A Random Forest Model for Post-Treatment Survival Prediction in Patients with Non-Squamous Cell Carcinoma of the Head and Neck
Abstract
:1. Introduction
2. Methods
2.1. Data Source
2.2. Endpoint and Variables
2.3. Prediction Model Establishment
2.4. Prediction Model Evaluation
2.5. Deployment
2.6. Statistical Analysis
3. Results
3.1. Characteristics and Regression Analysis
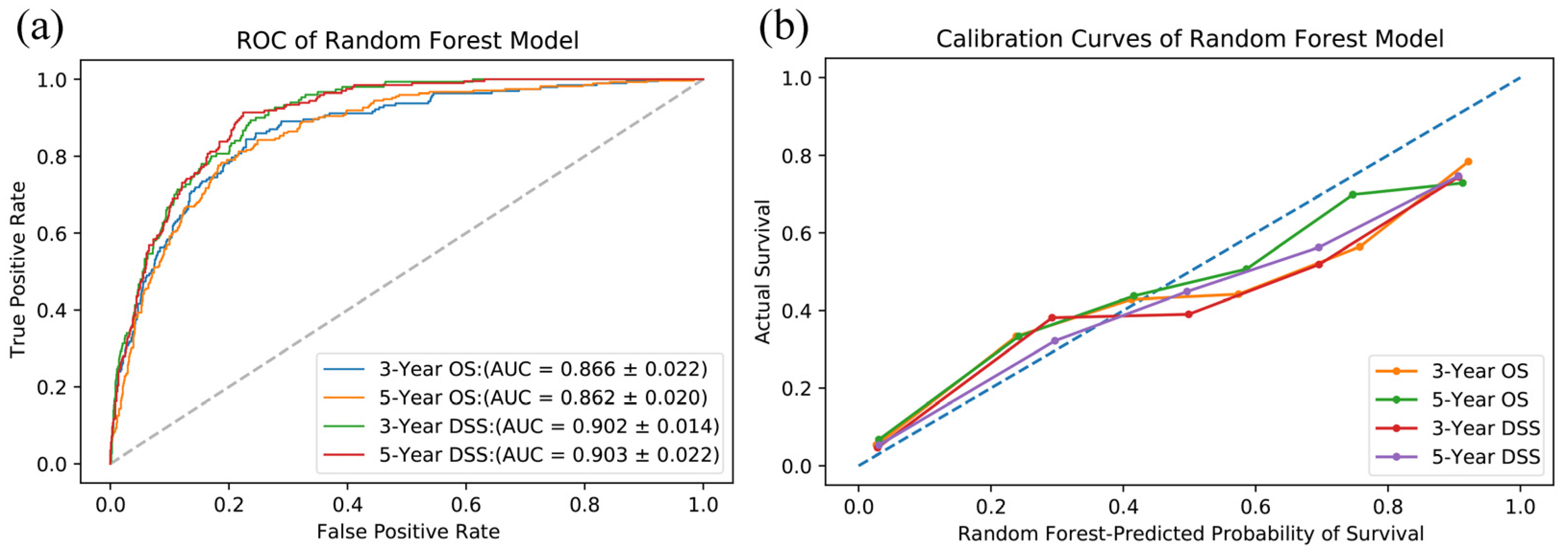
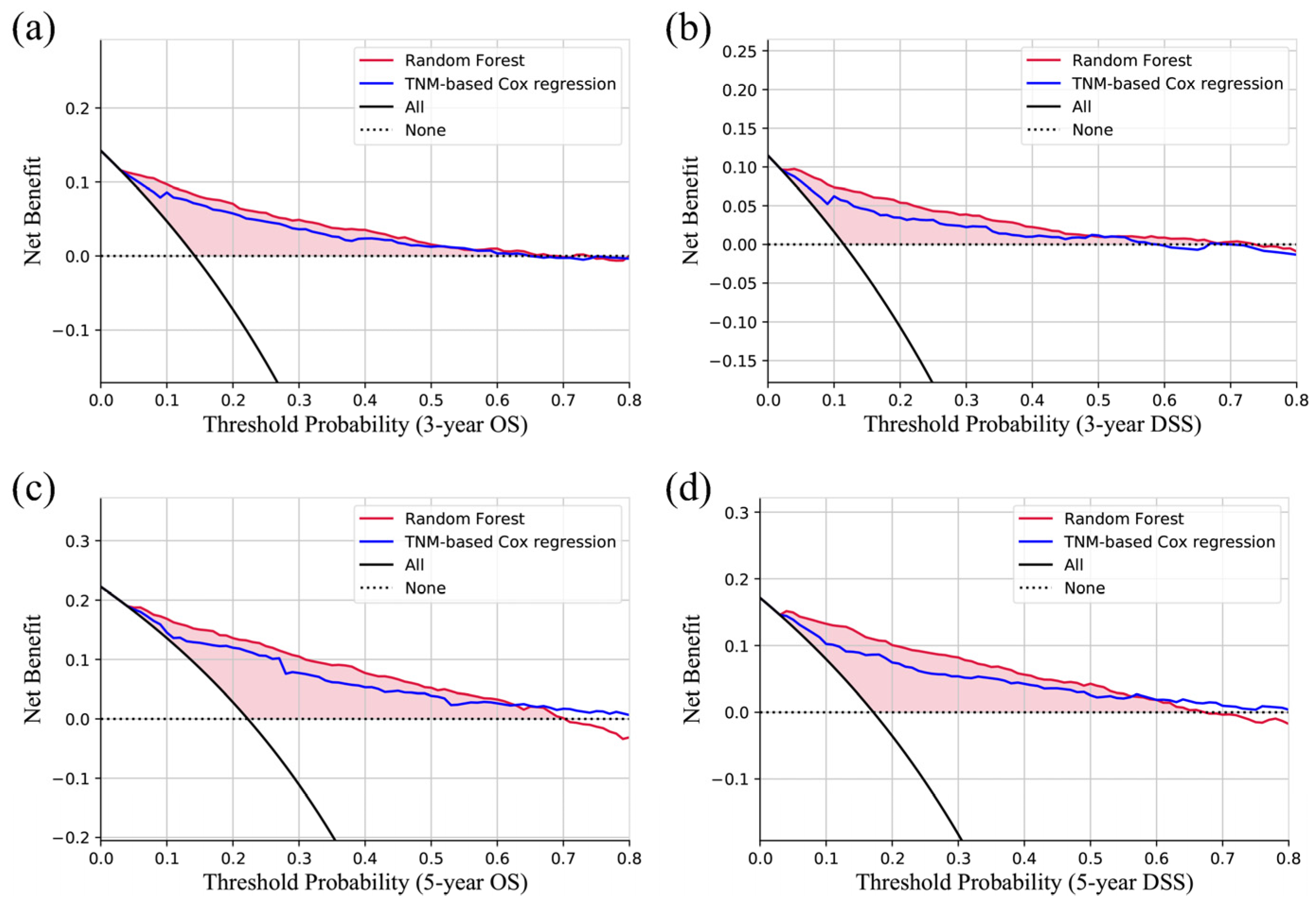
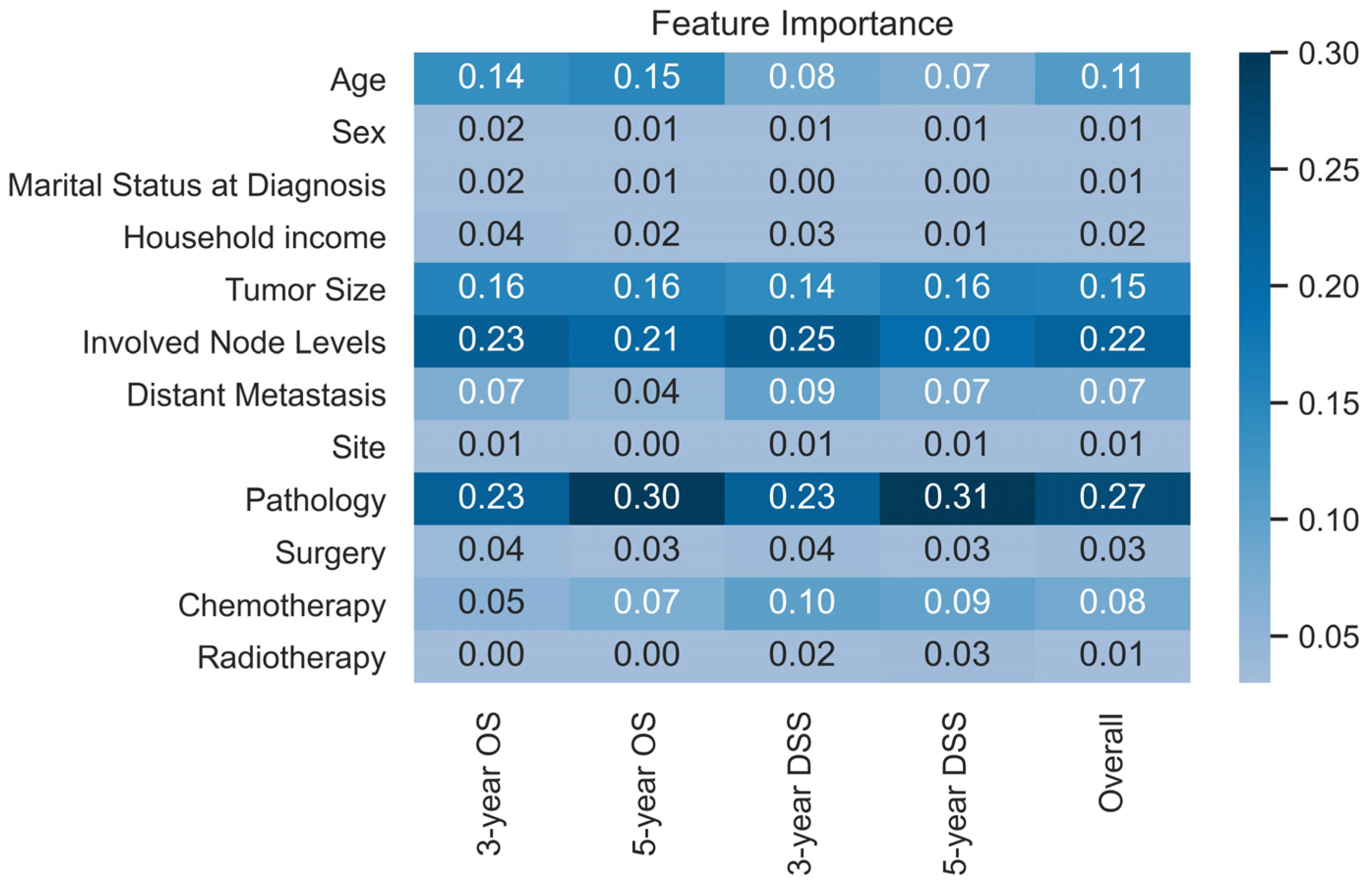
3.2. Model Evaluation
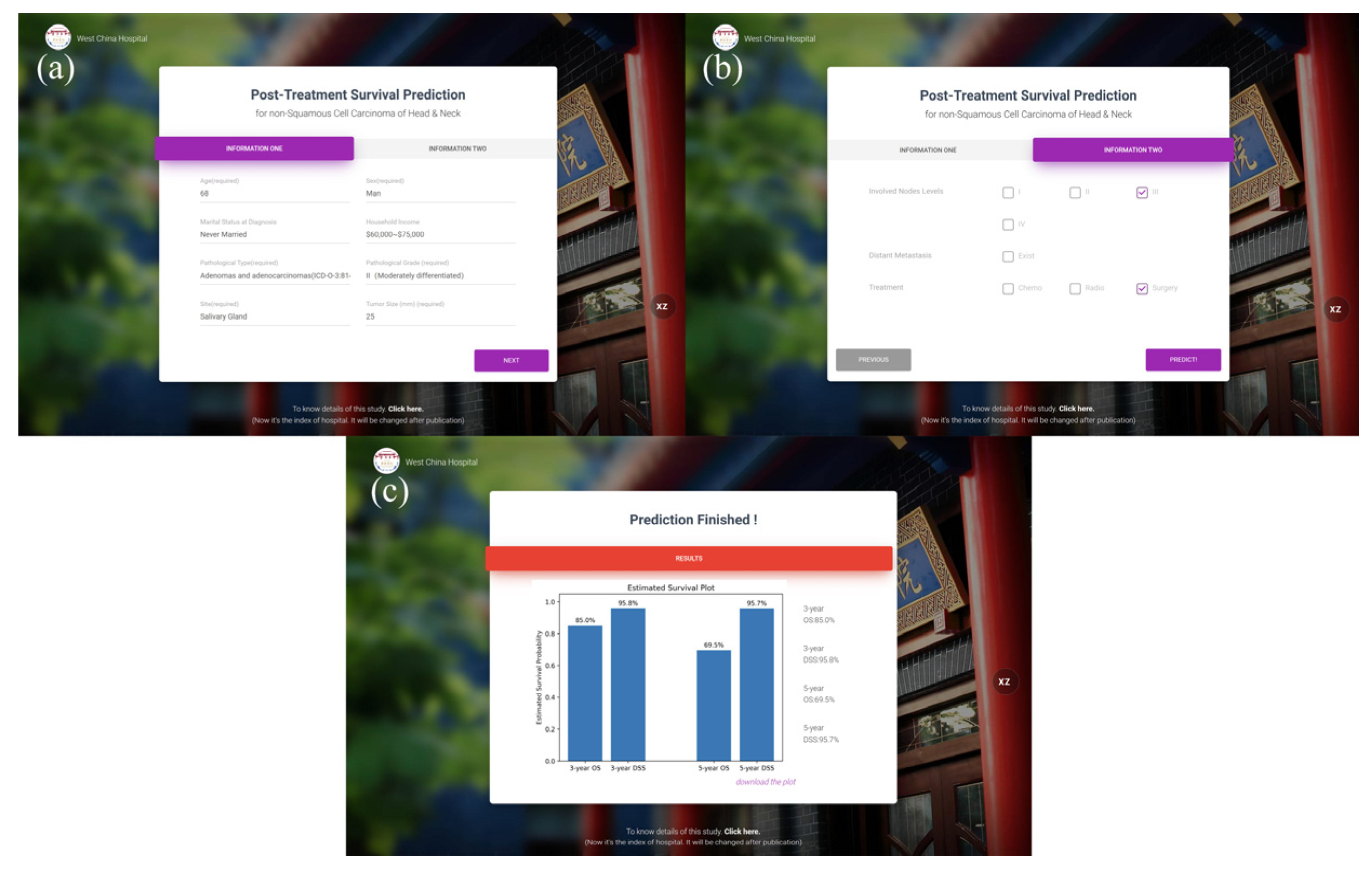
3.3. Prediction Website
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [PubMed]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [PubMed]
- Guo, K.; Xiao, W.; Chen, X.; Zhao, Z.; Lin, Y.; Chen, G. Epidemiological Trends of Head and Neck Cancer: A Population-Based Study. Biomed. Res. Int. 2021, 2021, 1738932. [Google Scholar] [PubMed]
- Imamura, Y.; Tanaka, K.; Kiyota, N.; Hayashi, H.; Ota, I.; Arai, A.; Iwae, S.; Minami, S.; Yane, K.; Yamazaki, T.; et al. Docetaxel plus cisplatin in recurrent and/or metastatic non-squamous-cell head and neck cancer: A multicenter phase II trial. Med. Oncol. 2021, 38, 128. [Google Scholar]
- Pulte, D.; Brenner, H. Changes in survival in head and neck cancers in the late 20th and early 21st century: A period analysis. Oncologist 2010, 15, 994–1001. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar]
- Li, W.; Zhao, K.; Wang, Z. Prognostic nomograms based on immune scores for head-neck squamous cell carcinoma patients. Eur. Arch. Otorhinolaryngol. 2021, 278, 2493–2500. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Li, J.; Huang, R.; Yao, Q.; Shi, Y.; Guo, S.; Wang, Y.; Cheng, J. Development of a novel head and neck squamous cell carcinoma prognostic signature by bulk/single-cell sequencing data integration. Oral Dis. 2022; in press. [Google Scholar] [CrossRef]
- Bobdey, S.; Balasubramaniam, G.; Mishra, P. Nomogram prediction for survival of patients with oral cavity squamous cell carcinoma. Head Neck 2016, 38, 1826–1831. [Google Scholar] [CrossRef]
- Yu, C.X.; Yibulayin, F.; Feng, L.; Wang, M.; Lu, M.M.; Luo, Y.; Liu, H.; Yang, Z.C.; Wushou, A. Clinicopathological characteristics, treatment and prognosis of head & neck small cell carcinoma: A SEER population-based study. BMC Cancer 2020, 20, 1208. [Google Scholar]
- Shuman, A.G.; Brennan, M.F.; Palmer, F.L.; Kuk, D.; Moraco, N.; Singer, S.; Shah, J.P.; Patel, S.G. Soft tissue sarcoma of the head & neck: Nomogram validation and analysis of staging systems. J. Surg. Oncol. 2015, 111, 690–695. [Google Scholar]
