Identification of a Novel Cuproptosis-Related Gene Signature and Integrative Analyses in Thyroid Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Clinical Samples
2.2. Differentially Expressed Genes (DEGs) Identification
2.3. Construction of the Protein–Protein Interaction (PPI) Network
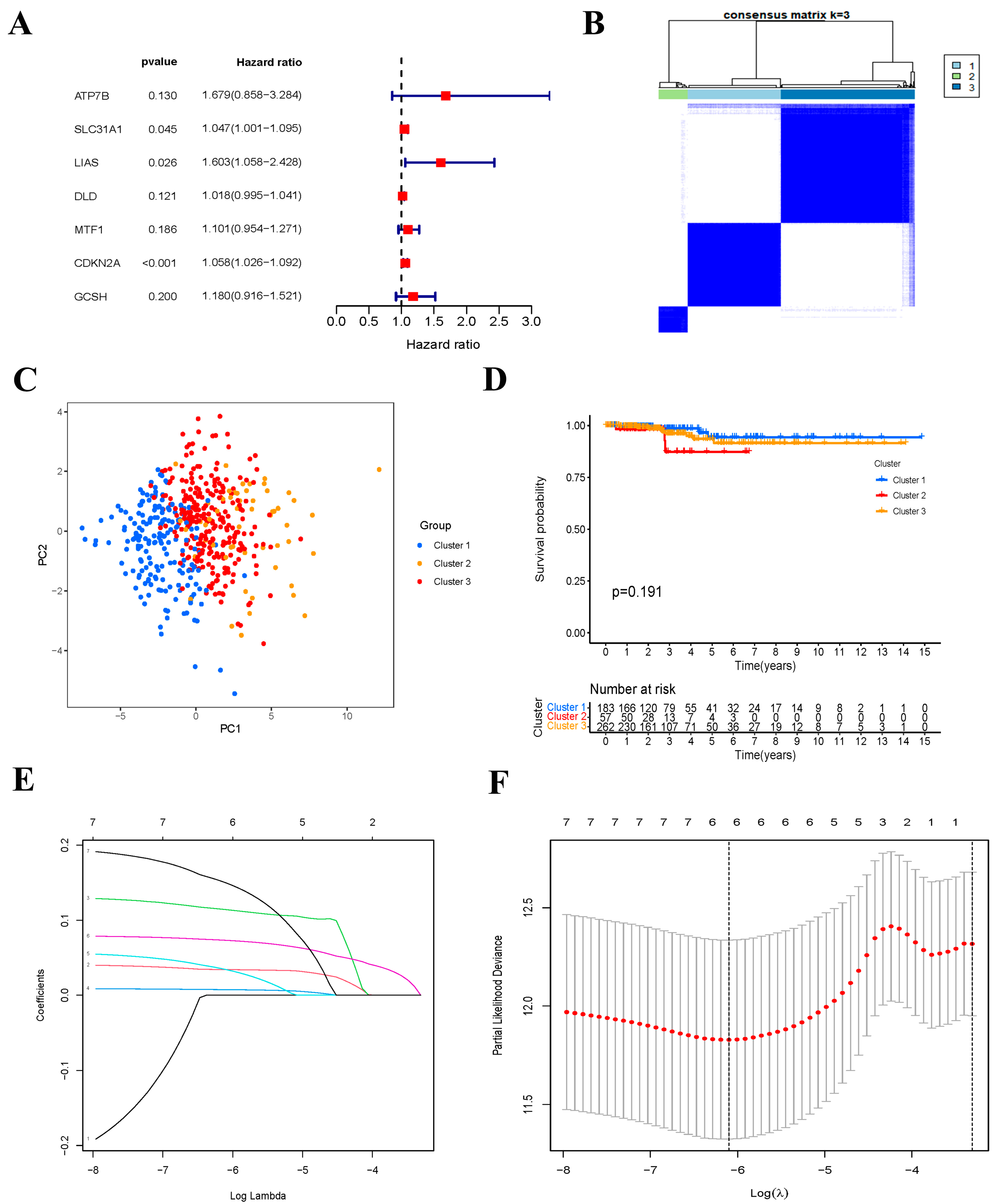
2.4. Univariate Cox Regression Analysis
2.5. Consensus Clustering of Prognostic CRGs Genes
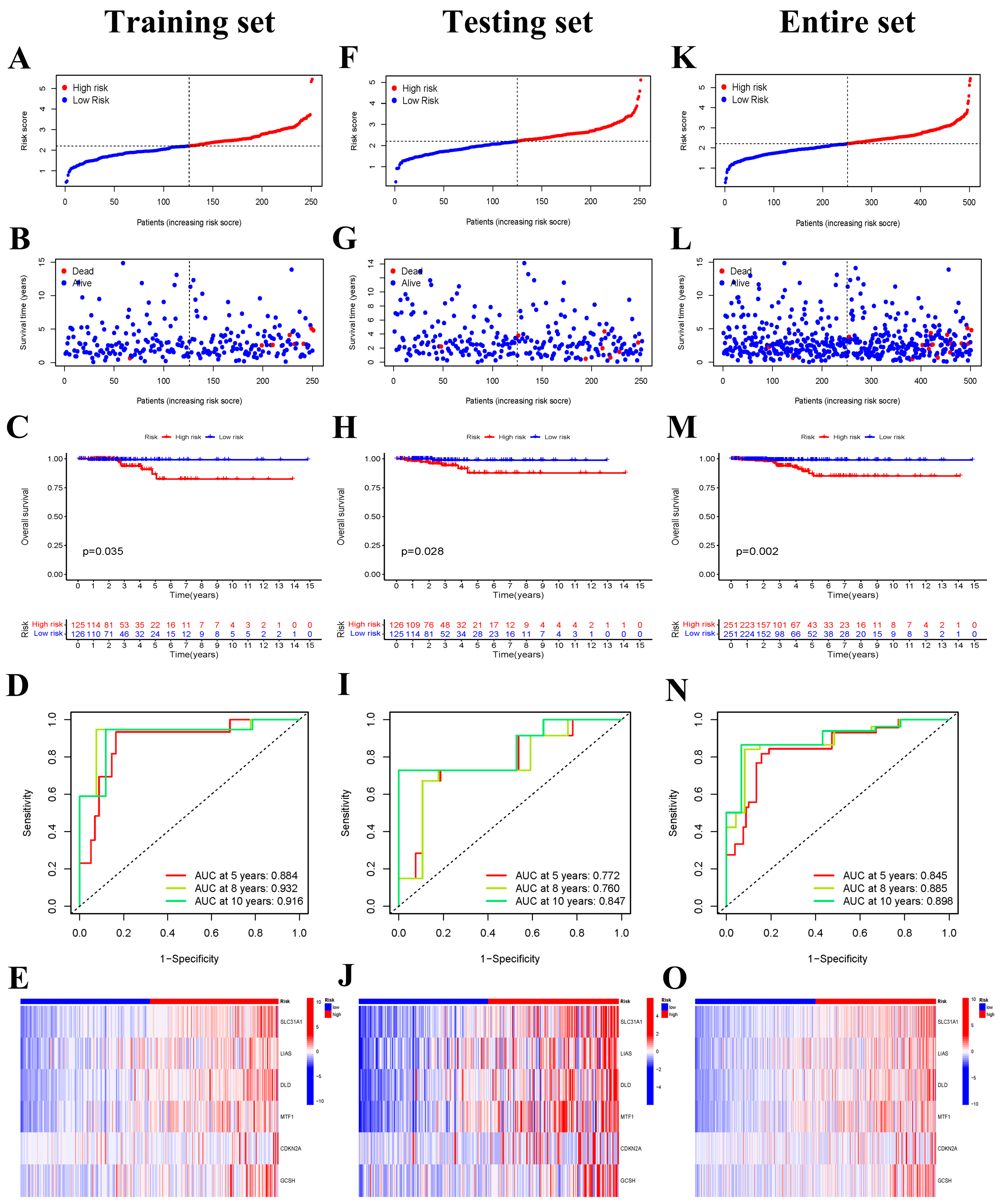
2.6. Construction of Cuproptosis-Related Prognostic Signature
2.7. The Clinicopathologic Features between the Low- and High-Risk Groups
2.8. Principal Component Analysis (PCA)
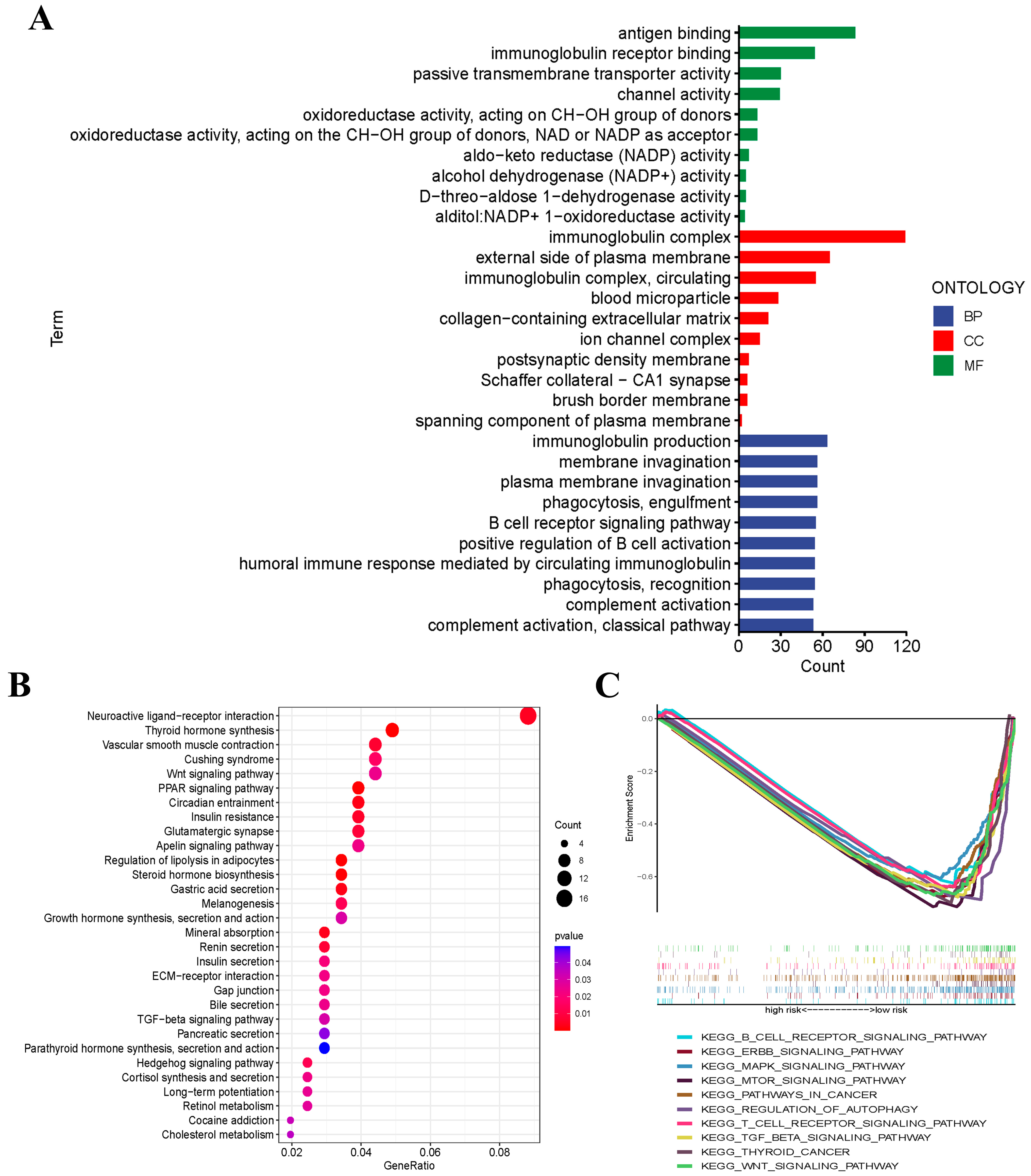
2.9. Functional Enrichment Analysis
2.10. Immune-Related Analysis
2.11. Immunophenoscore (IPS) Analysis
2.12. Potential Targeted Drug Sensitivity Prediction
2.13. Total RNA Extraction and qRT-PCR
2.14. Statistical Analysis
3. Results
3.1. Identification of DEGs in THCA from the TCGA Database
3.2. Consensus Clustering of Cuproptosis-Related Genes
3.3. Construction of Cuproptosis-Related Risk Signature
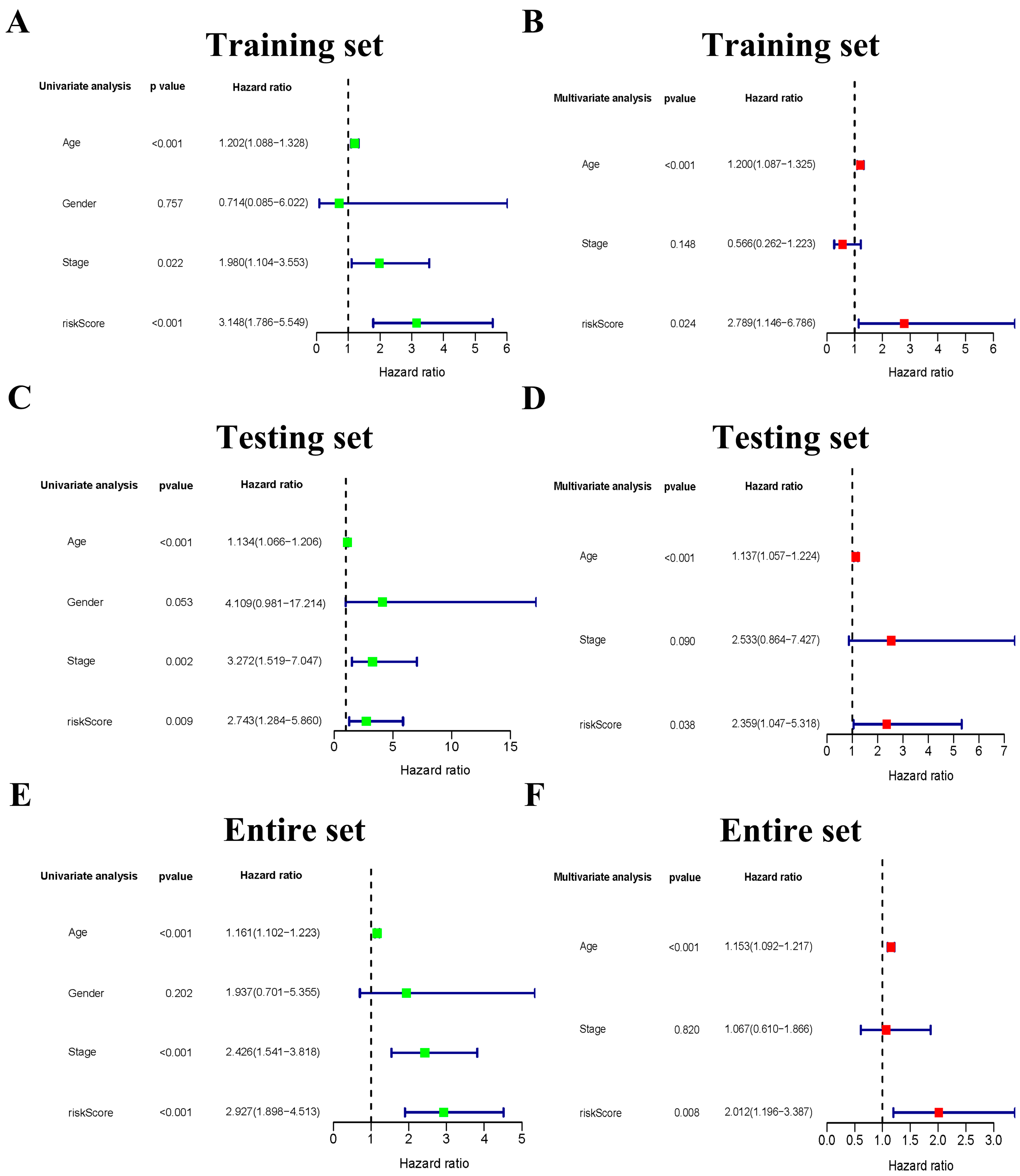
3.4. The Six-Gene Risk Signature Is an Independent Prognostic Indicator for OS
3.5. The Clinicopathological Stratified Analysis Validates the Risk Signature’s Prediction Performance
3.6. Functional Analyses Based on the Risk Signature
3.7. Gene Set Enrichment Analysis (GSEA)
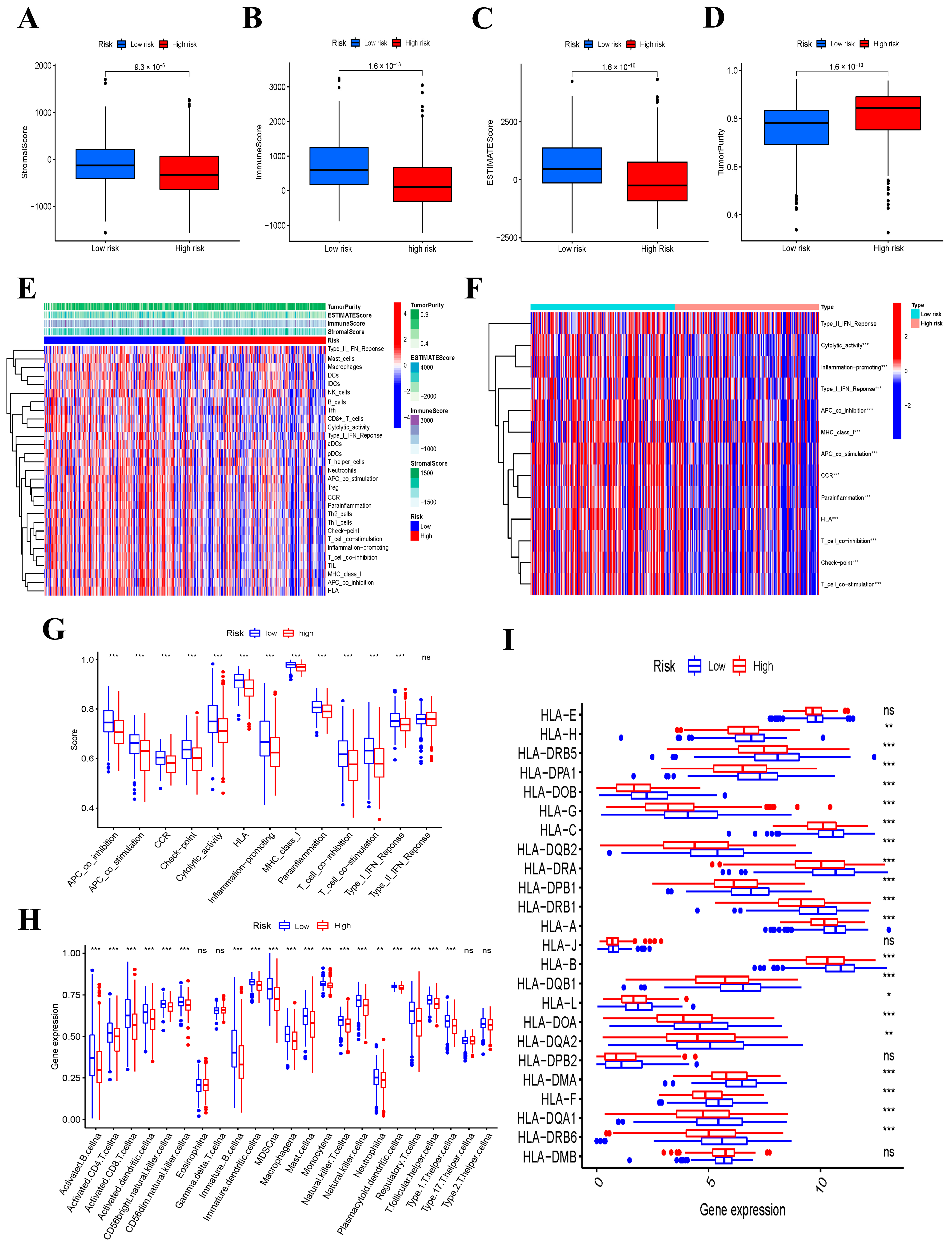
3.8. Immune Function and Immune Status Analyses between the Low- and High-Risk Groups
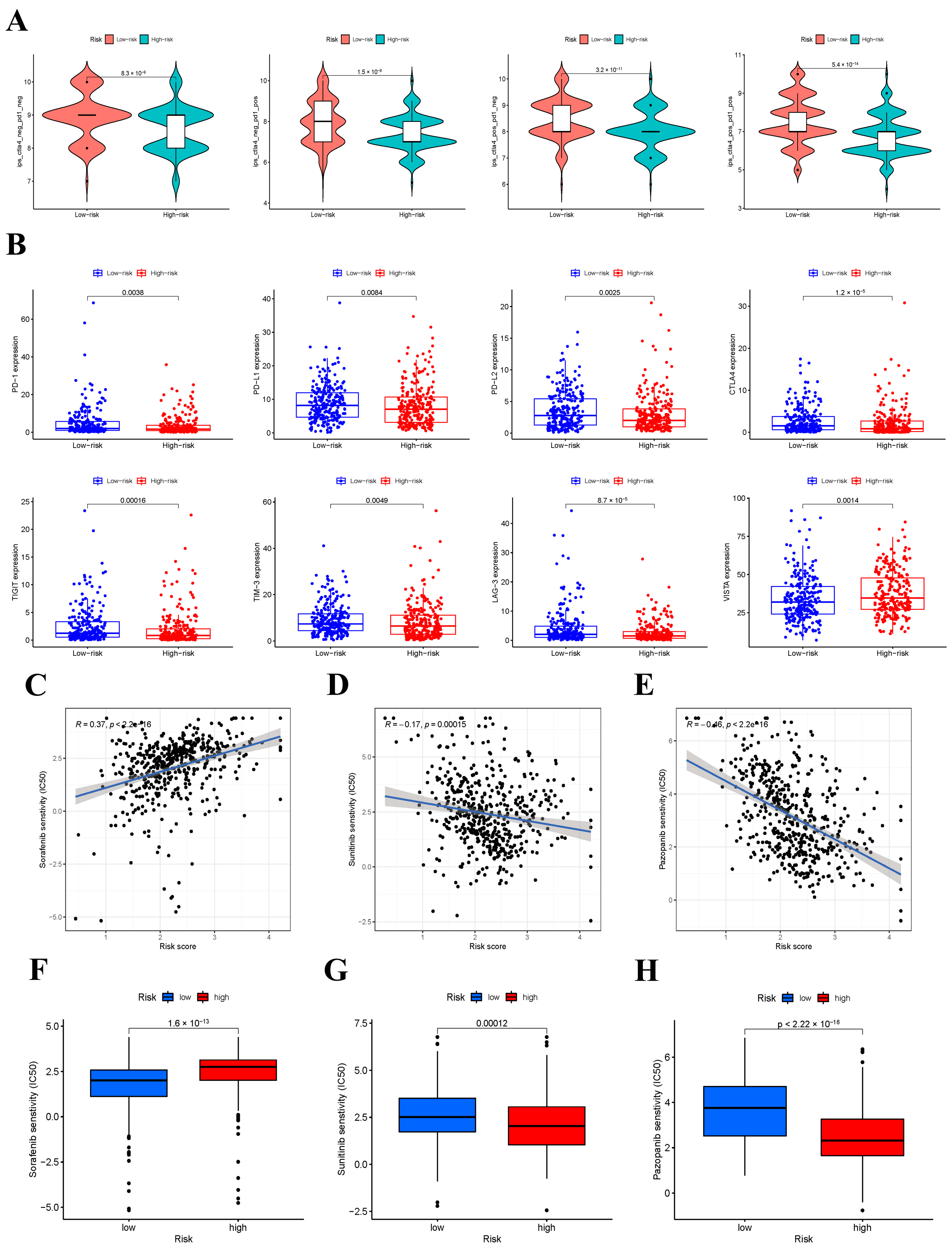
3.9. IPS Analysis and Response to ICIs
3.10. Response to Targeted Drugs
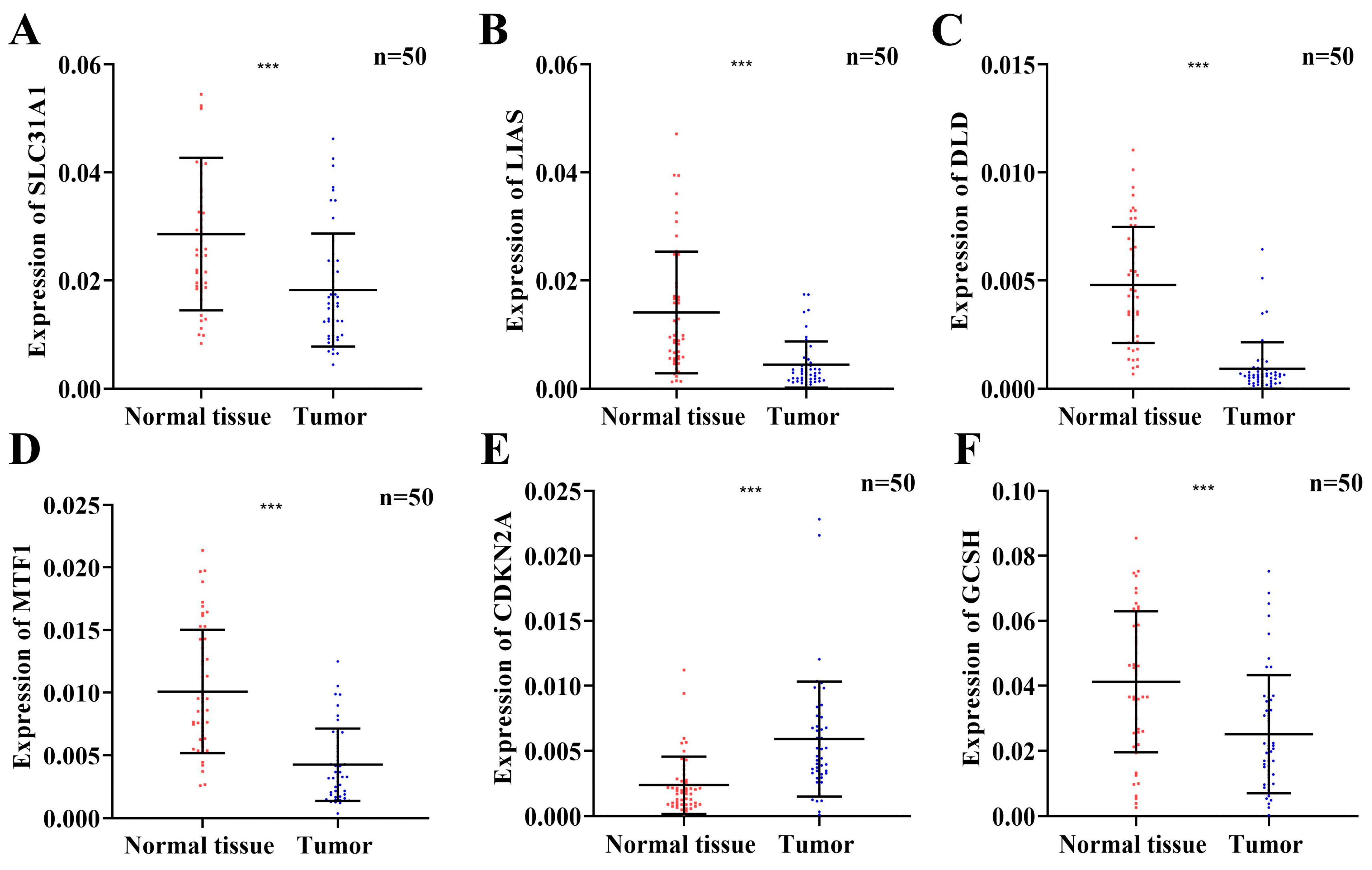
