The PD-L1 Expression and Tumor-Infiltrating Immune Cells Predict an Unfavorable Prognosis in Pancreatic Ductal Adenocarcinoma and Adenosquamous Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Involved Samples
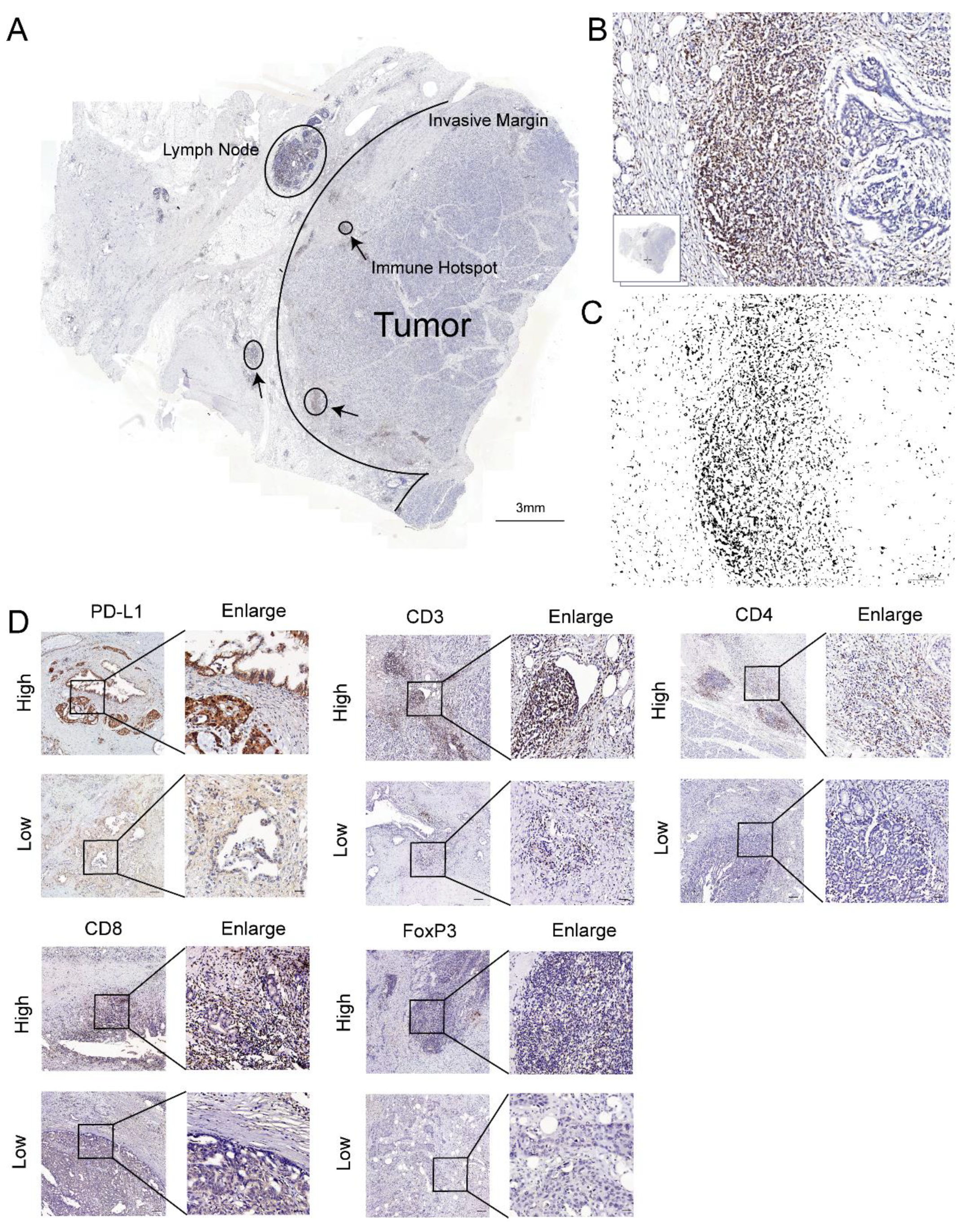
2.2. Immunohistochemistry (IHC)
2.3. Immunohistochemical Score
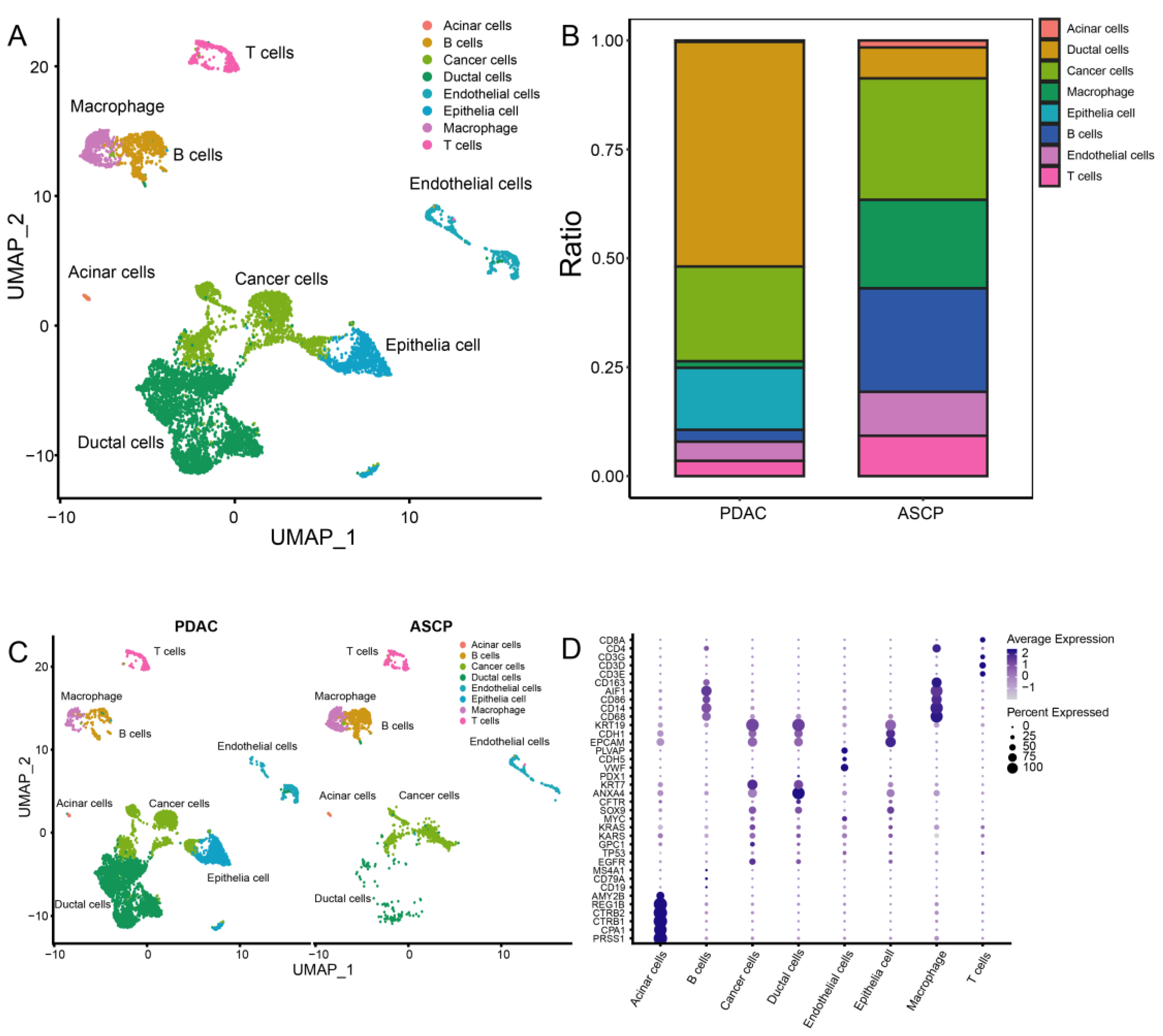
2.4. Integration, Clustering and Cell Type Identification
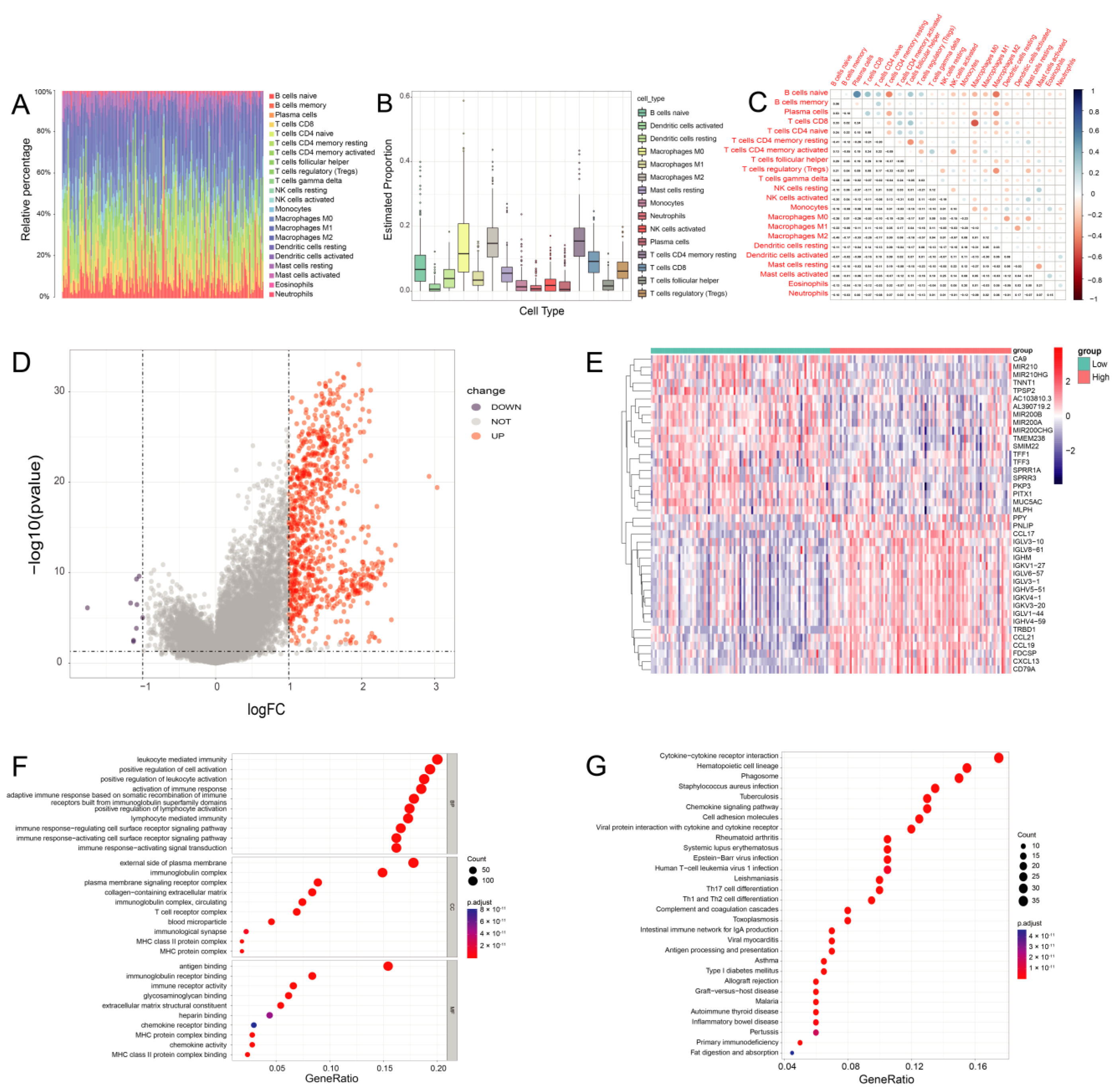
2.5. Assessment of Tumor-Infiltrating Immune Cells (TICs)
2.6. Assessment of TME Parameters (ImmuneScore, StromalScore, and ESTIMATEScore)
2.7. Identification of Differentially Expressed Genes (DEGs)
2.8. Statistical Analysis
3. Results
3.1. High PD-L1 Expression Is Associated with Shorter Overall Survival
3.2. CD3+, CD4+, CD8+, and FoxP3+ T Cells Infiltrate Pancreatic Tumors and Correlate with Survival
3.3. Cell Clustering of PDAC and ASCP
3.4. Dynamic Interaction between Tumor Cells and Immune Cells
3.5. The Proportion of TICs Was Rare in PC
3.6. DEGs Assessed by the ImmuneScore
3.7. PD-L1 Expression Is Related to the Proportion of TICs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.; Miller, K.; Fuchs, H.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.; Hong, T.; Bardeesy, N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014, 371, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Borazanci, E.; Millis, S.Z.; Korn, R.; Han, H.; Whatcott, C.J.; Gatalica, Z.; Barrett, M.T.; Cridebring, D.; Von Hoff, D.D. Adenosquamous carcinoma of the pancreas: Molecular characterization of 23 patients along with a literature review. World J. Gastrointest. Oncol. 2015, 7, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Boyd, C.A.; Benarroch-Gampel, J.; Sheffield, K.M.; Cooksley, C.D.; Riall, T.S. 415 Patients with Adenosquamous Carcinoma of the Pancreas: A Population-Based Analysis of Prognosis and Survival. J. Surg. Res. 2012, 174, 12–19. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhang, Z.; Xu, Y.; Zhu, Q. Pancreatic Adenosquamous Carcinoma: A Rare Pathological Subtype of Pancreatic Cancer. J. Clin. Med. 2022, 11, 7401. [Google Scholar] [CrossRef]
