The Microbiome in Inflammatory Bowel Disease
Abstract
:1. Introduction
2. Main Concepts Associated with the Human Gastrointestinal Microbiome
2.1. Microbiota and Microbiome
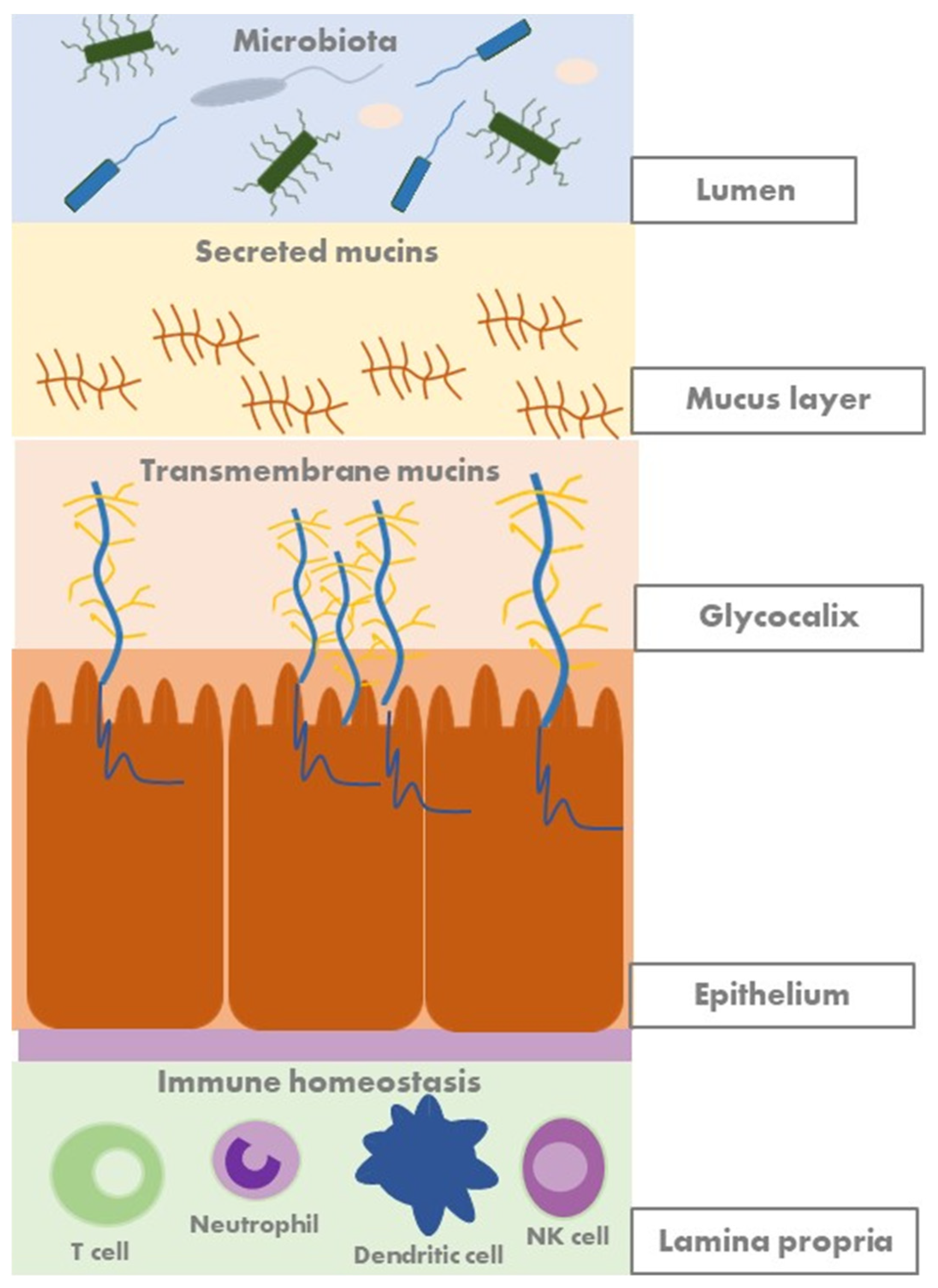
2.2. Mucosal Barrier and Mucins
2.3. Methods for the Analysis of the Microbiome: Multi-Omics
3. Changes in the Microbiome in Patients with Inflammatory Bowel Disease
3.1. Gut Dysbiosis
3.2. Microbiota as a Biomarker
4. Modulation of the Gut Microbiome in Inflammatory Bowel Disease
4.1. Early-Life Exposure
4.2. Physical Activity
4.3. Diet
4.4. Antibiotics
4.5. Probiotics
4.6. Prebiotics
4.7. Postbiotics
4.8. Faecal Microbiota Transplantation
5. Potential Microbiome-Based Therapies for Inflammatory Bowel Disease
5.1. Phagotherapie
5.2. Spore-Based Therapy
5.3. Next-Generation Probiotics and Live Biotherapeutics
5.4. Targeting Altered Microbiome Function: AhR Agonists
5.5. Bile Acids
6. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- De Souza, H.S.P.; Fiocchi, C. Immunopathogenesis of IBD: Current state of the art. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 13–27. [Google Scholar] [CrossRef]
- Guzzo, G.L.; Andrews, J.M.; Weyrich, L.S. The Neglected Gut Microbiome: Fungi, Protozoa, and Bacteriophages in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2022, 28, 1112–1122. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Ravel, J. The vocabulary of microbiome research: A proposal. Microbiome 2015, 3, 31. [Google Scholar] [CrossRef] [PubMed]
- Dave, M.; Higgins, P.D.; Middha, S.; Rioux, K.P. The human gut microbiome: Current knowledge, challenges, and future directions. Transl. Res. 2012, 160, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Liguori, G.; Lamas, B.; Richard, M.L.; Brandi, G.; da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J. Crohns Colitis 2016, 10, 296–305. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef]
- Manrique, P.; Dills, M.; Young, M.J. The human gut phage community and its implications for health and disease. Viruses 2017, 9, 141. [Google Scholar] [CrossRef]
- Breugelmans, T.; Van Spaendonk, H.; De Man, J.G.; De Schepper, H.; Jauregui-Amezaga, A.; Macken, E.; Lindén, S.; Pintelon, I.; Timmermans, J.P.; De Winter, B.Y.; et al. In-depth study of transmembrane mucins in association with intestinal barrier dysfunction during the course of t cell transfer and dss-induced colitis. J. Crohns Colitis 2020, 14, 974–994. [Google Scholar] [CrossRef] [PubMed]
- Breugelmans, T.; Oosterlinck, B.; Arras, W.; Ceuleers, H.; De Man, J.; Hold, G.L.; De Winter, B.Y.; Smet, A. The role of mucins in gastrointestinal barrier function during health and disease. Lancet Gastroenterol. Hepatol. 2022, 7, 455–471. [Google Scholar] [CrossRef] [PubMed]