3.11. The Expression Levels of Six Prognostic Risk Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Filho, A.; Lortet-Tieulent, J.; Bray, F.; Cao, B.; Franceschi, S.; Vaccarella, S.; Dal Maso, L. Thyroid cancer incidence trends by histology in 25 countries: A population-based study. Lancet Diabetes Endocrinol. 2021, 9, 225–234. [Google Scholar] [CrossRef]
- Schlumberger, M.; Leboulleux, S. Current practice in patients with differentiated thyroid cancer. Nat. Rev. Endocrinol. 2021, 17, 176–188. [Google Scholar] [CrossRef]
- Cabanillas, M.E.; McFadden, D.G.; Durante, C. Thyroid cancer. Lancet 2016, 388, 2783–2795. [Google Scholar] [CrossRef]
- Tsang, T.; Davis, C.I.; Brady, D.C. Copper biology. Curr. Biol. 2021, 31, R421–R427. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-E.; Nevitt, T.; Thiele, D.J. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 2008, 4, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Lutsenko, S. Human copper homeostasis: A network of interconnected pathways. Curr. Opin. Chem. Biol. 2010, 14, 211–217. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, Y.; Shi, H.; Peng, Y.; Fan, X.; Li, C. The molecular mechanisms of copper metabolism and its roles in human diseases. Pflugers. Arch. 2020, 472, 1415–1429. [Google Scholar] [CrossRef]
- Jiang, Y.; Huo, Z.; Qi, X.; Zuo, T.; Wu, Z. Copper-induced tumor cell death mechanisms and antitumor theragnostic applications of copper complexes. Nanomedicine 2022, 17, 303–324. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Ge, E.J.; Bush, A.I.; Casini, A.; Cobine, P.A.; Cross, J.R.; DeNicola, G.M.; Dou, Q.P.; Franz, K.J.; Gohil, V.M.; Gupta, S.; et al. Connecting copper and cancer: From transition metal signalling to metalloplasia. Nat. Rev. Cancer 2022, 22, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, V.C.; Gudekar, N.; Jasmer, K.; Papageorgiou, C.; Singh, K.; Petris, M.J. Copper metabolism as a unique vulnerability in cancer. Biochim. Biophys. Acta Mol. Cell. Res. 2021, 1868, 118893. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Zhou, C.; Lu, L.; Liu, B.; Ding, Y. Elesclomol: A copper ionophore targeting mitochondrial metabolism for cancer therapy. J. Exp. Clin. Cancer Res. 2022, 41, 271. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.-L.; Chien, P.-J.; Hsieh, H.-C.; Shen, H.-T.; Lee, H.-T.; Chen, S.-M.; Chang, W.-W. Disulfiram/Copper Suppresses Cancer Stem Cell Activity in Differentiated Thyroid Cancer Cells by Inhibiting BMI1 Expression. Int. J. Mol. Sci. 2022, 23, 13276. [Google Scholar] [CrossRef] [PubMed]
- He, J.-L.; Wu, H.-B.; Hu, W.-L.; Liu, J.-J.; Zhang, Q.; Xiao, W.; Hu, M.-J.; Wu, M.; Huang, F. Exposure to multiple trace elements and thyroid cancer risk in Chinese adults: A case-control study. Int. J. Hyg. Environ Health 2022, 246, 114049. [Google Scholar] [CrossRef]
- Wang, H.; Lengerich, B.J.; Aragam, B.; Xing, E.P. Precision Lasso: Accounting for correlations and linear dependencies in high-dimensional genomic data. Bioinformatics 2019, 35, 1181–1187. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Wu, P.; Sun, W.; Zhang, H. An immune-related prognostic signature for thyroid carcinoma to predict survival and response to immune checkpoint inhibitors. Cancer Immunol. Immunother. 2022, 71, 747–759. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell. Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Zhang, Y.; Xing, Z.; Liu, T.; Tang, M.; Mi, L.; Zhu, J.; Wu, W.; Wei, T. Targeted therapy and drug resistance in thyroid cancer. Eur. J. Med. Chem. 2022, 238, 114500. [Google Scholar] [CrossRef] [PubMed]
- Wiltshire, J.J.; Drake, T.M.; Uttley, L.; Balasubramanian, S.P. Systematic Review of Trends in the Incidence Rates of Thyroid Cancer. Thyroid 2016, 26, 1541–1552. [Google Scholar] [CrossRef] [PubMed]
