Alteration of m6A-Tagged RNA Profiles in Bone Originated from Periprosthetic Joint Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection and Sample Collection
2.2. Hematoxylin–Eosin (HE) Staining
2.3. Quatification of the Overall Level of m6A Methylation Modification
2.4. Quantification of m6A-Related Gene Expression by qPCR
2.5. Western Blot
2.6. m6A-Tagged RNA Immunoprecipitation, Microarray Hybridization and Data Analysis
2.7. Statistical Analysis
3. Results
3.1. PJI Patients
3.2. The Overall m6A Level of PJI Group Was Significantly Higher Than That of AF Group
3.3. The Expression of RNA Methylase METTL3 Was Significantly Upregulated in PJI Group
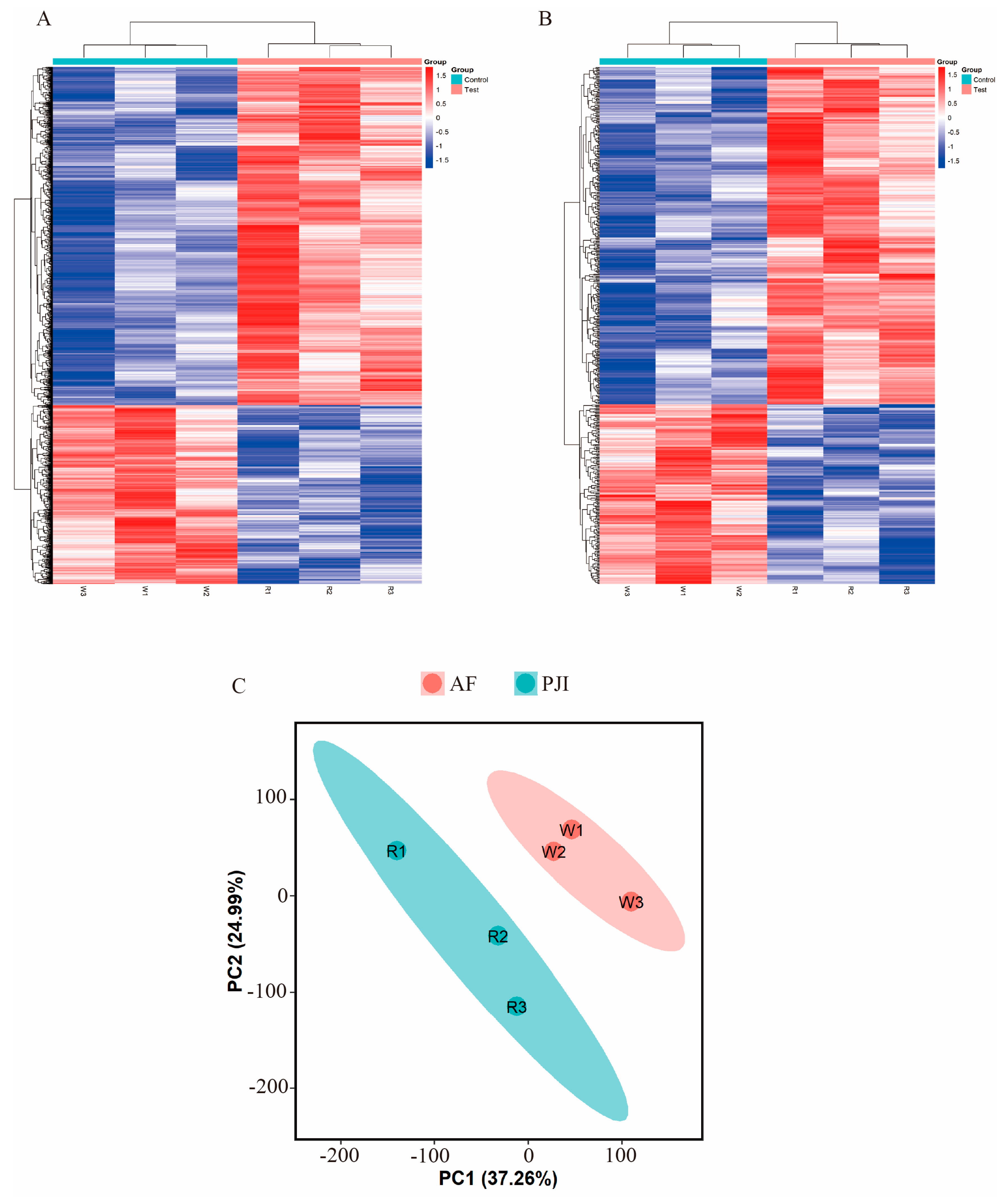
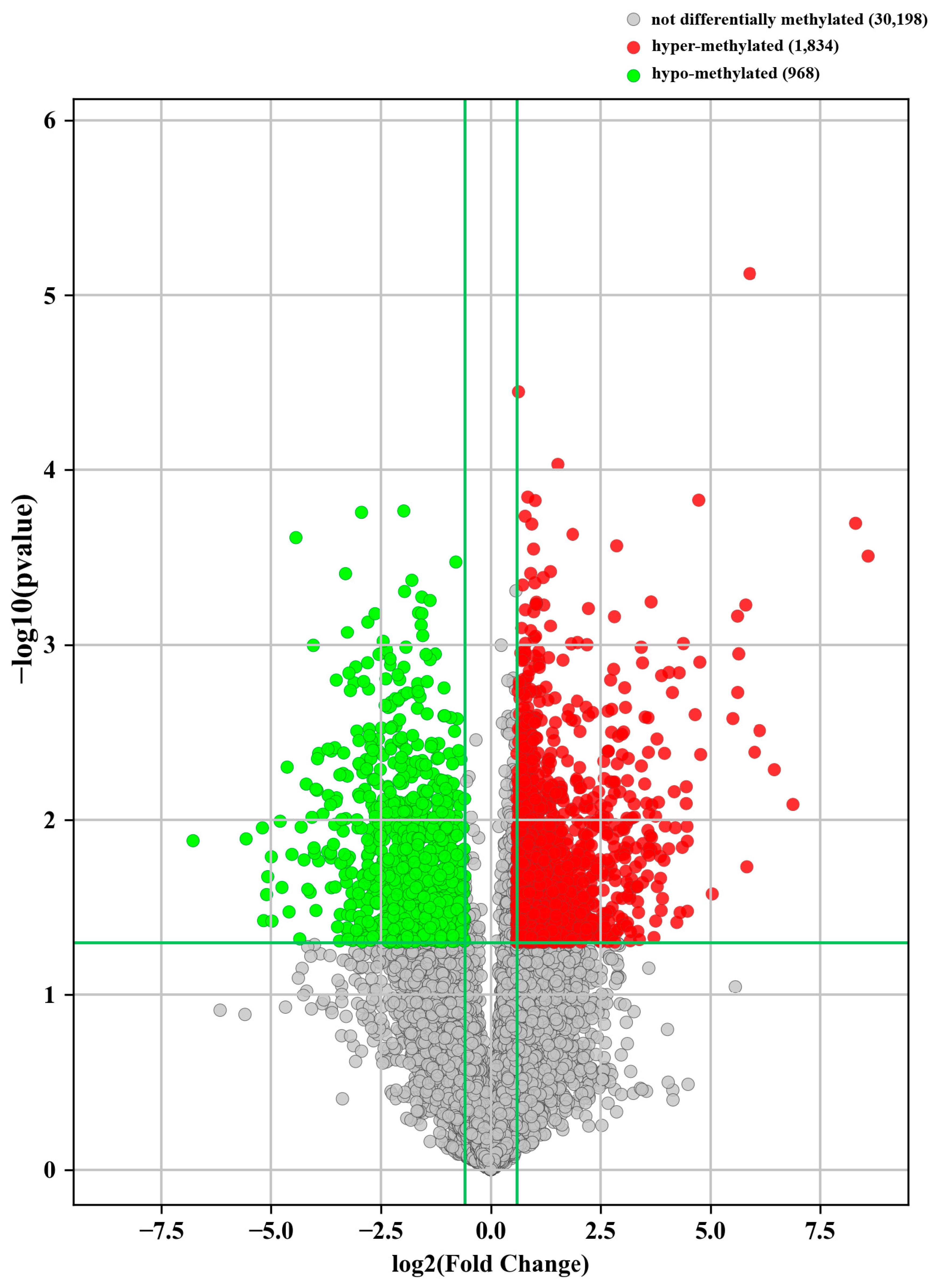
3.4. Alternations of m6A-Modified RNA Profiles in Bone Samples from PJI
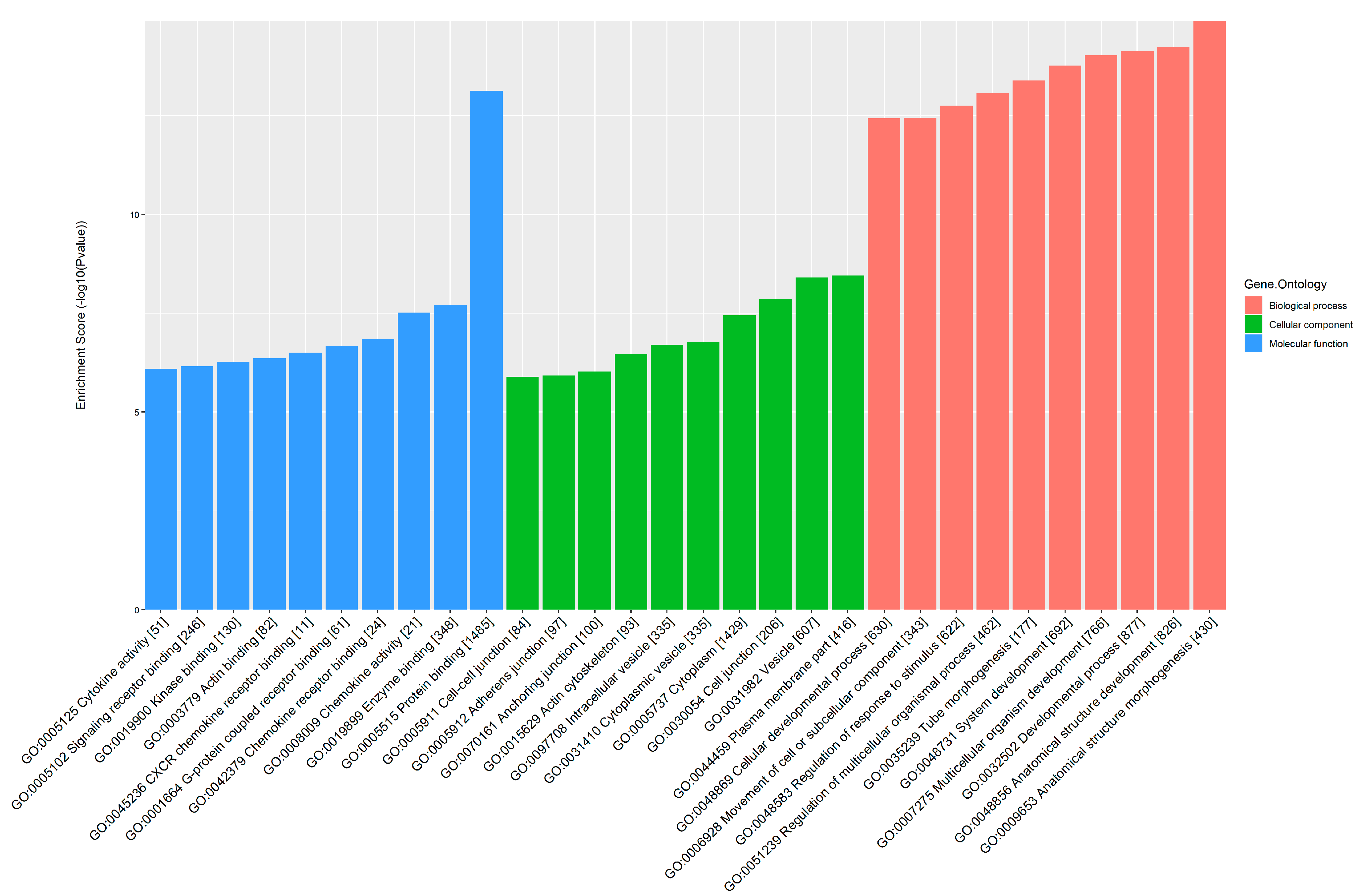
3.5. GO and KEGG Pathway Analysis of Differential m6A-Tagged mRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haddad, F.S.; Ngu, A.; Negus, J.J. Prosthetic Joint Infections and Cost Analysis? Adv. Exp. Med. Biol. 2017, 971, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Goswami, K.; Li, W.T.; Tan, T.L.; Yayac, M.; Wang, S.-H.; Parvizi, J. Is Treatment of Periprosthetic Joint Infection Improving Over Time? J. Arthroplast. 2020, 35, 1696–1702. [Google Scholar] [CrossRef] [PubMed]
- Petis, S.M.; Perry, K.I.; Mabry, T.M.; Hanssen, A.D.; Berry, D.J.; Abdel, M.P. Two-Stage Exchange Protocol for Periprosthetic Joint Infection Following Total Knee Arthroplasty in 245 Knees without Prior Treatment for Infection. J. Bone Jt. Surg. Am. 2019, 101, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Mussa, M.; Manciulli, T.; Corbella, M.; Mariani, B.; Cambieri, P.; Gipsz, N.; Scudeller, L.; Abbott, D.M.; Brunetti, E.; Mosconi, M.; et al. Epidemiology and Microbiology of Prosthetic Joint Infections: A Nine-Year, Single-Center Experience in Pavia, Northern Italy. Musculoskelet. Surg. 2020, 105, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Loss, G.; Simões, P.M.; Valour, F.; Cortês, M.F.; Gonzaga, L.; Bergot, M.; Trouillet-Assant, S.; Josse, J.; Diot, A.; Ricci, E.; et al. Staphylococcus Aureus Small Colony Variants (SCVs): News From a Chronic Prosthetic Joint Infection. Front. Cell. Infect. Microbiol. 2019, 9, 363. [Google Scholar] [CrossRef]
- Ricciardi, B.F.; Muthukrishnan, G.; Masters, E.; Ninomiya, M.; Lee, C.C.; Schwarz, E.M. Staphylococcus Aureus Evasion of Host Immunity in the Setting of Prosthetic Joint Infection: Biofilm and Beyond. Curr. Rev. Musculoskelet. Med. 2018, 11, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Tang, H.-F.; Kai, Y. N6-Methyladenine RNA Modification (M6A): An Emerging Regulator of Metabolic Diseases. Curr. Drug Targets 2020, 21, 1056–1067. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Fu, J.; Zhou, Y. A Review in Research Progress Concerning M6A Methylation and Immunoregulation. Front. Immunol. 2019, 10, 922. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic Transcriptomic M6A Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking M6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef]
- He, Y.; Xing, J.; Wang, S.; Xin, S.; Han, Y.; Zhang, J. Increased M6A Methylation Level Is Associated with the Progression of Human Abdominal Aortic Aneurysm. Ann. Transl. Med. 2019, 7, 797. [Google Scholar] [CrossRef]