- Breugelmans, T.; Arras, W.; Boen, L.E.; Borms, E.; Kamperdijk, L.; De Man, J.; Van de Vijver, E.; Van Gils, A.; De Winter, B.Y.; Moes, N.; et al. Aberrant Mucin Expression Profiles Associate with Pediatric Inflammatory Bowel Disease Presentation and Activity. Inflamm. Bowel Dis. 2023, 29, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, V.; Cassard, A.; Mas, E.; Barreau, F. Multi-omics analysis of gut microbiota in inflammatory bowel diseases: What benefits for diagnostic, prognostic and therapeutic tools? Int. J. Mol. Sci. 2021, 22, 11255. [Google Scholar] [CrossRef] [PubMed]
- Fiocchi, C.; Dragoni, G.; Iliopoulos, D.; Katsanos, K.; Hernandez Ramirez, V.; Suzuki, K. Scientific Workshop Steering Committee Results of the Seventh Scientific Workshop of ECCO: Precision Medicine in IBD-What, Why and How. J. Crohns Colitis 2021, 15, 1410–1430. [Google Scholar] [CrossRef] [PubMed]
- Aldars-García, L.; Chaparro, M.; Gisbert, J.P. Systematic review: The gut microbiome and its potential clinical application in inflammatory bowel disease. Microorganisms 2021, 9, 977. [Google Scholar] [CrossRef] [PubMed]
- Wensel, C.R.; Pluznick, J.L.; Salzberg, S.L.; Sears, C.L. Next-generation sequencing: Insights to advance clinical investigations of the microbiome. J. Clin. Investig. 2022, 132, e154944. [Google Scholar] [CrossRef] [PubMed]
- Boix-Amorós, A.; Monaco, H.; Sambataro, E.; Clemente, J.C. Novel technologies to characterize and engineer the microbiome in inflammatory bowel disease. Gut Microbes 2022, 14, 2107866. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.T. Pathophysiology of Inflammatory Bowel Diseases. N. Engl. J. Med. 2020, 383, 2652–2664. [Google Scholar] [CrossRef]
- Strachan, D.P. Hay fever, hygiene, and household size. BMJ 1989, 299, 1259–1260. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Pittayanon, R.; Lau, J.T.; Leontiadis, G.I.; Tse, F.; Yuan, Y.; Surette, M.; Moayyedi, P. Differences in Gut Microbiota in Patients with vs without Inflammatory Bowel Diseases: A Systematic Review. Gastroenterology 2020, 158, 930–946.e1. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed]
- Gophna, U.; Sommerfeld, K.; Gophna, S.; Doolittle, W.F.; Veldhuyzen Van Zanten, S.J.O. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J. Clin. Microbiol. 2006, 44, 4136–4141. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhu, X.; Huang, X.; Murff, H.J.; Ness, R.M.; Seidner, D.L.; Sorgen, A.A.; Blakley, I.C.; Yu, C.; Dai, Q.; et al. On the robustness of inference of association with the gut microbiota in stool, rectal swab and mucosal tissue samples. Sci. Rep. 2021, 11, 14828. [Google Scholar] [CrossRef] [PubMed]
- Limon, J.J.; Tang, J.; Li, D.; Wolf, A.J.; Michelsen, K.S.; Funari, V.; Gargus, M.; Nguyen, C.; Sharma, P.; Maymi, V.I.; et al. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 2019, 25, 377–388.e6. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Fan, D.; Sun, Y.; Huang, Z.; Li, Y.; Su, R.; Zhang, F.; Li, Q.; Yang, H.; Zhang, F.; et al. The gut ileal mucosal virome is disturbed in patients with Crohn’s disease and exacerbates intestinal inflammation in mice. Nat. Commun. 2024, 15, 1638. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.P.; Modos, D.; Rushbrook, S.M.; Powell, N.; Korcsmaros, T. The Emerging Role of Bile Acids in the Pathogenesis of Inflammatory Bowel Disease. Front. Immunol. 2022, 13, 829525. [Google Scholar] [CrossRef]