- Nixon, I.J.; Simo, R.; Newbold, K.; Rinaldo, A.; Suarez, C.; Kowalski, L.P.; Silver, C.; Shah, J.P.; Ferlito, A. Management of Invasive Differentiated Thyroid Cancer. Thyroid 2016, 26, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Póvoa, A.A.; Teixeira, E.; Bella-Cueto, M.R.; Melo, M.; Oliveira, M.J.; Sobrinho-Simões, M.; Maciel, J.; Soares, P. Clinicopathological Features as Prognostic Predictors of Poor Outcome in Papillary Thyroid Carcinoma. Cancers 2020, 12, 3186. [Google Scholar] [CrossRef]
- Tapiero, H.; Townsend, D.M.; Tew, K.D. Trace elements in human physiology and pathology. Copper. Biomed. Pharmacother. 2003, 57, 386–398. [Google Scholar] [CrossRef]
- Gaetke, L.M.; Chow-Johnson, H.S.; Chow, C.K. Copper: Toxicological relevance and mechanisms. Arch. Toxicol. 2014, 88, 1929–1938. [Google Scholar] [CrossRef]
- Aubert, L.; Nandagopal, N.; Steinhart, Z.; Lavoie, G.; Nourreddine, S.; Berman, J.; Saba-El-Leil, M.K.; Papadopoli, D.; Lin, S.; Hart, T.; et al. Copper bioavailability is a KRAS-specific vulnerability in colorectal cancer. Nat. Commun. 2020, 11, 3701. [Google Scholar] [CrossRef]
- Ren, X.; Li, Y.; Zhou, Y.; Hu, W.; Yang, C.; Jing, Q.; Zhou, C.; Wang, X.; Hu, J.; Wang, L.; et al. Overcoming the compensatory elevation of NRF2 renders hepatocellular carcinoma cells more vulnerable to disulfiram/copper-induced ferroptosis. Redox Biol. 2021, 46, 102122. [Google Scholar] [CrossRef]
- Cui, L.; Gouw, A.M.; LaGory, E.L.; Guo, S.; Attarwala, N.; Tang, Y.; Qi, J.; Chen, Y.-S.; Gao, Z.; Casey, K.M.; et al. Mitochondrial copper depletion suppresses triple-negative breast cancer in mice. Nat. Biotechnol. 2021, 39, 357–367. [Google Scholar] [CrossRef]
- Li, Y. Copper homeostasis: Emerging target for cancer treatment. IUBMB Life 2020, 72, 1900–1908. [Google Scholar] [CrossRef]