- Ma, S.; Chen, C.; Ji, X.; Liu, J.; Zhou, Q.; Wang, G.; Yuan, W.; Kan, Q.; Sun, Z. The Interplay between M6A RNA Methylation and Noncoding RNA in Cancer. J. Hematol. Oncol. 2019, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Liu, Z.; Xu, Y.; Liu, X.; Wang, D.; Li, F.; Wang, Y.; Bi, J. Abnormality of M6A MRNA Methylation Is Involved in Alzheimer’s Disease. Front. Neurosci. 2020, 14, 98. [Google Scholar] [CrossRef]
- Krüger, N.; Biwer, L.A.; Good, M.E.; Ruddiman, C.A.; Wolpe, A.G.; DeLalio, L.J.; Murphy, S.; Macal, E.H.; Ragolia, L.; Serbulea, V.; et al. Loss of Endothelial FTO Antagonizes Obesity-Induced Metabolic and Vascular Dysfunction. Circ. Res. 2020, 126, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the Human and Viral m(6)A RNA Methylomes during HIV-1 Infection of T Cells. Nat. Microbiol. 2016, 1, 16011. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Li, Q.; Meng, R.; Yi, B.; Xu, Q. METTL3 Regulates Alternative Splicing of MyD88 upon the Lipopolysaccharide-Induced Inflammatory Response in Human Dental Pulp Cells. J. Cell. Mol. Med. 2018, 22, 2558–2568. [Google Scholar] [CrossRef]
- Kiran, M.; Donnelly, T.D.; Armstrong, C.; Kapoor, B.; Kumar, G.; Peter, V. Diagnostic Utility of Fluorodeoxyglucose Positron Emission Tomography in Prosthetic Joint Infection Based on MSIS Criteria. Bone Jt. J. 2019, 101, 910–914. [Google Scholar] [CrossRef]
- Brocard, M.; Ruggieri, A.; Locker, N. M6A RNA Methylation, a New Hallmark in Virus-Host Interactions. J. Gen. Virol. 2017, 98, 2207–2214. [Google Scholar] [CrossRef]