- Vonderheide, R.H.; Bayne, L.J. Inflammatory networks and immune surveillance of pancreatic carcinoma. Curr. Opin. Immunol. 2013, 25, 200–205. [Google Scholar] [CrossRef]
- Kabacaoglu, D.; Ciecielski, K.J.; Ruess, D.A.; Algül, H. Immune Checkpoint Inhibition for Pancreatic Ductal Adenocarcinoma: Current Limitations and Future Options. Front. Immunol. 2018, 9, 1878. [Google Scholar] [CrossRef]
- Karakhanova, S.; Link, J.; Heinrich, M.; Shevchenko, I.; Yang, Y.; Hassenpflug, M.; Bunge, H.; Von Ahn, K.; Brecht, R.; Mathes, A.; et al. Characterization of myeloid leukocytes and soluble mediators in pancreatic cancer: Importance of myeloid-derived suppressor cells. Oncoimmunology 2015, 4, e998519. [Google Scholar] [CrossRef]
- Martinez-Bosch, N.; Vinaixa, J.; Navarro, P. Immune Evasion in Pancreatic Cancer: From Mechanisms to Therapy. Cancers 2018, 10, 6. [Google Scholar] [CrossRef]
- Orhan, A.; Vogelsang, R.P.; Andersen, M.B.; Madsen, M.T.; Hölmich, E.R.; Raskov, H.; Gögenur, I. The prognostic value of tumour-infiltrating lymphocytes in pancreatic cancer: A systematic review and meta-analysis. Eur. J. Cancer 2020, 132, 71–84. [Google Scholar] [CrossRef]
- Carstens, J.L.; de Sampaio, P.C.; Yang, D.; Barua, S.; Wang, H.; Rao, A.; Allison, J.P.; LeBleu, V.S.; Kalluri, R. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat. Commun. 2017, 8, 15095. [Google Scholar] [CrossRef]
- Tahkola, K.; Mecklin, J.-P.; Wirta, E.-V.; Ahtiainen, M.; Helminen, O.; Böhm, J.; Kellokumpu, I. High immune cell score predicts improved survival in pancreatic cancer. Virchows Arch. 2018, 472, 653–665. [Google Scholar] [CrossRef]
- Wang, W.-Q.; Liu, L.; Xu, H.-X.; Wu, C.-T.; Xiang, J.-F.; Xu, J.; Liu, C.; Long, J.; Ni, Q.-X.; Yu, X.-J. Infiltrating immune cells and gene mutations in pancreatic ductal adenocarcinoma. Br. J. Surg. 2016, 103, 1189–1199. [Google Scholar] [CrossRef]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.-S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Wirta, E.-V.; Seppälä, T.; Friman, M.; Väyrynen, J.; Ahtiainen, M.; Kautiainen, H.; Kuopio, T.; Kellokumpu, I.; Mecklin, J.-P.; Böhm, J. Immunoscore in mismatch repair-proficient and -deficient colon cancer. J. Pathol. Clin. Res. 2017, 3, 203–213. [Google Scholar] [CrossRef]
