Endostatin 33 Peptide Is a Deintegrin α6β1 Agent That Exerts Antitumor Activity by Inhibiting the PI3K-Akt Signaling Pathway in Prostate Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.3. Functional Enrichment Analysis
2.4. Protein–Protein Interaction (PPI) Network
2.5. Materials and Reagents
2.6. Cell Culture
2.7. MTT Cell Viability Assay
2.8. Cell Adhesion Assay
2.9. Transwell Assay
2.10. Cell Apoptosis Assay
2.11. Wound Healing Assay
2.12. SiRNA Transfections
2.13. Western Immunoblotting
2.14. Xenograft Tumor Model
2.15. Statistical Analysis
3. Results
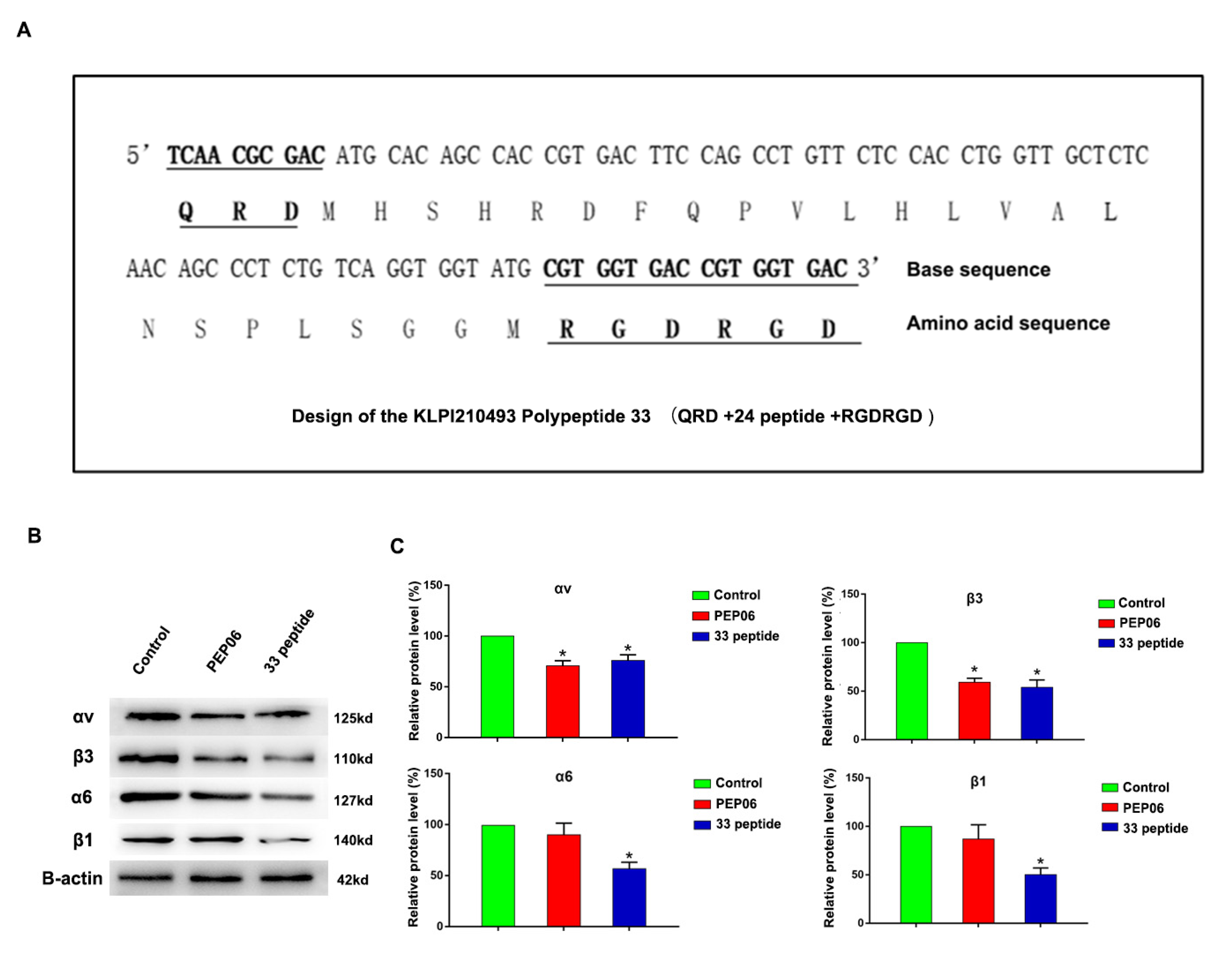
3.1. Synthesis of Endostatin 33 Peptide and Its Targeted Inhibitory Effect on Integrin α6β1 in Prostate Cancer
3.2. The 33 Polypeptide Can Inhibit the Proliferation and Promote Apoptosis of PCa
3.3. The 33 Polypeptide Can Inhibit the Invasion and Metastasis of PCa
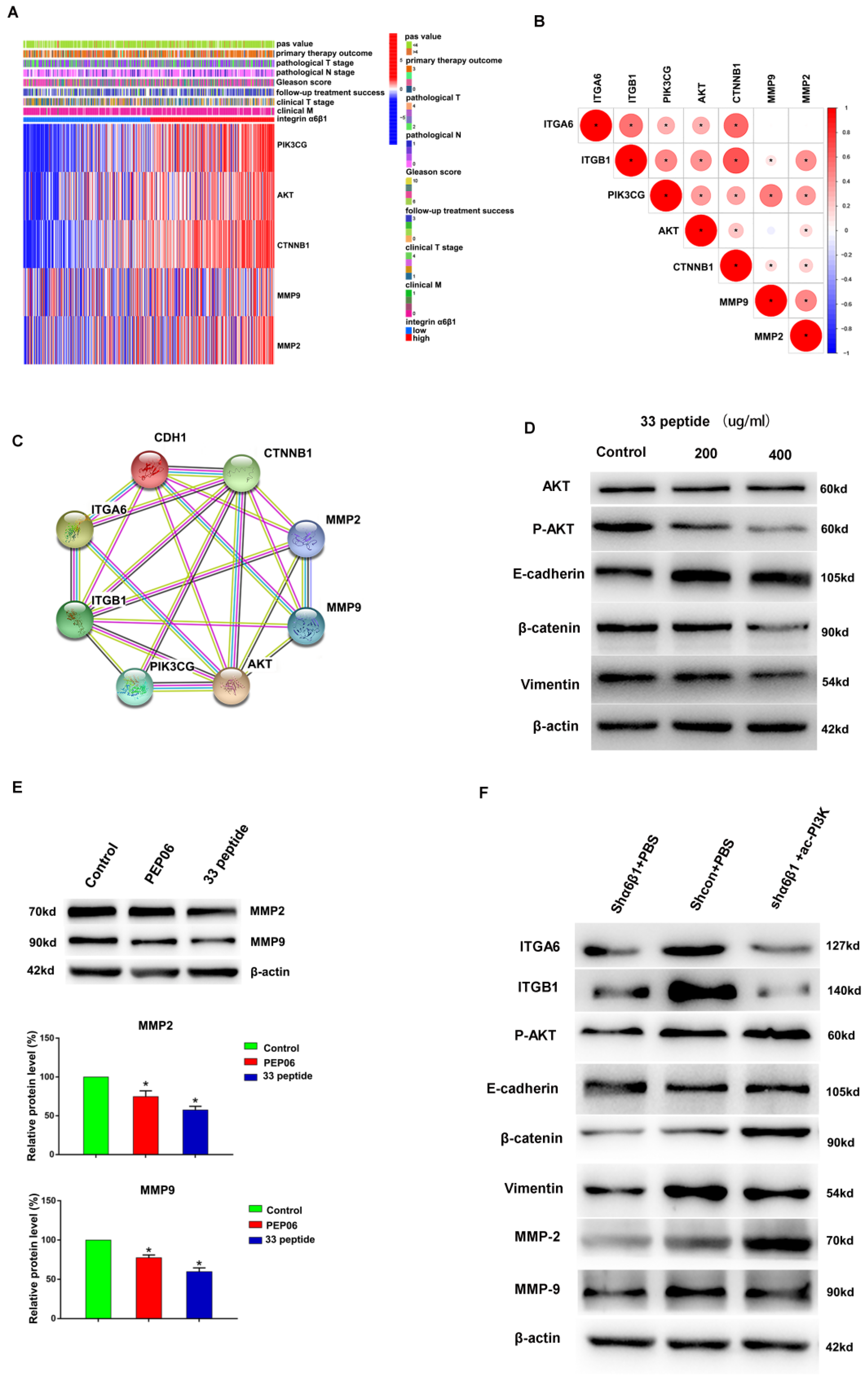
3.4. Integrin α6β1 Is Associated with Poor Prognosis and PI3K/Akt Pathway of PCa
3.5. The 33 Polypeptide Inhibits EMT and MMPs in PCa by Blocking the PI3K/AKT Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Psutka, S.P.; Frank, I.; Karnes, R.J. Risk Stratification in Hormone-sensitive Metastatic Prostate Cancer: More Questions than Answers. Eur. Urol. 2015, 68, 205–206. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, M.S.; Boehm, T.; Shing, Y.; Fukai, N.; Vasios, G.; Lane, W.S.; Flynn, E.; Birkhead, J.R.; Olsen, B.R.; Folkman, J. Endostatin: An Endogenous Inhibitor of Angiogenesis and Tumor Growth. Cell 1997, 88, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Huang, J.; Vue, M.; Alavian, M.R.; Goel, H.L.; Altieri, D.C.; Languino, L.R.; FitzGerald, T.J. αvβ3 Integrin Mediates Radioresistance of Prostate Cancer Cells through Regulation of Survivin. Mol. Cancer Res. 2019, 17, 398–408. [Google Scholar] [CrossRef]
- Nieberler, M.; Reuning, U.; Reichart, F.; Notni, J.; Wester, H.-J.; Schwaiger, M.; Weinmüller, M.; Räder, A.; Steiger, K.; Kessler, H. Exploring the Role of RGD-Recognizing Integrins in Cancer. Cancers 2017, 9, 116. [Google Scholar] [CrossRef]
