legghe, B.; Cochet, H.; De Backer, O.; Paulsen, R.R.; Camara, O. Deep learning framework for real-time estimation of in-silico thrombotic risk indices in the left atrial appendage. Front. Physiol. 2021, 12, 694945. [Google Scholar] [CrossRef] [PubMed]
- Heimann, T.; Meinzer, H.-P. Statistical shape models for 3D medical image segmentation: A review. Med. Image Anal. 2009, 13, 543–563. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Liu, M.; Martin, C.; Elefteriades, J.A.; Sun, W. A machine learning approach to investigate the relationship between shape features and numerically predicted risk of ascending aortic aneurysm. Biomech. Model. Mechanobiol. 2017, 16, 1519–1533. [Google Scholar] [CrossRef] [PubMed]
- Filipovic, N.; Ivanovic, M.; Krstajic, D.; Kojic, M. Hemodynamic flow modeling through an abdominal aorta aneurysm using data mining tools. Inf. Technol. Biomed. 2010, 15, 189–194. [Google Scholar] [CrossRef]
- Gharleghi, R.; Samarasinghe, G.; Sowmya, A.; Beier, S. Deep learning for time averaged wall shear stress prediction in left main coronary bifurcations. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1–4. [Google Scholar]
- Beier, S.; Ormiston, J.; Webster, M.; Cater, J.; Norris, S.; Medrano-Gracia, P.; Young, A.; Cowan, B. Impact of bifurcation angle and other anatomical characteristics on blood flow–A computational study of non-stented and stented coronary arteries. J. Biomech. 2016, 49, 1570–1582. [Google Scholar] [CrossRef]
- Jordanski, M.; Radovic, M.; Milosevic, Z.; Filipovic, N.; Obradovic, Z. Machine learning approach for predicting wall shear distribution for abdominal aortic aneurysm and carotid bifurcation models. IEEE J. Biomed. Health Inform. 2016, 22, 537–544. [Google Scholar] [CrossRef]
- Kojić, M.; Filipović, N.; Stojanović, B.; Kojić, N. Computer Modeling in Bioengineering: Theoretical Background, Examples and Software; John Wiley & Sons: New York, NY, USA, 2008. [Google Scholar]
- Balu, A.; Nallagonda, S.; Xu, F.; Krishnamurthy, A.; Hsu, M.-C.; Sarkar, S. A deep learning framework for design and analysis of surgical bioprosthetic heart valves. Sci. Rep. 2019, 9, 18560. [Google Scholar] [CrossRef] [Green Version]
- Oldenburg, J.; Renkewitz, J.; Stiehm, M.; Schmitz, K.-P. Contributions towards Data driven Deep Learning methods to predict Steady State Fluid Flow in mechanical Heart Valves. Curr. Dir. Biomed. Eng. 2021, 7, 625–628. [Google Scholar] [CrossRef]
- Qi, C.R.; Su, H.; Mo, K.; Guibas, L.J. Pointnet: Deep learning on point sets for 3d classification and segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 652–660. [Google Scholar]