- Babak, M.V.; Ahn, D. Modulation of Intracellular Copper Levels as the Mechanism of Action of Anticancer Copper Complexes: Clinical Relevance. Biomedicines 2021, 9, 852. [Google Scholar] [CrossRef] [PubMed]
- Voli, F.; Valli, E.; Lerra, L.; Kimpton, K.; Saletta, F.; Giorgi, F.M.; Mercatelli, D.; Rouaen, J.R.C.; Shen, S.; Murray, J.E.; et al. Intratumoral Copper Modulates PD-L1 Expression and Influences Tumor Immune Evasion. Cancer Res. 2020, 80, 4129–4144. [Google Scholar] [CrossRef] [PubMed]
- Brady, D.C.; Crowe, M.S.; Turski, M.L.; Hobbs, G.A.; Yao, X.; Chaikuad, A.; Knapp, S.; Xiao, K.; Campbell, S.L.; Thiele, D.J.; et al. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature 2014, 509, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Zimnicka, A.M.; Ivy, K.; Kaplan, J.H. Acquisition of dietary copper: A role for anion transporters in intestinal apical copper uptake. Am. J. Physiol. Cell. Physiol. 2011, 300, C588–C599. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Pang, X.; Guo, Z.; Guo, R.; Liu, L.; Chen, X. Dual Action of Acidic Microenvironment on the Enrichment of the Active Metabolite of Disulfiram in Tumor Tissues. Drug Metab. Dispos. 2021, 49, 434–441. [Google Scholar] [CrossRef]
- Li, X.; Ma, Z.; Mei, L. Cuproptosis-related gene SLC31A1 is a potential predictor for diagnosis, prognosis and therapeutic response of breast cancer. Am. J. Cancer Res. 2022, 12, 3561–3580. [Google Scholar] [PubMed]
- Wu, G.; Peng, H.; Tang, M.; Yang, M.; Wang, J.; Hu, Y.; Li, Z.; Li, J.; Li, Z.; Song, L. ZNF711 down-regulation promotes CISPLATIN resistance in epithelial ovarian cancer via interacting with JHDM2A and suppressing SLC31A1 expression. EBioMedicine 2021, 71, 103558. [Google Scholar] [CrossRef]