- Li, K.; Shi, M.; Qin, S. Current Status and Study Progress of Recombinant Human Endostatin in Cancer Treatment. Oncol. Ther. 2018, 6, 21–43. [Google Scholar] [CrossRef]
- Yu, S.; Li, L.; Tian, W.; Nie, D.; Mu, W.; Qiu, F.; Liu, Y.; Liu, X.; Wang, X.; Du, Z.; et al. PEP06 polypeptide 30 exerts antitumour effect in colorectal carcinoma via inhibiting epithelial-mesenchymal transition. Br. J. Pharmacol. 2018, 175, 3111–3130. [Google Scholar] [CrossRef]
- Mohammadi, R.; Shokri, B.; Shamshirian, D.; Zarghi, A.; Shahhosseini, S. Synthesis and biological evaluation of RGD conjugated with Ketoprofen/Naproxen and radiolabeled with [99mTc] via N4(GGAG) for αVβ3 integrin-targeted drug delivery. DARU 2020, 28, 87–96. [Google Scholar] [CrossRef]
- Krishn, S.R.; Singh, A.; Bowler, N.; Duffy, A.N.; Friedman, A.; Fedele, C.; Kurtoglu, S.; Tripathi, S.K.; Wang, K.; Hawkins, A.; et al. Prostate cancer sheds the αvβ3 integrin in vivo through exosomes. Matrix Biol. 2019, 77, 41–57. [Google Scholar] [CrossRef]
- Tang, L.; Xu, M.; Zhang, L.; Qu, L.; Liu, X. Role of αVβ3 in Prostate Cancer: Metastasis Initiator and Important Therapeutic Target. OncoTargets Ther. 2020, 13, 7411–7422. [Google Scholar] [CrossRef]
- Gajbhiye, K.R.; Gajbhiye, V.; Siddiqui, I.A.; Gajbhiye, J.M. cRGD functionalised nanocarriers for targeted delivery of bioactives. J. Drug Target. 2019, 27, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.-H.; Yu, J.-S.; Song, E.-L.; Zhang, C.; Ma, L.; Jiao, Z.-X.; Zhao, W.-M.; Shan, Y.-J.; Ni, S.-B. Antitumor effects of mutant endostatin are enhanced by Bcl-2 antisense oligonucleotides in UM-UC-3 bladder cancer cell line. Chin. Med. J. 2013, 126, 2834–2839. [Google Scholar] [PubMed]
- Tuguzbaeva, G.; Yue, E.; Chen, X.; He, L.; Li, X.; Ju, J.; Qin, Y.; Pavlov, V.; Lu, Y.; Jia, W.; et al. PEP06 polypeptide 30 is a novel cluster-dissociating agent inhibiting v integrin/FAK/Src signaling in oral squamous cell carcinoma cells. Acta Pharm. Sin. B 2019, 9, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wei, J.; Yuan, L.; Sun, H.; Liu, Y.; Zhang, Y.; Li, J.; Liu, X. RGD-Modified Endostatin Peptide 30 Derived from Endostatin Suppresses Invasion and Migration of HepG2 Cells Through the αvβ3 Pathway. Cancer Biother. Radiopharm. 2011, 26, 529–538. [Google Scholar] [CrossRef]