- Chou, W.C.; Chang, K.P.; Lu, C.H.; Chen, M.F.; Cheng, Y.F.; Yeh, K.Y.; Wang, C.H.; Lin, Y.C.; Yeh, T.S. Complementary role of the Memorial Sloan Kettering Cancer Center nomogram to the American Joint Committee on Cancer system for the prediction of relapse of major salivary gland carcinoma after surgery. Head Neck 2017, 39, 860–867. [Google Scholar] [PubMed]
- Lan, L.F.; Gao, C.K.; Ma, C.W. Prediction of Minor Salivary Gland Carcinoma: A Novel Nomogram and Risk Classification System for Overall Survival and Cancer-Specific Survival. Otolaryngol. Head Neck Surg. 2021, 164, 359–368. [Google Scholar] [PubMed]
- Alabi, R.O.; Mäkitie, A.A.; Pirinen, M.; Elmusrati, M.; Leivo, I.; Almangush, A. Comparison of nomogram with machine learning techniques for prediction of overall survival in patients with tongue cancer. Int. J. Med. Inform. 2021, 145, 104313. [Google Scholar] [CrossRef] [PubMed]
- Bean, M.B.; Liu, Y.; Jiang, R.; Steuer, C.E.; Patel, M.; McDonald, M.W.; Higgins, K.A.; Beitler, J.J.; Shin, D.M.; Saba, N.F. Small Cell and Squamous Cell Carcinomas of the Head and Neck: Comparing Incidence and Survival Trends Based on Surveillance, Epidemiology, and End Results (SEER) Data. Oncologist 2019, 24, 1562–1569. [Google Scholar] [PubMed] [Green Version]
- Sawhney, R.; Ahsanuddin, S.; Sheorey, L.; Wassef, D.W.; Baredes, S.; Park, R.C.W. Understanding giant cell sarcoma of the head and neck: A population-based study. Head Neck 2021, 43, 2786–2794. [Google Scholar] [PubMed]
- Olson, M.D.; Van Abel, K.M.; Wehrs, R.N.; Garcia, J.J.; Moore, E.J. Ewing sarcoma of the head and neck: The Mayo Clinic experience. Head Neck 2018, 40, 1999–2006. [Google Scholar]
- Cheng, N.M.; Kang, C.J.; Tsai, C.Y.; Lee, L.Y.; Lin, C.Y.; Hsueh, C.; Fan, K.H.; Wang, H.M.; Hsieh, C.H.; Ng, S.H.; et al. Improved prognostic stratification of patients with pN3b oral cavity cancer based on maximum standardized uptake value of metastatic nodes, lymph node ratio, and level of cervical nodal metastases. Oral Oncol. 2021, 123, 105593. [Google Scholar] [CrossRef]
- Liao, C.T.; Lee, L.Y.; Huang, S.F.; Chen, I.H.; Kang, C.J.; Lin, C.Y.; Fan, K.H.; Wang, H.M.; Ng, S.H.; Yen, T.C. Outcome analysis of patients with oral cavity cancer and extracapsular spread in neck lymph nodes. Int. J. Radiat. Oncol. Biol. Phys. 2011, 81, 930–937. [Google Scholar] [CrossRef]
- Klussmann, J.P.; Ponert, T.; Mueller, R.P.; Dienes, H.P.; Guntinas-Lichius, O. Patterns of lymph node spread and its influence on outcome in resectable parotid cancer. Eur. J. Surg. Oncol. 2008, 34, 932–937. [Google Scholar]