- Cheng, C.; Ding, Q.; Zhang, Z.; Wang, S.; Zhong, B.; Huang, X.; Shao, Z. PTBP1 modulates osteosarcoma chemoresistance to cisplatin by regulating the expression of the copper transporter SLC31A1. J. Cell. Mol. Med. 2020, 24, 5274–5289. [Google Scholar] [CrossRef]
- Yi, X.; Kim, K.; Yuan, W.; Xu, L.; Kim, H.-S.; Homeister, J.W.; Key, N.S.; Maeda, N. Mice with heterozygous deficiency of lipoic acid synthase have an increased sensitivity to lipopolysaccharide-induced tissue injury. J. Leukoc. Biol. 2009, 85, 146–153. [Google Scholar] [CrossRef]
- Cai, Y.; He, Q.; Liu, W.; Liang, Q.; Peng, B.; Li, J.; Zhang, W.; Kang, F.; Hong, Q.; Yan, Y.; et al. Comprehensive analysis of the potential cuproptosis-related biomarker LIAS that regulates prognosis and immunotherapy of pan-cancers. Front. Oncol. 2022, 12, 952129. [Google Scholar] [CrossRef]
- Wang, S.; Xing, N.; Meng, X.; Xiang, L.; Zhang, Y. Comprehensive bioinformatics analysis to identify a novel cuproptosis-related prognostic signature and its ceRNA regulatory axis and candidate traditional Chinese medicine active ingredients in lung adenocarcinoma. Front. Pharmacol. 2022, 13, 971867. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Lee, J.; You, J.H.; Kim, D.; Roh, J.-L. Dihydrolipoamide dehydrogenase regulates cystine deprivation-induced ferroptosis in head and neck cancer. Redox Biol. 2020, 30, 101418. [Google Scholar] [CrossRef] [PubMed]
- Tavera-Montañez, C.; Hainer, S.J.; Cangussu, D.; Gordon, S.J.V.; Xiao, Y.; Reyes-Gutierrez, P.; Imbalzano, A.N.; Navea, J.G.; Fazzio, T.G.; Padilla-Benavides, T. The classic metal-sensing transcription factor MTF1 promotes myogenesis in response to copper. FASEB J. 2019, 33, 14556–14574. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin, W.; Yang, Y.; Shao, J.; Zhao, H.; Wang, G.; Shen, A. Role of cuproptosis-related gene in lung adenocarcinoma. Front. Oncol. 