- Ino, Y.; Yamazaki-Itoh, R.; Shimada, K.; Iwasaki, M.; Kosuge, T.; Kanai, Y.; Hiraoka, N. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br. J. Cancer 2013, 108, 914–923. [Google Scholar] [CrossRef]
- Hiraoka, N.; Onozato, K.; Kosuge, T.; Hirohashi, S. Prevalence of FOXP3+ Regulatory T Cells Increases During the Progression of Pancreatic Ductal Adenocarcinoma and Its Premalignant Lesions. Clin. Cancer Res. 2006, 12, 5423–5434. [Google Scholar] [CrossRef]
- Hou, Y.; Chao, Y.; Hsieh, M.; Tung, H.; Wang, H.; Shan, Y. Low CD8(+) T Cell Infiltration and High PD-L1 Expression Are Associated with Level of CD44(+)/CD133(+) Cancer Stem Cells and Predict an Unfavorable Prognosis in Pancreatic Cancer. Cancers 2019, 11, 541. [Google Scholar] [CrossRef]
- Yu, J.; Wang, X.; Teng, F.; Kong, L. PD-L1 expression in human cancers and its association with clinical outcomes. OncoTargets Ther. 2016, 9, 5023–5039. [Google Scholar] [CrossRef]
- Sideras, K.; Biermann, K.; Yap, K.; Mancham, S.; Boor, P.P.; Hansen, B.E.; Stoop, H.J.; Peppelenbosch, M.; Van Eijck, C.H.; Sleijfer, S.; et al. Tumor cell expression of immune inhibitory molecules and tumor-infiltrating lymphocyte count predict cancer-specific survival in pancreatic and ampullary cancer. Int. J. Cancer 2017, 141, 572–582. [Google Scholar] [CrossRef]
- Tahkola, K.; Leppänen, J.; Ahtiainen, M.; Väyrynen, J.; Haapasaari, K.-M.; Karttunen, T.; Kellokumpu, I.; Helminen, O.; Böhm, J. Immune cell score in pancreatic cancer—Comparison of hotspot and whole-section techniques. Virchows Arch. 2019, 474, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. Review The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Moncada, R.; Barkley, D.; Wagner, F.; Chiodin, M.; Devlin, J.C.; Baron, M.; Hajdu, C.H.; Simeone, D.M.; Yanai, I. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 2020, 38, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, H.; Lyu, S.; Zhai, J.; Ji, Z.; Zhang, Z.; Zhang, X.; Liu, Z.; Wang, H.; Xu, J.; et al. Single-cell transcriptomics reveals heterogeneous progression and EGFR activation in pancreatic adenosquamous carcinoma. Int. J. Biol. Sci. 2021, 17, 2590–2605. [Google Scholar] [CrossRef]