- Wu, Y.; Wang, L.; Ma, X.; Guo, W.; Ren, G. The existence of early stage oral mucosal melanoma: A 10-year retrospective analysis of 170 patients in a single institute. Oral Oncol. 2018, 87, 70–76. [Google Scholar] [CrossRef]
- Haase, K.; Piwonski, I.; Stromberger, C.; Thieme, N.; Heiland, M.; Beck-Broichsitter, B.; Hofmann, V.M.; Kofla, G.; Sander, S.; Keilholz, U.; et al. Incidence and survival of HNSCC patients living with HIV compared with HIV-negative HNSCC patients. Eur. Arch. Otorhinolaryngol. 2021, 278, 3941–3953. [Google Scholar] [CrossRef]
- Massa, S.T.; Cass, L.M.; Challapalli, S.; Zahirsha, Z.; Simpson, M.; Ward, G.; Osazuwa-Peters, N. Demographic predictors of head and neck cancer survival differ in the elderly. Laryngoscope 2019, 129, 146–153. [Google Scholar] [CrossRef]
- Peng, G.; Chi, H.; Gao, X.; Zhang, J.; Song, G.; Xie, X.; Su, K.; Song, B.; Yang, J.; Gu, T.; et al. Identification and validation of neurotrophic factor-related genes signature in HNSCC to predict survival and immune landscapes. Front. Genet. 2022, 13, 1010044. [Google Scholar] [CrossRef] [PubMed]
- Van Der Kamp, M.F.; Muntinghe, F.O.W.; Iepsma, R.S.; Plaat, B.E.C.; van der Laan, B.F.A.M.; Algassab, A.; Steenbakkers, R.J.H.M.; Witjes, M.J.H.; van Dijk, B.A.C.; de Bock, G.H.; et al. Predictors for distant metastasis in head and neck cancer, with emphasis on age. Eur. Arch. Otorhinolaryngol. 2021, 278, 181–190. [Google Scholar] [CrossRef]
- Zhu, R.Q.; Zhang, Y.M.; Luo, X.Y.; Shen, W.Y.; Zhu, H.Y. A novel nomogram and risk classification system for predicting overall survival in head and neck squamous cell cancer with distant metastasis at initial diagnosis. Eur. Arch. Otorhinolaryngol. 2023, 280, 1467–1478. [Google Scholar] [CrossRef]
- Afshar, N.; English, D.R.; Milne, R.L. Factors Explaining Socio-Economic Inequalities in Cancer Survival: A Systematic Review. Cancer Control 2021, 28, 10732748211011956. [Google Scholar] [CrossRef]
- Braaten, T.; Weiderpass, E.; Lund, E. Socioeconomic differences in cancer survival: The Norwegian Women and Cancer Study. BMC Public Health 2009, 9, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Wang, L.; Dai, S.; Chen, M.; Li, F.; Sun, J.; Luo, F. Epidemiologic Trends of and Factors Associated with Overall Survival for Patients with Gastroenteropancreatic Neuroendocrine Tumors in the United States. JAMA Netw. Open 2021, 4, e2124750. [Google Scholar] [CrossRef]
- Ding, Z.; Yu, D.; Li, H.; Ding, Y. Effects of marital status on overall and cancer-specific survival in laryngeal cancer patients: A population-based study. Sci. Rep. 2021, 11, 723. [Google Scholar] [CrossRef] [PubMed]