- Wang, S.; van Schooten, F.J.; Jin, H.; Jonkers, D.; Godschalk, R. The Involvement of Intestinal Tryptophan Metabolism in Inflammatory Bowel Disease Identified by a Meta-Analysis of the Transcriptome and a Systematic Review of the Metabolome. Nutrients 2023, 15, 2886. [Google Scholar] [CrossRef]
- Lamas, B.; Natividad, J.M.; Sokol, H. Aryl hydrocarbon receptor and intestinal immunity review-article. Mucosal Immunol. 2018, 11, 1024–1038. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Clooney, A.G.; Eckenberger, J.; Laserna-Mendieta, E.; Sexton, K.A.; Bernstein, M.T.; Vagianos, K.; Sargent, M.; Ryan, F.J.; Moran, C.; Sheehan, D.; et al. Ranking microbiome variance in inflammatory bowel disease: A large longitudinal intercontinental study. Gut 2021, 70, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, A.J.; Colburn, W.A.; DeGruttola, V.G.; DeMets, D.L.; Downing, G.J.; Hoth, D.F.; Oates, J.A.; Peck, C.C.; Schooley, R.T.; Spilker, B.A.; et al. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef]
- Papa, E.; Docktor, M.; Smillie, C.; Weber, S.; Preheim, S.P.; Gevers, D.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Ingram, J.; et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS ONE 2012, 7, e39242. [Google Scholar] [CrossRef] [PubMed]
- Estevinho, M.M.; Rocha, C.; Correia, L.; Lago, P.; Ministro, P.; Portela, F.; Trindade, E.; Afonso, J.; Peyrin-Biroulet, L.; Magro, F.; et al. Features of Fecal and Colon Microbiomes Associate with Responses to Biologic Therapies for Inflammatory Bowel Diseases: A Systematic Review. Clin. Gastroenterol. Hepatol. 2020, 18, 1054–1069. [Google Scholar] [CrossRef] [PubMed]
- Vich Vila, A.; Imhann, F.; Collij, V.; Jankipersadsing, S.A.; Gurry, T.; Mujagic, Z.; Kurilshikov, A.; Bonder, M.J.; Jiang, X.; Tigchelaar, E.F.; et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl. Med. 2018, 10, eaap8914. [Google Scholar] [CrossRef] [PubMed]
- Ryan, F.J.; Ahern, A.M.; Fitzgerald, R.S.; Laserna-Mendieta, E.J.; Power, E.M.; Clooney, A.G.; O’Donoghue, K.W.; McMurdie, P.J.; Iwai, S.; Crits-Christoph, A.; et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat. Commun. 2020, 11, 1512. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Ricciuto, A.; Lewis, A.; D’Amico, F.; Dhaliwal; Griffiths, A.M.; Bettenworth, D.; Sandborn, W.J.; Sands, B.E.; Reinisch, W.; et al. STRIDE-II: An Update on the Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE) Initiative of the International Organization for the Study of IBD (IOIBD): Determining Therapeutic Goals for Treat-to-Target strategies in IBD. Gastroenterology 2021, 160, 1570–1583. [Google Scholar] [CrossRef]
- Torres, J.; Bonovas, S.; Doherty, G.; Kucharzik, T.; Gisbert, J.P.; Raine, T.; Adamina, M.; Armuzzi, A.; Bachmann, O.; Bager, P.; et al. ECCO guidelines on therapeutics in Crohn’s disease: Medical treatment. J. Crohns Colitis 2020, 14, 4–22. [Google Scholar] [CrossRef]