- Slipsager, J.M.; Juhl, K.A.; Sigvardsen, P.E.; Kofoed, K.F.; Backer, O.D.; Olivares, A.L.; Camara, O.; Paulsen, R.R. Statistical shape clustering of left atrial appendages. In Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, Granada, Spain, 16 September 2018; pp. 32–39. [Google Scholar]
- Wang, Z.H.; Lahoti, G.; Wang, K.; Liu, S.; Zhang, C.; Wang, B.; Wu, C.-W.; Vannan, M.; Qian, Z. Prediction of paravalvular leak post transcatheter aortic valve replacement using a convolutional neural network. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 1088–1091. [Google Scholar]
- Jia, Y.; Luosang, G.; Li, Y.; Wang, J.; Li, P.; Xiong, T.; Li, Y.; Liao, Y.; Zhao, Z.; Peng, Y.; et al. Deep Learning in Prediction of Late Major Bleeding After Transcatheter Aortic Valve Replacement. Clin. Epidemiol. 2022, 14, 9. [Google Scholar] [CrossRef]
- Penso, M.; Pepi, M.; Fusini, L.; Muratori, M.; Cefalù, C.; Mantegazza, V.; Gripari, P.; Ali, S.G.; Fabbiocchi, F.; Bartorelli, A.; et al. Predicting long-term mortality in TAVI patients using machine learning techniques. J. Cardiovasc. Dev. Dis. 2021, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Agasthi, P.; Ashraf, H.; Pujari, S.H.; Girardo, M.E.; Tseng, A.; Mookadam, F.; Venepally, N.R.; Buras, M.; Khetarpal, B.K.; Allam, M.; et al. Artificial Intelligence Trumps TAVI2-SCORE and CoreValve Score in Predicting 1-Year Mortality Post-Transcatheter Aortic Valve Replacement. Cardiovasc. Revascularization Med. 2021, 24, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Galli, V.; Loncaric, F.; Rocatello, G.; Astudillo, P.; Sanchis, L.; Regueiro, A.; De Backer, O.; Swaans, M.; Bosmans, J.; Ribeiro, J.; et al. Towards patient-specific prediction of conduction abnormalities induced by transcatheter aortic valve implantation: A combined mechanistic modelling and machine learning approach. Eur. Heart J.-Digit. Health 2021, 2, 606–615. [Google Scholar] [CrossRef]
- Astudillo, P.; Mortier, P.; Bosmans, J.; De Backer, O.; de Jaegere, P.; De Beule, M.; Dambre, J. Enabling Automated Device Size Selection for Transcatheter Aortic Valve Implantation. J. Interv. Cardiol. 2019, 2019, 3591314. [Google Scholar] [CrossRef] [PubMed]
- Kiranyaz, S.; Malik, J.; Abdallah, H.B.; Ince, T.; Iosifidis, A.; Gabbouj, M. Self-organized operational neural networks with generative neurons. Neural Netw. 2021, 140, 294–308. [Google Scholar] [CrossRef] [PubMed]
- Malik, J.; Kiranyaz, S.; Gabbouj, M. Self-organized operational neural networks for severe image restoration problems. Neural Netw. 2021, 135, 201–211. [Google Scholar] [CrossRef]