- Krajc, K.; Miroševič, Š.; Sajovic, J.; Klemenc Ketiš, Z.; Spiegel, D.; Drevenšek, G.; Drevenšek, M. Marital status and survival in cancer patients: A systematic review and meta-analysis. Cancer Med. 2023, 12, 1685–1708. [Google Scholar] [CrossRef]
- Kiecolt-Glaser, J.K.; Newton, T.L. Marriage and health: His and hers. Psychol. Bull. 2001, 127, 472–503. [Google Scholar] [CrossRef]
- Fugmann, D.; Boeker, M.; Holsteg, S.; Steiner, N.; Prins, J.; Karger, A. A Systematic Review: The Effect of Cancer on the Divorce Rate. Front. Psychol. 2022, 13, 828656. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, A.C.; Yi, J.; Wright, J.; Warner, E.L.; Smith, K.R. Marriage and divorce among young adult cancer survivors. J. Cancer Surviv. 2012, 6, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Zhu, H.; Zheng, X.; Jiang, M.; Chen, L.; Lan, L.; Ren, J.; Luo, X.; Zheng, J.; Zheng, Z.; et al. Machine Learning Models for the Diagnosis and Prognosis Prediction of High-Grade B-Cell Lymphoma. Front. Immunol. 2022, 13, 919012. [Google Scholar] [CrossRef]
- Marquardt, A.; Landwehr, L.S.; Ronchi, C.L.; di Dalmazi, G.; Riester, A.; Kollmannsberger, P.; Altieri, B.; Fassnacht, M.; Sbiera, S. Identifying New Potential Biomarkers in Adrenocortical Tumors Based on mRNA Expression Data Using Machine Learning. Cancers 2021, 13, 4671. [Google Scholar] [CrossRef]
- Fernandez-Delgado, M.; Cernadas, E.; Barro, S.; Amorim, D. Do we Need Hundreds of Classifiers to Solve Real World Classification Problems? J. Mach. Learn. Res. 2014, 15, 3133–3181. [Google Scholar]

| Characteristic | Cohort, No. (%) | |||||
|---|---|---|---|---|---|---|
| Train & Validation | Test | Total | ||||
| Age, y | ||||||
| <45 | 895 | 28.8% | 391 | 28.9% | 1286 | 28.8% |
| 45–59 | 884 | 28.5% | 407 | 30.1% | 1291 | 29.0% |
| 60–74 | 884 | 28.5% | 385 | 28.4% | 1269 | 28.5% |
| >74 | 441 | 14.2% | 171 | 12.6% | 612 | 13.7% |
| Sex | ||||||
| Male | 1507 | 48.6% | 672 | 49.6% | 2179 | 48.9% |
| Female | 1597 | 51.4% | 682 | 50.4% | 2279 | 51.1% |
| Marital Status at Diagnosis | ||||||
| Married | 1763 | 56.8% | 760 | 56.1% | 2523 | 56.6% |
| Never Married | 702 | 22.6% | 313 | 23.1% | 1015 | 22.8% |
| Widowed/Separated/Others | 639 | 20.6% | 281 | 20.8% | 920 | 20.6% |
| Race | ||||||
| White | 2370 | 76.4% | 1484 | 109.6% | 3854 | 86.5% |
| Black | 369 | 11.9% | 217 | 16.0% | 586 | 13.1% |
| Asian | 327 | 10.5% | 202 | 14.9% | 529 | 11.9% |
| Others | 38 | 1.2% | 28 | 2.1% | 66 | 1.5% |
| Median household income | ||||||
| USD 0~45,000 | 238 | 7.7% | 93 | 6.9% | 331 | 7.4% |