- Varghese, F.; Bukhari, A.B.; Malhotra, R.; De, A. IHC Profiler: An Open Source Plugin for the Quantitative Evaluation and Automated Scoring of Immunohistochemistry Images of Human Tissue Samples. PLoS ONE 2014, 9, e96801. [Google Scholar] [CrossRef]
- Väyrynen, J.P.; Vornanen, J.O.; Sajanti, S.; Böhm, J.P.; Tuomisto, A.; Mäkinen, M.J. An improved image analysis method for cell counting lends credibility to the prognostic significance of T cells in colorectal cancer. Virchows Arch. 2012, 460, 455–465. [Google Scholar] [CrossRef]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Zhang, X.; Lan, Y.; Xu, J.; Quan, F.; Zhao, E.; Deng, C.; Luo, T.; Xu, L.; Liao, G.; Yan, M.; et al. CellMarker: A manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019, 47, D721–D728. [Google Scholar] [CrossRef]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Finn, O.J. Immuno-oncology: Understanding the function and dysfunction of the immune system in cancer. Ann. Oncol. 2012, 23 (Suppl. S8), viii6–viii9. [Google Scholar] [CrossRef]
- Cai, J.; Wang, D.; Zhang, G.; Guo, X. The Role Of PD-1/PD-L1 Axis in Treg Development and Function: Implications for Cancer Immunotherapy. OncoTargets Ther. 2019, 12, 8437–8445. [Google Scholar] [CrossRef]
- Nikolova, M.; Wiedemann, A.; Muhtarova, M.; Achkova, D.; Lacabaratz, C.; Lévy, Y. Subset- and Antigen-Specific Effects of Treg on CD8+ T Cell Responses in Chronic HIV Infection. PLOS Pathog. 2016, 12, e1005995. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Marchesi, F.; Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 2022, 21, 799–820. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Wei, X.; Jiang, H.; Lan, C.; Yang, S.; Wang, H.; Yang, Y.; Tian, C.; Xu, Z.; et al. PD-L1 is a direct target of cancer-FOXP3 in pancreatic ductal adenocarcinoma (PDAC), and combined immunotherapy with antibodies against PD-L1 and CCL5 is effective in the treatment of PDAC. Signal Transduct. Target. Ther. 2020, 5, 38. [Google Scholar] [CrossRef]
- Peng, W.; Liu, C.; Xu, C.; Lou, Y.; Chen, J.; Yang, Y.; Yagita, H.; Overwijk, W.; Lizee, G.; Radvanyi, L.; et al. PD-1 blockade enhances T-cell migration to tumors by elevating IFN-gamma inducible chemokines. Cancer Res. 2012, 72, 5209–5218. [Google Scholar] [CrossRef]