- Ho, J.; Jain, A.; Abbeel, P. Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 2020, 33, 6840–6851. [Google Scholar]
- Khader, F.; Mueller-Franzes, G.; Arasteh, S.T.; Han, T.; Haarburger, C.; Schulze-Hagen, M.; Schad, P.; Engelhardt, S.; Baessler, B.; Foersch, S.; et al. Medical Diffusion—Denoising Diffusion Probabilistic Models for 3D Medical Image Generation. arXiv 2022, arXiv:2211.03364. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Singh, N.K.; Raza, K. Medical image generation using generative adversarial networks: A review. In Health Informatics: A Computational Perspective in Healthcare; Springer: Singapore, 2021; pp. 77–96. [Google Scholar]
- Ferdian, E.; Suinesiaputra, A.; Dubowitz, D.J.; Zhao, D.; Wang, A.; Cowan, B.; Young, A.A. 4DFlowNet: Super-resolution 4D flow MRI using deep learning and computational fluid dynamics. Front. Phys. 2020, 8, 138. [Google Scholar] [CrossRef]
- Strönisch, S.; Sander, M.; Meyer, M.; Knüpfer, A. Multi-GPU Approach for Training of Graph ML Models on large CFD Meshes. In Proceedings of the AIAA SCITECH 2023 Forum, National Harbor, MI, USA, 23–27 January 2023; p. 1203. [Google Scholar]
- Mariathas, M.; Rawlins, J.; Curzen, N. Transcatheter aortic valve implantation: Where are we now? Future Cardiol. 2017, 13, 551–566. [Google Scholar] [CrossRef] [PubMed]
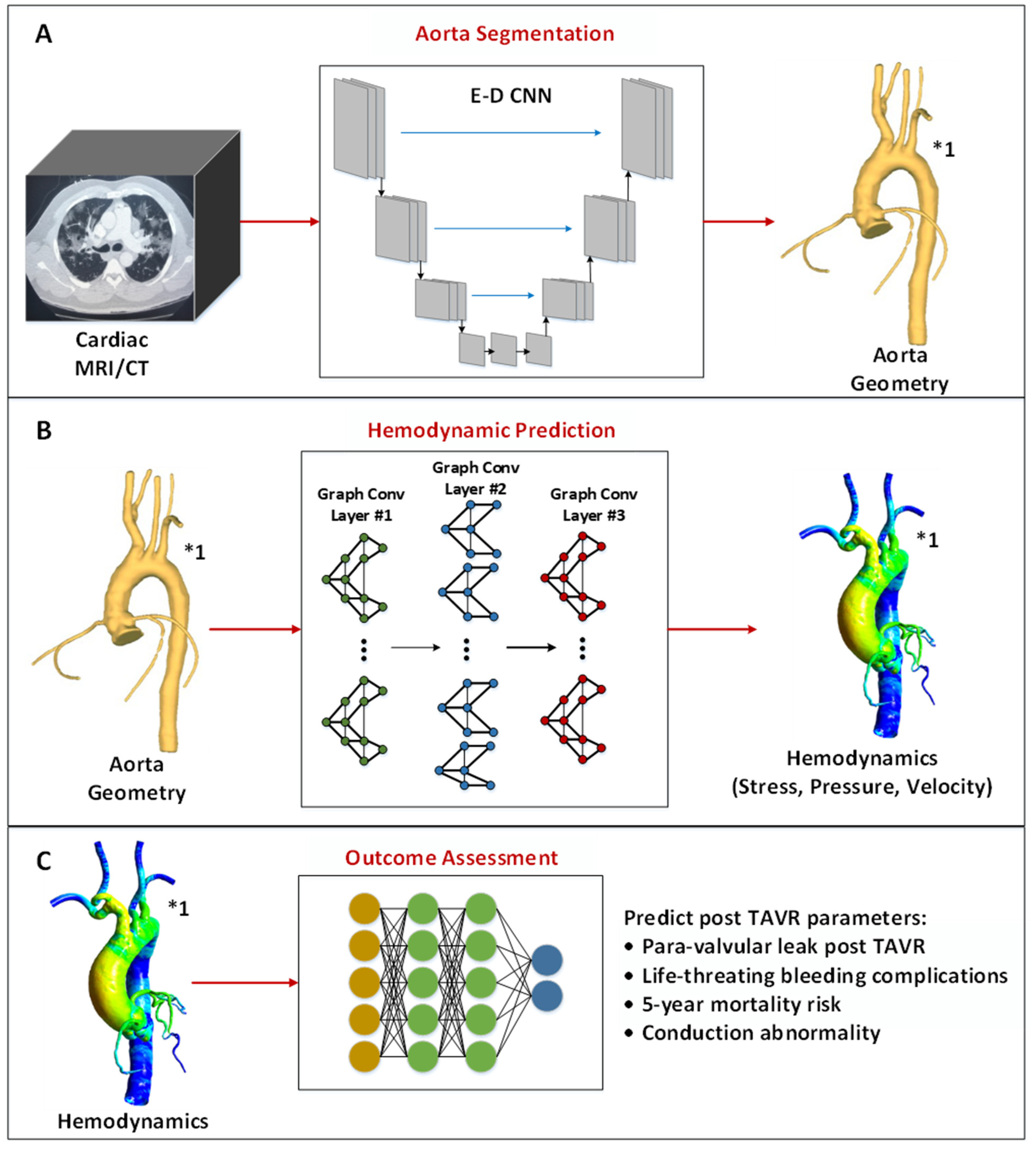

| Ref. | Dataset | DL Model | Results | |
|---|---|---|---|---|
| 3D CT/MRI Scans | 2D Slices | |||
| [78] | 69 CT Scans | 14,597 | 2D U-Net | DSC 91.2% |
| [79] | 70 CT Scans | 11,200 | Attention Gate 3D U-Net | DSC 90.5% |
| [81] | 20 CT and 20 MRI (MM-WHS Challenge 2017) | _ | Two-Stage 3D U-Net | DSC 95.5% |
| [82] | MM-WHS Challenge 2017 | _ | Multi-Depth Fusion U-Net | DSC 96.7% |
| [83] | MM-WHS Challenge 2017 | _ | AG-UCNet | DSC 96.8% |
| [84] | MM-WHS Challenge 2017 | _ | UAD | DSC 81.3% |
| [62] | MM-WHS Challenge 2017 | _ | Few-Shot Learning Framework | DSC 94.3% |
| Ref. | Dataset | Synthetic Dataset | Availability |
|---|---|---|---|
| [85] | 25 CT scans from AsAA patients | 729 geometries created using SSM based on PCA [92,93] | √ |
| [86] | Similar to [85] | Similar to [85] | √ |
| [94] | NA | 6000 geometries based on different parameters (i.e., AsAA length and curvature) | |
| [87] | 103 cardiac CT scans | 300 LAA geometries created using SSM based on PCA | √ |
| [88] | Similar to [87] | Similar to [87] | √ |
| [95] | 127 coronary artery CT scans | 3302 modified bifurcation geometries based on generic shape change [96] | |
| [89] | - 143 3D MRI scans from CoA Patients - 85 healthy 3D MRI scans - 87 4D MRI scans to obtain flow boundary conditions | 3000 aortic geometries and inlet vector fields created using SDM | |