2022, 12, 1080985. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Wu, Y.; Liu, Z.; Yu, Y.; Sun, S.; Guo, M.; Sun, B.; Huang, C. CDKN2A-mediated molecular subtypes characterize the hallmarks of tumor microenvironment and guide precision medicine in triple-negative breast cancer. Front. Immunol. 2022, 13, 970950. [Google Scholar] [CrossRef]
- Luo, J.-P.; Wang, J.; Huang, J.-H. CDKN2A is a prognostic biomarker and correlated with immune infiltrates in hepatocellular carcinoma. Biosci. Rep. 2021, 41, BSR20211103. [Google Scholar] [CrossRef]
- Adamus, A.; Müller, P.; Nissen, B.; Kasten, A.; Timm, S.; Bauwe, H.; Seitz, G.; Engel, N. GCSH antisense regulation determines breast cancer cells’ viability. Sci. Rep. 2018, 8, 15399. [Google Scholar] [CrossRef]
- Gan, X.; Guo, M.; Chen, Z.; Li, Y.; Shen, F.; Feng, J.; Cai, W.; Xu, B. Development and validation of a three-immune-related gene signature prognostic risk model in papillary thyroid carcinoma. J. Endocrinol. Invest. 2021, 44, 2153–2163. [Google Scholar] [CrossRef]
- Jarzab, B.; Wiench, M.; Fujarewicz, K.; Simek, K.; Jarzab, M.; Oczko-Wojciechowska, M.; Wloch, J.; Czarniecka, A.; Chmielik, E.; Lange, D.; et al. Gene expression profile of papillary thyroid cancer: Sources of variability and diagnostic implications. Cancer Res. 2005, 65, 1587–1597. [Google Scholar] [CrossRef]

| Variables | Type | The Entire Set (n = 502) | The Training Set (n = 251) | The Testing Set (n = 251) | p Value |
|---|---|---|---|---|---|
| Age | <55 | 336 (66.93%) | 170 (67.73%) | 166 (66.14%) | 0.7043 |
| ≥55 | 166 (33.07%) | 81 (32.27%) | 85 (33.86%) | ||
| Gender | Female | 367 (73.11%) | 181 (72.11%) | 186 (74.10%) | 0.6148 |
| Male | 135 (26.89%) | 70 (27.89%) | 65 (25.90%) | ||
| Stage | Stage I-II | 335 (66.73%) | 171 (68.13%) | 164 (65.34%) | 0.5073 |