- Raine, T.; Bonovas, S.; Burisch, J.; Kucharzik, T.; Adamina, M.; Annese, V.; Bachmann, O.; Bettenworth, D.; Chaparro, M.; Czuber-Dochan, W.; et al. ECCO Guidelines on Therapeutics in Ulcerative Colitis: Medical Treatment. J. Crohns Colitis 2022, 16, 2–17. [Google Scholar] [CrossRef]
- Spinelli, A.; Bonovas, S.; Burisch, J.; Kucharzik, T.; Adamina, M.; Annese, V.; Bachmann, O.; Bettenworth, D.; Chaparro, M.; Czuber-Dochan, W.; et al. ECCO Guidelines on Therapeutics in Ulcerative Colitis: Surgical Treatment. J. Crohns Colitis 2022, 16, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Van Rheenen, P.F.; Aloi, M.; Assa, A.; Bronsky, J.; Escher, J.C.; Fagerberg, U.L.; Gasparetto, M.; Gerasimidis, K.; Griffiths, A.; Henderson, P.; et al. The Medical Management of Paediatric Crohn’s Disease: An ECCO-ESPGHAN Guideline Update. J. Crohns Colitis 2021, 15, 171–194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Agrawal, M.; Ng, S.C.; Jess, T. Early-life exposures and the microbiome: Implications for IBD prevention. Gut 2024, 73, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Rajagopalan, V.; Kaur, S.; Reimer, R.A.; Ma, C.; Ghosh, S.; Vallance, J. Physical Activity in Patients with Inflammatory Bowel Disease: A Narrative Review. Inflamm. Bowel Dis. 2022, 28, 1100–1111. [Google Scholar] [CrossRef]
- Borrelli, O.; Cordischi, L.; Cirulli, M.; Paganelli, M.; Labalestra, V.; Uccini, S.; Russo, P.M.; Cucchiara, S. Polymeric Diet Alone Versus Corticosteroids in the Treatment of Active Pediatric Crohn’s Disease: A Randomized Controlled Open-Label Trial. Clin. Gastroenterol. Hepatol. 2006, 4, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Narula, N.; Dhillon, A.; Zhang, D.; Sherlock, M.E.; Tondeur, M.; Zachos, M. Enteral nutritional therapy for induction of remission in Crohn’s disease. Cochrane Database Syst. Rev. 2018, 2018, CD000542. [Google Scholar] [CrossRef]
- Gatti, S.; Galeazzi, T.; Franceschini, E.; Annibali, R.; Albano, V.; Verma, A.K.; De Angelis, M.; Lionetti, M.E.; Catassi, C. Effects of the exclusive enteral nutrition on the microbiota profile of patients with crohn’s disease: A systematic review. Nutrients 2017, 9, 832. [Google Scholar] [CrossRef]
- Pigneur, B.; Lepage, P.; Mondot, S.; Schmitz, J.; Goulet, O.; Doré, J.; Ruemmele, F.M. Mucosal Healing and Bacterial Composition in Response to Enteral Nutrition Vs Steroid-based Induction Therapy—A Randomised Prospective Clinical Trial in Children with Crohn’s Disease. J. Crohns Colitis 2019, 13, 846–855. [Google Scholar] [CrossRef]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.P.; Michel, M.L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. ARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef]