| [90] | 120 coronary arterial geometries (with/without stenosis) | ||
| [97] | NA | 4000 AsAA and carotid bifurcation 2D geometries created using in-house software [98] | |
| [77] | 110 CT scans from patients with LAD stenosis | 1110 geometries | |
| [91] | - 256 synthetic and real LAA geometries - 114 real LAA geometries | ||
| [99] | 90,941 valve closure simulations | ||
| [100] | 3500 mechanical aortic valves with varying opening angles in randomly generated aortic root geometries |
| Ref | Computational Modeling | Input Geometry | Output | DL Model | Predicted Hemodynamic Results | Derived Parameter Results |
|---|---|---|---|---|---|---|
| [85] | FEA | Aorta geometries | Aortic wall stress distributions | PCA + MLP | Estimated stress distribution: NMAES11 of 0.492%, NMAES22 of 0.492%, and NMAES12 of 0.492% - Estimated peak stress value: NMAES11 of 0.891%, NMAES22 of 0.891%, and NMAES12 of 0.891% | Estimated stress distribution: NMAEVon Mises of 0.492% - Estimated peak stress value: NAEVon Mises of 0.891% |
| [86] | CFD | Aorta geometries | Pressure field and velocity field magnitudes | MLP | - Pressure field: NMAE of 1.427% - Velocity magnitude: NMAE of 1.961% | |
| [94] | CFD | Aorta geometries | MWSS | (1) MLR (2) PLS (3) RPART (4) MLP | - MLP: MAE of 0.001% | |
| [87] | CFD | LAA geometry | ECAP map | (1) MLP (2) PCA + MLP | - MLP: MAE of 0.646% and NMAE of 4.720% - PCA + MLP: MAE of 0.649% and NMAE of 5.756% | Binary classification based on ECAP values (subject at risk or safe) |
| [88] | CFD | LAA geometry | ECAP map | (1) PCA + MLP (2) ED-CNN | - PCA + MLP: MAE of 0.73% - E-D CNN: MAE of 0.63% | Binary classification based on ECAP values: - PCA + MLP MAE 81.7% - E-D CNN MAE 87.9% |
| [95] | CFD | Coronary artery | TAWSS | CNN | NMAE of 10.38% | |
| [89] | CFD | Center-line based shape model and flow boundary conditions | WSS, SFD, and KE along the centerline | Encoder NN +LSTM +1D DenseNet | ||
| [90] | CFD | Hand-crafted features for each node in the coronary artery geometry | Pressure field and velocity field magnitude at a specific node | MLP | - Pressure field accuracy of 98.7% - Velocity magnitude accuracy of 93.2% | |
| [97] | CFD | 2D geometry of AsAA and carotid bifurcation | WSS | GCRF | - AsAA coefficient of determination of 0.93 - Carotid bifurcation coefficient of determination of 0.95 | |
| [77] | CFD | Geometry containing aorta, coronary arteries, and bypass graft | 3D pre-operative and post-operative velocity and pressure field | PointNet | Aorta and superior aortic branch artery - Pre-operative values: NMAEPressure of 4.30% and NMAEVelocity of 6.01% - Post-operative values: NMAEPressure of 4.28% and NMAEVelocity of 6.02% | |