- Mammadova-Bach, E.; Zigrino, P.; Brucker, C.; Bourdon, C.; Freund, M.; De Arcangelis, A.; Abrams, S.I.; Orend, G.; Gachet, C.; Mangin, P.H. Platelet integrin α6β1 controls lung metastasis through direct binding to cancer cell–derived ADAM9. JCI Insight 2016, 1, e88245. [Google Scholar] [CrossRef]
- Nollet, E.A.; Cardo-Vila, M.; Ganguly, S.S.; Tran, J.D.; Schulz, V.V.; Cress, A.; Corey, E.; Miranti, C.K. Androgen receptor-induced integrin α6β1 and Bnip3 promote survival and resistance to PI3K inhibitors in castration-resistant prostate cancer. Oncogene 2020, 39, 5390–5404. [Google Scholar] [CrossRef]
- Sroka, I.C.; Anderson, T.A.; McDaniel, K.M.; Nagle, R.B.; Gretzer, M.B.; Cress, A.E. The laminin binding integrin α6β1 in prostate cancer perineural invasion. J. Cell Physiol. 2010, 224, 283–288. [Google Scholar] [CrossRef]
- Demetriou, M.C.; Pennington, M.; Nagle, R.B.; Cress, A. Extracellular alpha 6 integrin cleavage by urokinase-type plasminogen activator in human prostate cancer. Exp. Cell Res. 2004, 294, 550–558. [Google Scholar] [CrossRef]
- Lamb, L.E.; Zarif, J.C.; Miranti, C.K. The Androgen Receptor Induces Integrin α6β1 to Promote Prostate Tumor Cell Survival via NF-κB and Bcl-xL Independently of PI3K Signaling. Cancer Res. 2011, 71, 2739–2749. [Google Scholar] [CrossRef]
- Serru, V.; Le Naour, F.; Billard, M.; Azorsa, D.; Lanza, F.; Boucheix, C.; Rubinstein, E. Selective tetraspan-integrin complexes (CD81/alpha4beta1, CD151/alpha3beta1, CD151/alpha6beta1) under conditions disrupting tetraspan interactions. Biochem. J. 1999, 340 Pt 1, 103–111. [Google Scholar] [CrossRef]