| USD 45,000~60,000 | 653 | 21.0% | 296 | 21.9% | 949 | 21.3% |
| USD 60,000~75,000 | 1287 | 41.5% | 533 | 39.4% | 1820 | 40.8% |
| USD 75,000+ | 926 | 29.8% | 432 | 31.9% | 1358 | 30.5% |
| Living Area | ||||||
| Urban | 2752 | 88.7% | 1215 | 89.7% | 3967 | 89.0% |
| Rural | 352 | 11.3% | 139 | 10.3% | 491 | 11.0% |
| TNM stage | ||||||
| I | 1218 | 39.2% | 558 | 41.2% | 1776 | 39.8% |
| II | 669 | 21.6% | 284 | 21.0% | 953 | 21.4% |
| III | 467 | 15.0% | 180 | 13.3% | 647 | 14.5% |
| IV | 750 | 24.2% | 332 | 24.5% | 1082 | 24.3% |
| T stage | ||||||
| T0 | 2 | 0.1% | 0 | 0.0% | 2 | 0.0% |
| T1 | 1329 | 42.8% | 600 | 44.3% | 1929 | 43.3% |
| T2 | 813 | 26.2% | 357 | 26.4% | 1170 | 26.2% |
| T3 | 481 | 15.5% | 189 | 14.0% | 670 | 15.0% |
| T4 | 479 | 15.4% | 208 | 15.4% | 687 | 15.4% |
| N stage | ||||||
| N0 | 2484 | 80.0% | 1067 | 78.8% | 3551 | 79.7% |
| N1 | 257 | 8.3% | 116 | 8.6% | 373 | 8.4% |
| N2 | 348 | 11.2% | 165 | 12.2% | 513 | 11.5% |
| N3 | 15 | 0.5% | 6 | 0.4% | 21 | 0.5% |
| M stage | ||||||
| M0 | 3014 | 97.1% | 1319 | 97.4% | 4333 | 97.2% |
| M1 | 90 | 2.9% | 35 | 2.6% | 125 | 2.8% |
| Tumor site | ||||||
| Salivary gland | 2091 | 67.4% | 912 | 67.4% | 3003 | 67.4% |
| Oral cavity | 747 | 24.1% | 341 | 25.2% | 1088 | 24.4% |
| Nasal cavity/paranasal sinus | 168 | 5.4% | 53 | 3.9% | 221 | 5.0% |
| Larynx/hypopharynx | 37 | 1.2% | 13 | 1.0% | 50 | 1.1% |
| Nasopharynx | 27 | 0.9% | 16 | 1.2% | 43 | 1.0% |
| Oropharynx | 34 | 1.1% | 19 | 1.4% | 53 | 1.2% |
| Histopathologic type | ||||||
| Acinar cell neoplasms | 252 | 8.1% | 118 | 8.7% | 370 | 8.3% |
| Adenomas and adenocarcinomas | 908 | 29.3% | 359 | 26.5% | 1267 | 28.4% |
| Complex mixed and stromal neoplasms | 173 | 5.6% | 77 | 5.7% | 250 | 5.6% |
| Ductal and lobular neoplasms | 98 | 3.2% | 43 | 3.2% | 141 | 3.2% |
| Mucoepidermoid neoplasms | 1363 | 43.9% | 617 | 45.6% | 1980 | 44.4% |
| Others | 310 | 10.0% | 140 | 10.3% | 450 | 10.1% |
| Histopathologic Grade | ||||||
| I (Well differentiated) | 820 | 26.4% | 355 | 26.2% | 1175 | 26.4% |
| II (Moderately differentiated) | 1335 | 43.0% | 573 | 42.3% | 1908 | 42.8% |
| III (Poorly differentiated) | 565 | 18.2% | 265 | 19.6% | 830 | 18.6% |
| IV (Undifferentiated) | 384 | 12.4% | 161 | 11.9% | 545 | 12.2% |
| Surgery | ||||||
| Yes | 2964 | 95.5% | 1300 | 96.0% | 4264 | 95.6% |
| No | 140 | 4.5% | 54 | 4.0% | 194 | 4.4% |
| Radiotherapy | ||||||
| Yes | 1541 | 49.6% | 680 | 50.2% | 2221 | 49.8% |
| No evidence | 1563 | 50.4% | 674 | 49.8% | 2237 | 50.2% |
| Chemotherapy | ||||||
| Yes | 331 | 10.7% | 179 | 13.2% | 510 | 11.4% |