| [91] | CFD | LAA geometry | ECAP | (1) PCA + MLP (2) E-D CNN (3) Geometric PointNet | - PCA-MLP MAE of 0.608% - ED-CNN with Cartesian mapping input: MAE of 0.651% - ED-CNN with Bull’s eye mapping input: MAE of 0.654% - Geometric CNN: MAE of 0.521% | |
| [99] | FEA | - Aorta geometry with undeformed heart valve - Aortic pressure - Valve material property | - Deformed, closed shape of the heart valve - Coaptation area | Autoencoder-CNN | - Valve deformation: Euclidean distance of 0.0649 cm - Coaptation area: CC of 0.933 - RMSE of 0.117 cm2 | |
| [100] | CFD | Aortic root with mechanical valve | Pressure field and velocity field magnitude | U-Net | MSE < 0.06 |
| Ref. | Dataset | Targeted Problem | DL Model | Results |
|---|---|---|---|---|
| [103] | 168 CT scans | PVL prediction | 3-layer CNN | Sens. 76.9% Spec. 86.9% Acc. 78.6% |
| [104] | 668 imaging scans and clinical data | MLBC prediction | 2-layer FCN (BLeNet) | AU 0.84 3-Year Sens. 67% 3-Year Spec. 89% |
| [105] | 471 ECHO scans and clinical data | 5-year mortality prediction | FCN | AU 0.79 Sens. 71% |
| [106] | Demographics, ECG, CT scans, and ECHO data from 1055 patients | 1-year mortality prediction | Gradient boosting | AUC 0.72 |
| [107] | 151 CT scans | Conduction abnormality prediction and best valve design recommendation | Ensemble of ML models | AU 0.84 Sens. 100% Spec. 62% |
| [108] | 453 CT scans | Aortic annulus perimeter and area | U-Net | Area MSE 0.1089 cm2 Perimeter MSE 0.6 cm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tahir, A.M.; Mutlu, O.; Bensaali, F.; Ward, R.; Ghareeb, A.N.; Helmy, S.M.H.A.; Othman, K.T.; Al-Hashemi, M.A.; Abujalala, S.; Chowdhury, M.E.H.; et al. Latest Developments in Adapting Deep Learning for Assessing TAVR Procedures and Outcomes. J. Clin. Med. 2023, 12, 4774. https://doi.org/10.3390/jcm12144774
Tahir AM, Mutlu O, Bensaali F, Ward R, Ghareeb AN, Helmy SMHA, Othman KT, Al-Hashemi MA, Abujalala S, Chowdhury MEH, et al. Latest Developments in Adapting Deep Learning for Assessing TAVR Procedures and Outcomes. Journal of Clinical Medicine. 2023; 12(14):4774. https://doi.org/10.3390/jcm12144774
Chicago/Turabian StyleTahir, Anas M., Onur Mutlu, Faycal Bensaali, Rabab Ward, Abdel Naser Ghareeb, Sherif M. H. A. Helmy, Khaled T. Othman, Mohammed A. Al-Hashemi, Salem Abujalala, Muhammad E. H. Chowdhury, and et al. 2023. "Latest Developments in Adapting Deep Learning for Assessing TAVR Procedures and Outcomes" Journal of Clinical Medicine 12, no. 14: 4774. https://doi.org/10.3390/jcm12144774