- Winkler, R.; Gillis, E.; Lasman, L.; Safra, M.; Geula, S.; Soyris, C.; Nachshon, A.; Tai-Schmiedel, J.; Friedman, N.; Le-Trilling, V.T.K.; et al. M 6 A Modification Controls the Innate Immune Response to Infection by Targeting Type I Interferons. Nat. Immunol. 2019, 20, 173–182. [Google Scholar] [CrossRef]
- Alp, E.; Cevahir, F.; Ersoy, S.; Guney, A. Incidence and Economic Burden of Prosthetic Joint Infections in a University Hospital: A Report from a Middle-Income Country. J. Infect. Public Health 2016, 9, 494–498. [Google Scholar] [CrossRef]
- Sethi, S.; Chakraborty, T. Role of TLR-/NLR-Signaling and the Associated Cytokines Involved in Recruitment of Neutrophils in Murine Models of Staphylococcus Aureus Infection. Virulence 2011, 2, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-P.; Cao, A.T.; Feng, T.; Li, Q.; Zhang, W.; Yao, S.; Dann, S.M.; Elson, C.O.; Cong, Y. TGF-β Converts Th1 Cells into Th17 Cells through Stimulation of Runx1 Expression. Eur. J. Immunol. 2015, 45, 1010–1018. [Google Scholar] [CrossRef]
- Qin, X.-H.; Ma, X.; Fang, S.-F.; Zhang, Z.-Z.; Lu, J.-M. IL-17 Produced by Th17 Cells Alleviates the Severity of Fungal Keratitis by Suppressing CX43 Expression in Corneal Peripheral Vascular Endothelial Cells. Cell Cycle 2019, 18, 274–287. [Google Scholar] [CrossRef]
- Muthukrishnan, G.; Masters, E.A.; Daiss, J.L.; Schwarz, E.M. Mechanisms of Immune Evasion and Bone Tissue Colonization That Make Staphylococcus Aureus the Primary Pathogen in Osteomyelitis. Curr. Osteoporos. Rep. 2019, 17, 395–404. [Google Scholar] [CrossRef]