| No evidence | 2773 | 89.3% | 1175 | 86.8% | 3948 | 88.6% |
| Tumor size | ||||||
| ≤20 mm | 1488 | 47.9% | 664 | 49.0% | 2152 | 48.3% |
| >20 mm, ≤40 mm | 1195 | 38.5% | 500 | 36.9% | 1695 | 38.0% |
| >40 mm | 421 | 13.6% | 190 | 14.0% | 611 | 13.7% |
| Involved lymph nodes | ||||||
| Levels I | 271 | 8.7% | 121 | 8.9% | 392 | 8.8% |
| Levels II | 319 | 10.3% | 166 | 12.3% | 485 | 10.9% |
| Levels III | 174 | 5.6% | 88 | 6.5% | 262 | 5.9% |
| Levels IV | 91 | 2.9% | 41 | 3.0% | 132 | 3.0% |
| Levels V | 83 | 2.7% | 42 | 3.1% | 125 | 2.8% |
| Levels Parotid | 143 | 4.6% | 51 | 3.8% | 194 | 4.4% |
| Levels Others | 46 | 1.5% | 20 | 1.5% | 66 | 1.5% |
| Characteristic | Univariate Analysis | Multivariate Analysis | |
|---|---|---|---|
| p Value | HR (95%CI) | p Values | |
| Age, y | <0.001 | <0.001 | |
| <45 | Reference | ||
| 45–59 | 1.948 (1.508, 2.517) | <0.001 | |
| 60–74 | 2.524 (1.964, 3.243) | <0.001 | |
| >74 | 6.698 (5.159, 8.694) | <0.001 | |
| Sex | <0.001 | ||
| Female | Reference | ||
| Male | 1.281 (1.108, 1.480) | 0.001 | |
| Marital Status at Diagnosis | <0.001 | 0.010 | |
| Married | Reference | ||
| Never Married | 1.062 (0.964, 1.206) | 0.113 | |
| Widowed/Separated/Others | 1.291 (1.095, 1.521) | 0.002 | |
| Race | <0.001 | 0.823 | |
| White | Reference | ||
| Black | 1.047 (0.830, 1.321) | 0.698 | |
| Asian | 1.036 (0.799, 1.342) | 0.792 | |
| Others | 1.474 (0.605, 3.596) | 0.393 | |
| Median household income (adj to 2019) | <0.001 | 0.001 | |
| USD 0~45,000 | 1.720 (1.267, 2.334) | 0.001 | |
| USD 45,000~60,000 | 1.138 (0.919, 1.408) | 0.235 | |
| USD 60,000~75,000 | 1.272 (1.074, 1.507) | 0.005 | |
| USD 75,000+ | Reference | ||
| Living in Rural Area | 0.009 | 1.177 (0.923, 1.503) | 0.189 |
| Tumor size | <0.001 | <0.001 | |
| ≤20 mm | Reference | ||
| >20 mm, ≤40 mm | 1.625 (1.371, 1.927) | <0.001 | |
| >40 mm | 2.648 (2.165, 3.238) | <0.001 | |
| Involved lymph nodes | |||
| Levels I | <0.001 | 1.247 (1.029, 1.510) | 0.024 |
| Levels II | <0.001 | 1.322 (1.051, 1.664) | 0.017 |
| Levels III | <0.001 | 1.409 (1.048, 1.893) | 0.023 |
| Levels IV | <0.001 | 1.493 (1.038, 2.147) | 0.031 |
| Levels V | <0.001 | 1.384 (0.984, 1.946) | 0.062 |
| Levels Parotid | <0.001 | 1.016 (0.786, 1.312) | 0.906 |
| Levels Others | <0.001 | 1.046 (0.676, 1.618) | 0.840 |
| Distant Metastasis | <0.001 | 3.406 (2.589, 4.481) | <0.001 |
| Tumor site | <0.001 | 0.016 | |
| Salivary gland | Reference | ||
| Oral cavity | 0.990 (0.812, 1.207) | 0.923 | |
| Nasal cavity/paranasal sinus | 1.084 (0.827, 1.419) | 0.560 | |
| Larynx/hypopharynx | 2.209 (1.419, 3.437) | <0.001 | |