- Hwang, S.; Takimoto, T.; Hemler, M.E. Integrin-independent support of cancer drug resistance by tetraspanin CD151. Cell. Mol. Life Sci. 2019, 76, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E.; Pierschbacher, M.D. New Perspectives in Cell Adhesion: RGD and Integrins. Science 1987, 238, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Toth, R.K.; Tran, J.D.; Muldong, M.T.; Nollet, E.; Schulz, V.V.; Jensen, C.C.; Hazlehurst, L.; Corey, E.; Durden, D.; Jamieson, C.; et al. Hypoxia-induced PIM kinase and laminin-activated integrin α6 mediate resistance to PI3K inhibitors in bone-metastatic CRPC. Am. J. Clin. Exp. Urol. 2019, 7, 297–312. [Google Scholar] [PubMed]
- Ranjan, A.; Bane, S.M.; Kalraiya, R.D. Glycosylation of the laminin receptor (α3β1) regulates its association with tetraspanin CD151: Impact on cell spreading, motility, degradation and invasion of basement membrane by tumor cells. Exp. Cell Res. 2014, 322, 249–264. [Google Scholar] [CrossRef]
- Lochter, A.; Bissell, M.J. An odyssey from breast to bone: Multi-step control of mammary metastases and osteolysis by matrix metalloproteinases. Apmis 1999, 107, 128–136. [Google Scholar] [CrossRef]
- Wang, K.; Ruan, H.; Xu, T.; Liu, L.; Liu, D.; Yang, H.; Zhang, X.; Chen, K. Recent advances on the progressive mechanism and therapy in castration-resistant prostate cancer. OncoTargets Ther. 2018, 11, 3167–3178. [Google Scholar] [CrossRef]
- Howe, A.; Aplin, A.; Alahari, S.K.; Juliano, R. Integrin signaling and cell growth control. Curr. Opin. Cell Biol. 1998, 10, 220–231. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, Y.; Yang, G.; Lan, Y.; Han, J.; Wang, J.; Yin, D.; Song, R.; Zheng, T.; Zhang, S.; et al. Tetraspanin 1 promotes epithelial-to-mesenchymal transition and metastasis of cholangiocarcinoma via PI3K/AKT signaling. J. Exp. Clin. Cancer Res. 2018, 37, 300. [Google Scholar] [CrossRef]
- Chetty, C.; Lakka, S.S.; Bhoopathi, P.; Rao, J.S. MMP-2 alters VEGF expression via αVβ3 integrin-mediated PI3K/AKT signaling in A549 lung cancer cells. Int. J. Cancer 2010, 127, 1081–1095. [Google Scholar] [CrossRef]