- Wedrychowicz, A.; Kowalska-Duplaga, K.; Jedynak-Wasowicz, U.; Pieczarkowski, S.; Sladek, M.; Tomasik, P.; Fyderek, K. Serum concentrations of VEGF and TGF-β1 during exclusive enteral nutrition in IBD. J. Pediatr. Gastroenterol. Nutr. 2011, 53, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Wine, E.; Assa, A.; Sigall Boneh, R.; Shaoul, R.; Kori, M.; Cohen, S.; Peleg, S.; Shamaly, H.; On, A.; et al. Crohn’s Disease Exclusion Diet Plus Partial Enteral Nutrition Induces Sustained Remission in a Randomized Controlled Trial. Gastroenterology 2019, 157, 440.e8–450.e8. [Google Scholar] [CrossRef] [PubMed]
- Limketkai, B.N.; Godoy-Brewer, G.; Parian, A.M.; Noorian, S.; Krishna, M.; Shah, N.D.; White, J.; Mullin, G.E. Dietary Interventions for the Treatment of Inflammatory Bowel Diseases: An Updated Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2023, 21, 2508–2525.e10. [Google Scholar] [CrossRef] [PubMed]
- Nieva, C.; Pryor, J.; Williams, G.M.; Hoedt, E.C.; Burns, G.L.; Eslick, G.D.; Talley, N.J.; Duncanson, K.; Keely, S. The Impact of Dietary Interventions on the Microbiota in Inflammatory Bowel Disease: A Systematic Review. JCC 2024, 18, 920–942. [Google Scholar] [CrossRef] [PubMed]
- Federici, S.; Kredo-Russo, S.; Valdés-Mas, R.; Kviatcovsky, D.; Weinstock, E.; Matiuhin, Y.; Silberberg, Y.; Atarashi, K.; Furuichi, M.; Oka, A.; et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 2022, 185, 2879–2898.e24. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.H.; Khandpur, N.; Rossato, S.L.; Lochhead, P.; Lopes, E.W.; Burke, K.E.; Richter, J.M.; Song, M.; Ardisson Korat, A.V.; Sun, Q.; et al. Ultra-processed Foods and Risk of Crohn’s Disease and Ulcerative Colitis: A Prospective Cohort Study. Clin. Gastroenterol. Hepatol. 2022, 20, e1323–e1337. [Google Scholar] [CrossRef] [PubMed]
- Narula, N.; Wong, E.C.L.; Dehghan, M.; Mente, A.; Rangarajan, S.; Lanas, F.; Lopez-Jaramillo, P.; Rohatgi, P.; Lakshmi, P.V.M.; Varma, R.P.; et al. Association of ultra-processed food intake with risk of inflammatory bowel disease: Prospective cohort study. BMJ 2021, 374, n1554. [Google Scholar] [CrossRef] [PubMed]
- Logan, M.; Gkikas, K.; Svolos, V.; Nichols, B.; Milling, S.; Gaya, D.R.; Seenan, J.P.; Macdonald, J.; Hansen, R.; Ijaz, U.Z.; et al. Analysis of 61 exclusive enteral nutrition formulas used in the management of active Crohn’s disease—New insights into dietary disease triggers. Aliment. Pharmacol. Ther. 2020, 51, 935–947. [Google Scholar] [CrossRef]
- Fenneman, A.C.; Weidner, M.; Chen, L.A.; Nieuwdorp, M.; Blaser, M.J. Antibiotics in the pathogenesis of diabetes and inflammatory diseases of the gastrointestinal tract. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 81–100. [Google Scholar] [CrossRef]
- Kronman, M.P.; Zaoutis, T.E.; Haynes, K.; Feng, R.; Coffin, S.E. Antibiotic exposure and IBD development among children: A population-based cohort study. Pediatrics 2012, 130, e794–e803. [Google Scholar] [CrossRef]