- Winograd, R.; Byrne, K.T.; Evans, R.A.; Odorizzi, P.M.; Meyer, A.R.; Bajor, D.L.; Clendenin, C.; Stanger, B.Z.; Furth, E.E.; Wherry, E.J.; et al. Induction of T-cell Immunity Overcomes Complete Resistance to PD-1 and CTLA-4 Blockade and Improves Survival in Pancreatic Carcinoma. Cancer Immunol. Res. 2015, 3, 399–411. [Google Scholar] [CrossRef]
- Butte, M.J.; Keir, M.E.; Phamduy, T.B.; Sharpe, A.H.; Freeman, G.J. Programmed Death-1 Ligand 1 Interacts Specifically with the B7-1 Costimulatory Molecule to Inhibit T Cell Responses. Immunity 2007, 27, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Nomi, T.; Sho, M.; Akahori, T.; Hamada, K.; Kubo, A.; Kanehiro, H.; Nakamura, S.; Enomoto, K.; Yagita, H.; Azuma, M.; et al. Clinical Significance and Therapeutic Potential of the Programmed Death-1 Ligand/Programmed Death-1 Pathway in Human Pancreatic Cancer. Clin. Cancer Res. 2007, 13, 2151–2157. [Google Scholar] [CrossRef] [PubMed]
- Moral, J.A.; Leung, J.; Rojas, L.A.; Ruan, J.; Zhao, J.; Sethna, Z.; Ramnarain, A.; Gasmi, B.; Gururajan, M.; Redmond, D.; et al. ILC2s amplify PD-1 blockade by activating tissue-specific cancer immunity. Nature 2020, 579, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.; Fearon, D. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015, 348, 74–80. [Google Scholar] [CrossRef]
- Mirji, G.; Worth, A.; Bhat, S.A.; El Sayed, M.; Kannan, T.; Goldman, A.R.; Tang, H.-Y.; Liu, Q.; Auslander, N.; Dang, C.V.; et al. The microbiome-derived metabolite TMAO drives immune activation and boosts responses to immune checkpoint blockade in pancreatic cancer. Sci. Immunol. 2022, 7, e7. [Google Scholar] [CrossRef]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; Lucas, A.S.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Routy, B.; le Chatelier, E.; DeRosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Peng, Z.; Cheng, S.; Kou, Y.; Wang, Z.; Jin, R.; Hu, H.; Zhang, X.; Gong, J.; Li, J.; Lu, M.; et al. The Gut Microbiome Is Associated with Clinical Response to Anti-PD-1/PD-L1 Immunotherapy in Gastrointestinal Cancer. Cancer Immunol. Res. 2020, 8, 1251–1261. [Google Scholar] [CrossRef]