| Nasopharynx | 0.850 (0.466, 1.552) | 0.598 | |
| Oropharynx | 0.864 (0.480, 1.554) | 0.625 | |
| Histopathologic type | <0.001 | <0.001 | |
| Acinar cell neoplasms | 1.322 (0.932.1.875) | 0.118 | |
| Adenomas and adenocarcinomas | 1.480 (1.246, 1.757) | <0.001 | |
| Complex mixed and stromal neoplasms | 0.800 (0.599, 1.068) | 0.130 | |
| Ductal and lobular neoplasms | 1.167 (0.847, 1.607) | 0.344 | |
| Mucoepidermoid neoplasms | Reference | ||
| Others | 1.520 (1.187, 1.946) | <0.001 | |
| Histopathologic Grade | <0.001 | <0.001 | |
| I (Well differentiated) | Reference | ||
| II (Moderately differentiated) | 1.589 (1.246, 2.026) | <0.001 | |
| III (Poorly differentiated) | 3.443 (2.655, 4.465) | <0.001 | |
| IV (Undifferentiated) | 3.836 (2.932, 5.017) | <0.001 | |
| Treatment | |||
| Surgery | <0.001 | 0.675 (0.519, 0.877) | 0.003 |
| Radiotherapy | <0.001 | 1.091 (0.929, 1.281) | 0.287 |
| Chemotherapy | <0.001 | 1.271 (1.049, 1.540) | 0.014 |
| Characteristic | Univariate Analysis | Multivariate Analysis | |
|---|---|---|---|
| p Value | HR (95%CI) | p Values | |
| Age, y | <0.001 | <0.001 | |
| <45 | Reference | ||
| 45–59 | 1.514 (1.137, 2.014) | 0.004 | |
| 60–74 | 1.636 (1.234, 2.170) | 0.001 | |
| >74 | 2.705 (1.978, 3.699) | <0.001 | |
| Sex | <0.001 | ||
| Female | Reference | ||
| Male | 1.221 (1.018, 1.464) | 0.031 | |
| Marital Status at Diagnosis | 0.011 | 0.187 | |
| Married | Reference | ||
| Never Married | 0.861 (0.668, 1.108) | 0.244 | |
| Widowed/Separated/Others | 1.137 (0.919, 1.406) | 0.236 | |
| Race | 0.008 | 0.525 | |
| White | Reference | ||
| Black | 1.107 (0.827, 1.482) | 0.492 | |
| Asian | 1.148 (0.839, 1.572) | 0.389 | |
| Others | 1.754 (0.642, 4.795) | 0.273 | |
| Median household income (adj to 2019) | <0.001 | 0.015 | |
| USD 0~45,000 | 1.570 (1.084, 2.274) | 0.017 | |
| USD 45,000~60,000 | 1.039 (0.796, 1.355) | 0.779 | |
| USD 60,000~75,000 | 1.275 (1.033, 1.573) | 0.024 | |
| USD 75,000+ | Reference | ||
| Living in Rural Area | 0.016 | 1.143 (0.846, 1.544) | 0.384 |
| Tumor size | <0.001 | <0.001 | |
| ≤20 mm | Reference | ||
| >20 mm, ≤40 mm | 1.818 (1.450, 2.281) | <0.001 | |
| >40 mm | 3.101 (2.408, 3.994) | <0.001 | |
| Involved lymph nodes | |||
| Levels I | <0.001 | 1.548 (1.252, 1.914) | <0.001 |
| Levels II | <0.001 | 1.353 (1.044, 1.753) | 0.021 |
| Levels III | <0.001 | 1.341 (0.963, 1.867) | 0.083 |
| Levels IV | <0.001 | 1.530 (1.059, 2.211) | 0.024 |
| Levels V | <0.001 | 1.392 (0.970, 1.998) | 0.073 |
| Levels Parotid | <0.001 | 1.185 (0.895, 1.569) | 0.238 |
| Levels Others | <0.001 | 0.843 (0.516, 1.377) | 0.495 |