| Stage III–IV | 167 (33.27%) | 80 (31.87%) | 87 (34.66%) | ||
| T | T1-2 | 307 (61.15%) | 157 (62.55%) | 150 (59.76%) | 0.5202 |
| T3-4 | 193 (38.45%) | 93 (37.05%) | 100 (39.84%) | ||
| Unknown | 2 (0.40%) | 1 (0.40%) | 1 (0.40%) | ||
| M | M0 | 281 (55.98%) | 137 (54.58%) | 144 (57.37%) | 0.6879 |
| M1 | 9 (1.79%) | 5 (1.99%) | 4 (1.59%) | ||
| Unknown | 212 (42.23%) | 109 (43.43%) | 103 (41.04%) | ||
| N | N0 | 228 (45.42%) | 114 (45.42%) | 114 (45.42%) | 0.6028 |
| N1 | 225 (44.82%) | 107 (42.63%) | 118 (47.01%) | ||
| Unknown | 49 (9.76%) | 30 (11.95%) | 19 (7.57%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Shi, J.; Wu, P.; Sun, W.; Zhang, D.; Wang, Z.; Ji, X.; Lv, C.; Zhang, T.; Zhang, P.; et al. Identification of a Novel Cuproptosis-Related Gene Signature and Integrative Analyses in Thyroid Cancer. J. Clin. Med. 2023, 12, 2014. https://doi.org/10.3390/jcm12052014
Huang J, Shi J, Wu P, Sun W, Zhang D, Wang Z, Ji X, Lv C, Zhang T, Zhang P, et al. Identification of a Novel Cuproptosis-Related Gene Signature and Integrative Analyses in Thyroid Cancer. Journal of Clinical Medicine. 2023; 12(5):2014. https://doi.org/10.3390/jcm12052014
Chicago/Turabian StyleHuang, Jiapeng, Jinyuan Shi, Pu Wu, Wei Sun, Dalin Zhang, Zhihong Wang, Xiaoyu Ji, Chengzhou Lv, Ting Zhang, Ping Zhang, and et al. 2023. "Identification of a Novel Cuproptosis-Related Gene Signature and Integrative Analyses in Thyroid Cancer" Journal of Clinical Medicine 12, no. 5: 2014. https://doi.org/10.3390/jcm12052014
APA StyleHuang, J., Shi, J., Wu, P., Sun, W., Zhang, D., Wang, Z., Ji, X., Lv, C., Zhang, T., Zhang, P., & Zhang, H. (2023). Identification of a Novel Cuproptosis-Related Gene Signature and Integrative Analyses in Thyroid Cancer. Journal of Clinical Medicine, 12(5), 2014. https://doi.org/10.3390/jcm12052014