- Krauss, J.L.; Roper, P.M.; Ballard, A.; Shih, C.-C.; Fitzpatrick, J.A.J.; Cassat, J.E.; Ng, P.Y.; Pavlos, N.J.; Veis, D.J. Staphylococcus Aureus Infects Osteoclasts and Replicates Intracellularly. MBio 2019, 10, e02447-19. [Google Scholar] [CrossRef] [PubMed]
- Selan, L.; Papa, R.; Ermocida, A.; Cellini, A.; Ettorre, E.; Vrenna, G.; Campoccia, D.; Montanaro, L.; Arciola, C.R.; Artini, M. Serratiopeptidase Reduces the Invasion of Osteoblasts by Staphylococcus Aureus. Int. J. Immunopathol. Pharmacol. 2017, 30, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Karner, C.M.; Long, F. Wnt Signaling and Cellular Metabolism in Osteoblasts. Cell. Mol. Life Sci. 2017, 74, 1649–1657. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, J.; Li, Y.; Zhou, Y.; Cui, Y.; Yang, G.; Hong, Y. Rap1A Regulates Osteoblastic Differentiation via the ERK and P38 Mediated Signaling. PLoS ONE 2015, 10, e0143777. [Google Scholar] [CrossRef]
- Yang, W.; Han, W.; Qin, A.; Wang, Z.; Xu, J.; Qian, Y. The Emerging Role of Hippo Signaling Pathway in Regulating Osteoclast Formation. J. Cell. Physiol. 2018, 233, 4606–4617. [Google Scholar] [CrossRef]
- Josse, J.; Velard, F.; Gangloff, S.C. Staphylococcus Aureus vs. Osteoblast: Relationship and Consequences in Osteomyelitis. Front. Cell. Infect. Microbiol. 2015, 5, 85. [Google Scholar] [CrossRef]
- Bergsbaken, T.; Fink, S.L.; Cookson, B.T. Pyroptosis: Host Cell Death and Inflammation. Nat. Rev. Microbiol. 2009, 7, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Zhu, H.; Santo, A.; Jia, Z.; Li, R.Y. GPx4 in Bacterial Infection and Polymicrobial Sepsis: Involvement of Ferroptosis and Pyroptosis. React. Oxyg. Species 2019, 7, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Miao, E.A.; Leaf, I.A.; Treuting, P.M.; Mao, D.P.; Dors, M.; Sarkar, A.; Warren, S.E.; Wewers, M.D.; Aderem, A. Caspase-1-Induced Pyroptosis Is an Innate Immune Effector Mechanism against Intracellular Bacteria. Nat. Immunol. 