- June, C.; O’Connor, R.; Kawalekar, O.; Ghassemi, S.; Milone, M. CAR T cell immunotherapy for human cancer. Science 2018, 359, 1361–1365. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Martinez, A.B.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef]
- Catanzaro, E.; Feron, O.; Skirtach, A.G.; Krysko, D.V. Immunogenic Cell Death and Role of Nanomaterials Serving as Therapeutic Vaccine for Personalized Cancer Immunotherapy. Front. Immunol. 2022, 13, 925290. [Google Scholar] [CrossRef]



| ASCP (n = 29) | PDAC (n = 54) | Total (n = 83) | p-Value | |
|---|---|---|---|---|
| Age (years) | 60.90 ± 9.43 | 60.26 ± 9.35 | 60.48 ± 9.38 | 0.77 |
| Sex (N, %) | 0.76 | |||
| Male | 20 (69.87) | 34 (62.96) | 54 (65.06) | |
| Female | 9 (31.03) | 20 (37.04) | 29 (34.94) | |
| T Stage (N, %) | 0.04 | |||
| I | 0 (0.00) | 1 (1.85) | 1 (1.20) | |
| II | 4 (13.79) | 2 (3.70) | 6 (7.23) | |
| III | 21 (72.41) | 50 (92.59) | 71 (85.54) | |
| IV | 4 (13.79) | 1 (1.85) | 5 (6.02) | |
| N Stage (N, %) | 0.54 | |||
| 0 | 23 (79.31) | 47 (87.04) | 70 (84.34) | |
| 1 | 6 (20.69) | 7 (12.96) | 13 (15.66) | |
| M Stage (N, %) | 0.91 | |||
| 0 | 27 (93.10) | 52 (96.30) | 79 (95.18) | |
| 1 | 2 (6.90) | 2 (3.70) | 4 (4.82) | |
| TNM Stage (N, %) | 0.13 | |||
| I | 4 (13.79) | 3 (5.56) | 7 (8.43) | |
| II | 20 (68.97) | 48 (88.89) | 68 (81.93) | |
| III | 3 (10.34) | 1 (1.85) | 4 (4.82) | |
| IV | 2 (6.90) | 2 (3.70) | 4 (4.82) | |
| Histologic Grading (N, %) | 0.14 | |||
| Well differentiated | 1 (3.45) | 1 (1.85) | 2 (2.41) | |
| Moderately differentiated | 19 (65.52) | 24 (44.44) | 43 (51.81) | |
| Poorly differentiated | 9 (31.03) | 29 (53.70) | 38 (45.78) | |
| Perineural Invasion (N, %) | 0.35 | |||
| Yes | 15 (51.72) | 35 (64.81) | 50 (60.24) | |
| No | 14 (48.28) | 19 (35.19) | 33 (39.76) | |
| Hypertension (N, %) | 1.00 | |||
| Yes | 5 (17.24) | 10 (18.52) | 15 (18.07) | |
| No | 24 (82.76) | 44 (81.48) | 68 (81.93) | |
| Diabetes (N, %) | 0.64 | |||
| Yes | 8 (27.59) | 11 (20.37) | 19 (22.89) | |
| No | 21 (72.41) | 43 (79.63) | 64 (77.11) | |
| Drinking (N, %) | 0.03 | |||
| Yes | 16 (55.17) | 15 (27.78) | 31 (37.35) | |
| No | 13 (44.83) | 39 (72.22) | 52 (62.65) | |
| Smoking (N, %) | 0.64 | |||
| Yes | 9 (31.03) | 21 (38.89) | 30 (36.14) | |