- Liu, J.; Chen, P.; Chang, T.; Hou, C. Thrombospondin-2 stimulates MMP-9 production and promotes osteosarcoma metastasis via the PLC, PKC, c-Src and NF-κB activation. J. Cell. Mol. Med. 2020, 24, 12826–12839. [Google Scholar] [CrossRef]
- Jiang, Q.; Pan, Y.; Cheng, Y.; Li, H.; Liu, D.; Li, H. Lunasin suppresses the migration and invasion of breast cancer cells by inhibiting matrix metalloproteinase-2/-9 via the FAK/Akt/ERK and NF-κB signaling pathways. Oncol. Rep. 2016, 36, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Li, J.; Wang, Z.; Zhang, T.; Han, Y.; Liu, Y.; Chu, W.; Liu, Y.; Yang, B. HYD-PEP06 suppresses hepatocellular carcinoma metastasis, epithelial–mesenchymal transition and cancer stem cell-like properties by inhibiting PI3K/AKT and WNT/β-catenin signaling activation. Acta Pharm. Sin. B 2021, 11, 1592–1606. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Wang, C.-L.; Pang, Z.-Q.; Gao, K.; Shen, L.-K.; Xu, W.-H.; Ren, M.-H. Endostatin 33 Peptide Is a Deintegrin α6β1 Agent That Exerts Antitumor Activity by Inhibiting the PI3K-Akt Signaling Pathway in Prostate Cancer. J. Clin. Med. 2023, 12, 1861. https://doi.org/10.3390/jcm12051861
Liu Y, Wang C-L, Pang Z-Q, Gao K, Shen L-K, Xu W-H, Ren M-H. Endostatin 33 Peptide Is a Deintegrin α6β1 Agent That Exerts Antitumor Activity by Inhibiting the PI3K-Akt Signaling Pathway in Prostate Cancer. Journal of Clinical Medicine. 2023; 12(5):1861. https://doi.org/10.3390/jcm12051861
Chicago/Turabian StyleLiu, Yang, Chang-Lin Wang, Zhong-Qi Pang, Ke Gao, Lin-Kun Shen, Wan-Hai Xu, and Ming-Hua Ren. 2023. "Endostatin 33 Peptide Is a Deintegrin α6β1 Agent That Exerts Antitumor Activity by Inhibiting the PI3K-Akt Signaling Pathway in Prostate Cancer" Journal of Clinical Medicine 12, no. 5: 1861. https://doi.org/10.3390/jcm12051861
APA StyleLiu, Y., Wang, C.-L., Pang, Z.-Q., Gao, K., Shen, L.-K., Xu, W.-H., & Ren, M.-H. (2023). Endostatin 33 Peptide Is a Deintegrin α6β1 Agent That Exerts Antitumor Activity by Inhibiting the PI3K-Akt Signaling Pathway in Prostate Cancer. Journal of Clinical Medicine, 12(5), 1861. https://doi.org/10.3390/jcm12051861