2010, 11, 1136–1142. [Google Scholar] [CrossRef]
- Matsushita, M.; Freigang, S.; Schneider, C.; Conrad, M.; Bornkamm, G.W.; Kopf, M. T Cell Lipid Peroxidation Induces Ferroptosis and Prevents Immunity to Infection. J. Exp. Med. 2015, 212, 555–568. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Y.; Chen, X.; Huang, C.; Chen, Y.; Zhang, C.; Huang, Z.; Zhang, W.; Tang, Y.; Fang, X. Alteration of m6A-Tagged RNA Profiles in Bone Originated from Periprosthetic Joint Infection. J. Clin. Med. 2023, 12, 2863. https://doi.org/10.3390/jcm12082863
Cai Y, Chen X, Huang C, Chen Y, Zhang C, Huang Z, Zhang W, Tang Y, Fang X. Alteration of m6A-Tagged RNA Profiles in Bone Originated from Periprosthetic Joint Infection. Journal of Clinical Medicine. 2023; 12(8):2863. https://doi.org/10.3390/jcm12082863
Chicago/Turabian StyleCai, Yuanqing, Xiaoqing Chen, Changyu Huang, Yang Chen, Chaofan Zhang, Zida Huang, Wenming Zhang, Yusen Tang, and Xinyu Fang. 2023. "Alteration of m6A-Tagged RNA Profiles in Bone Originated from Periprosthetic Joint Infection" Journal of Clinical Medicine 12, no. 8: 2863. https://doi.org/10.3390/jcm12082863
APA StyleCai, Y., Chen, X., Huang, C., Chen, Y., Zhang, C., Huang, Z., Zhang, W., Tang, Y., & Fang, X. (2023). Alteration of m6A-Tagged RNA Profiles in Bone Originated from Periprosthetic Joint Infection. Journal of Clinical Medicine, 12(8), 2863. https://doi.org/10.3390/jcm12082863