| No | 20 (68.97) | 33 (61.11) | 53 (63.86) | |
| Recurrent Status (N, %) | 0.84 | |||
| Yes | 10 (34.48) | 16 (29.63) | 26 (31.33) | |
| No | 19 (65.52) | 38 (70.37) | 57 (68.67) |
| OS of PDAC (n = 54) | OS of ASCP (n = 29) | |||
|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Age (years) (≥65 vs. <65) | 1.20 (0.62–2.10) | 0.650 | 1.00 (0.40–2.70) | 0.960 |
| Sex (female vs. male) | 1.30 (0.73–2.50) | 0.350 | 0.78 (0.34–1.80) | 0.550 |
| T Stage (T4+T3 vs. T2+T1) | 2.50 (0.59–11.00) | 0.210 | 0.26 (0.08–0.83) | 0.023 |
| N Stage (N1 vs. N0) | 1.00 (0.43–2.50) | 0.950 | 0.79 (0.31–2.00) | 0.620 |
| M Stage (M1 vs. M0) | 16.00 (2.90–86.00) | 0.001 | 1.70 (0.39–7.80) | 0.470 |
| TNM Stage (III+IV vs. I+II) | 1.30 (0.39–4.30) | 0.680 | 2.00 (0.70–5.60) | 0.200 |
| Histologic grading (poorly differentiated vs. well- and moderately differentiated) | 2.40 (1.30–4.50) | 0.006 | 2.00 (0.70–5.60) | 0.200 |
| Perineural invasion (yes vs. no) | 1.20 (0.67–2.30) | 0.490 | 0.61 (0.27–1.4) | 0.230 |
| Hypertension (yes vs. no) | 1.70 (0.83–3.50) | 0.150 | 0.81 (0.27–2.4) | 0.700 |
| Diabetes (yes vs. no) | 1.70 (0.83–3.50) | 0.150 | 0.52 (0.20–1.30) | 0.180 |
| Drinking (yes vs. no) | 1.80 (0.93–3.30) | 0.083 | 0.98 (0.44–2.20) | 0.960 |
| Smoking (yes vs. no) | 1.40 (0.81–2.60) | 0.220 | 0.51 (0.18–1.40) | 0.190 |
| Recurrent status (no vs. yes) | 0.80 (0.43–1.50) | 0.490 | 0.55 (0.24–1.30) | 0.160 |
| CD3 (high expression vs. low expression) | 0.39 (0.20–0.76) | 0.006 | 0.37 (0.15–0.92) | 0.033 |
| CD4 (high expression vs. low expression) | 0.62 (0.34–1.10) | 0.120 | 0.66 (0.28–1.60) | 0.350 |
| CD8 (high expression vs. low expression) | 0.48 (0.25–0.93) | 0.030 | 0.47 (0.19–1.20) | 0.099 |
| FoxP3 (high expression vs. low expression) | 1.10 (0.58–2.00) | 0.840 | 1.30 (0.54–3.00) | 0.590 |
| PD-L1 (high expression vs. low expression) | 1.80 (1.06–4.15) | 0.047 | 4.20 (1.70–10.00) | 0.002 |
| OS of PDAC (n = 54) | OS of ASCP (n = 29) | |||
|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Age (years) (≥65 vs. <65) | 1.50 (0.65–3.30) | 0.370 | 1 (0.21–5) | 0.970 |
| Sex (female vs. male) | 1.10 (0.46–2.60) | 0.830 | 0.34 (0.063–1.8) | 0.210 |