| Distant Metastasis | <0.001 | 3.758 (2.792, 5.057) | <0.001 |
| Tumor site | <0.001 | 0.001 | |
| Salivary gland | Reference | ||
| Oral cavity | 1.113 (0.862, 1.438) | 0.412 | |
| Nasal cavity/paranasal sinus | 1.143 (0.823, 1.587) | 0.425 | |
| Larynx/hypopharynx | 2.608 (1.603, 4.245) | <0.001 | |
| Nasopharynx | 0.751 (0.364, 1.548) | 0.437 | |
| Oropharynx | 0.852 (0.420, 1.730) | 0.658 | |
| Histopathologic type | <0.001 | <0.001 | |
| Acinar cell neoplasms | 1.867 (1.218, 2.863) | 0.004 | |
| Adenomas and adenocarcinomas | 1.782 (1.432, 2.219) | <0.001 | |
| Complex mixed and stromal neoplasms | 0.858 (0.602, 1.223) | 0.398 | |
| Ductal and lobular neoplasms | 1.228 (0.846, 1.783) | 0.279 | |
| Mucoepidermoid neoplasms | Reference | ||
| Others | 1.414 (1.003, 1.993) | 0.048 | |
| Histopathologic Grade | <0.001 | <0.001 | |
| I (Well differentiated) | Reference | ||
| II (Moderately differentiated) | 3.040 (1.990, 4.644) | <0.001 | |
| III (Poorly differentiated) | 8.372 (5.440, 12.885) | <0.001 | |
| IV (Undifferentiated) | 9.006 (5.818, 13.940) | <0.001 | |
| Treatment | |||
| Surgery | <0.001 | 0.668 (0.493, 0.906) | 0.010 |
| Radiotherapy | <0.001 | 1.343 (1.085, 1.662) | 0.007 |
| Chemotherapy | <0.001 | 1.212 (0.976, 1.505) | 0.014 |
| Survival | Test Cohort | ||
|---|---|---|---|
| AUC | 95%CI | p Value | |
| OS-3 year | <0.001 | ||
| Random Forest | 0.866 | 0.844–0.888 | |
| TNM-based Cox | 0.831 | 0.802–0.860 | |
| OS-5 year | <0.001 | ||
| Random Forest | 0.862 | 0.842–0.882 | |
| TNM-based Cox | 0.836 | 0.808–0.864 | |
| DSS-3 year | <0.001 | ||
| Random Forest | 0.902 | 0.888–0.916 | |
| TNM-based Cox | 0.861 | 0.825–0.897 | |
| DSS-5 year | <0.001 | ||
| Random Forest | 0.903 | 0.881–0.925 | |
| TNM-based Cox | 0.872 | 0.846–0.902 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Liu, G.; Peng, X. A Random Forest Model for Post-Treatment Survival Prediction in Patients with Non-Squamous Cell Carcinoma of the Head and Neck. J. Clin. Med. 2023, 12, 5015. https://doi.org/10.3390/jcm12155015
Zhang X, Liu G, Peng X. A Random Forest Model for Post-Treatment Survival Prediction in Patients with Non-Squamous Cell Carcinoma of the Head and Neck. Journal of Clinical Medicine. 2023; 12(15):5015. https://doi.org/10.3390/jcm12155015
Chicago/Turabian StyleZhang, Xin, Guihong Liu, and Xingchen Peng. 2023. "A Random Forest Model for Post-Treatment Survival Prediction in Patients with Non-Squamous Cell Carcinoma of the Head and Neck" Journal of Clinical Medicine 12, no. 15: 5015. https://doi.org/10.3390/jcm12155015