| T Stage (T4+T3 vs. T2+T1) | 2.20 (0.28–16.00) | 0.460 | 0.47 (0.084–2.7) | 0.400 |
| N Stage (N1 vs. N0) | 3.70 (1.00–13.00) | 0.049 | 0.087 (0.011–0.7) | 0.022 |
| TNM Stage (III+IV vs. I+II) | 1.50 (0.38–6.20) | 0.550 | 0.93 (0.22–3.8) | 0.920 |
| Histologic grading (poorly differentiated vs. well- and moderately differentiated) | 2.30 (0.95–5.60) | 0.064 | 3.21(0.25–10.89) | 0.370 |
| Perineural invasion (yes vs. no) | 1.40 (0.59–3.50) | 0.430 | 0.35 (0.09–1.3) | 0.120 |
| CD3 (high expression vs. low expression) | 0.70 (0.25–2.00) | 0.500 | 0.64 (0.098–4.2) | 0.640 |
| CD4 (high expression vs. low expression) | 0.48 (0.21–1.10) | 0.084 | 1.3 (0.29–6) | 0.720 |
| CD8 (high expression vs. low expression) | 0.43 (0.20–0.89) | 0.023 | 0.15 (0.037–0.63) | 0.010 |
| FoxP3 (high expression vs. low expression) | 2.10 (0.92–4.80) | 0.077 | 1.5 (0.46–4.7) | 0.520 |
| PD-L1 (high expression vs. low expression) | 2.70 (1.10–7.00) | 0.036 | 19 (3.5–110) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Xiong, Q.; Xu, Y.; Cai, X.; Zhang, L.; Zhu, Q. The PD-L1 Expression and Tumor-Infiltrating Immune Cells Predict an Unfavorable Prognosis in Pancreatic Ductal Adenocarcinoma and Adenosquamous Carcinoma. J. Clin. Med. 2023, 12, 1398. https://doi.org/10.3390/jcm12041398
Zhang Z, Xiong Q, Xu Y, Cai X, Zhang L, Zhu Q. The PD-L1 Expression and Tumor-Infiltrating Immune Cells Predict an Unfavorable Prognosis in Pancreatic Ductal Adenocarcinoma and Adenosquamous Carcinoma. Journal of Clinical Medicine. 2023; 12(4):1398. https://doi.org/10.3390/jcm12041398
Chicago/Turabian StyleZhang, Zhiwei, Qunli Xiong, Yongfeng Xu, Xuebin Cai, Lisha Zhang, and Qing Zhu. 2023. "The PD-L1 Expression and Tumor-Infiltrating Immune Cells Predict an Unfavorable Prognosis in Pancreatic Ductal Adenocarcinoma and Adenosquamous Carcinoma" Journal of Clinical Medicine 12, no. 4: 1398. https://doi.org/10.3390/jcm12041398
APA StyleZhang, Z., Xiong, Q., Xu, Y., Cai, X., Zhang, L., & Zhu, Q. (2023). The PD-L1 Expression and Tumor-Infiltrating Immune Cells Predict an Unfavorable Prognosis in Pancreatic Ductal Adenocarcinoma and Adenosquamous Carcinoma. Journal of Clinical Medicine, 12(4), 1398. https://doi.org/10.3390/jcm12041398