- Hviid, A.; Svanström, H.; Frisch, M. Antibiotic use and inflammatory bowel diseases in childhood. Gut 2011, 60, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Townsend, C.M.; Parker, C.E.; MacDonald, J.K.; Nguyen, T.M.; Jairath, V.; Feagan, B.G.; Khanna, R. Antibiotics for induction and maintenance of remission in Crohn’s disease. Cochrane Database Syst. Rev. 2019, 2019, CD012730. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.; Sinopoulou, V.; Grafton-Clarke, C.; Akobeng, A.K. Antibiotics for the induction and maintenance of remission in ulcerative colitis. Cochrane Database Syst. Rev. 2022, 2022, CD013743. [Google Scholar] [CrossRef]
- Verburgt, C.M.; Heutink, W.P.; Kuilboer, L.I.M.; Dickmann, J.D.; van Etten-Jamaludin, F.S.; Benninga, M.A.; de Jonge, W.J.; Van Limbergen, J.E.; Tabbers, M.M. Antibiotics in pediatric inflammatory bowel diseases: A systematic review. Expert. Rev. Gastroenterol. Hepatol. 2021, 15, 891–908. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, S.C.; Bager, P.; Escher, J.; Forbes, A.; Hébuterne, X.; Hvas, C.L.; Joly, F.; Klek, S.; Krznaric, Z.; Ockenga, J.; et al. ESPEN guideline on Clinical Nutrition in inflammatory bowel disease. Clin. Nutr. 2023, 42, 352–379. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F.; Sanders, M.E.; Szajewska, H.; Cohen, H.M.; Eliakim, R.; Herrera-Deguise, C.; Karakan, T.; Merenstein, D.; Piscoya, A.M.; Ramakrishna, B.; et al. World Gastroenterology Organisation Global Guidelines: Probiotics and Prebiotics. J. Clin. Gastroenterol. 2024, 58, 533–553. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Bermudez-Brito, M.; Plaza Diaz, J.; Muñoz-Quezada, S.; Gil, A. Sources, isolation, characterisation and evaluation of probiotics. Br. J. Nutr. 2013, 109 (Suppl. 2), S35–S50. [Google Scholar] [CrossRef] [PubMed]
- Limketkai, B.N.; Akobeng, A.K.; Gordon, M.; Adepoju, A.A. Probiotics for induction of remission in Crohn’s disease. Cochrane Database Syst. Rev. 2020, 2020, CD006634. [Google Scholar] [CrossRef]
- Ganji-Arjenaki, M.; Rafieian-Kopaei, M. Probiotics are a good choice in remission of inflammatory bowel diseases: A meta analysis and systematic review. J. Cell Physiol. 2018, 233, 2091–2103. [Google Scholar] [CrossRef]
- Zhang, X.F.; Guan, X.X.; Tang, Y.J.; Sun, J.F.; Wang, X.K.; Wang, W.D.; Fan, J.M. Clinical effects and gut microbiota changes of using probiotics, prebiotics or synbiotics in inflammatory bowel disease: A systematic review and meta-analysis. Eur. J. Nutr. 2021, 60, 2855–2875. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef] [PubMed]
- Van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.; Tijssen, J.G.; et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Eiseman, B.; Silen, W.; Bascom, G.S.; Kauvar, A.J. Fecal enema as an adjunct in the treatment of pseudomembranous. Surgery 1958, 44, 854–859. [Google Scholar] [PubMed]
- Lopetuso, L.R.; Deleu, S.; Godny, L.; Petito, V.; Puca, P.; Facciotti, F.; Sokol, H.; Ianiro, G.; Masucci, L.; Abreu, M.; et al. The first international Rome consensus conference on gut microbiota and faecal microbiota transplantation in inflammatory bowel disease. Gut 2023, 72, 1642–1650. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J.; Waetzig, G.H.; Rehman, A.; Moltzau-Anderson, J.; Bharti, R.; Grasis, J.A.; Cassidy, L.; Tholey, A.; Fickenscher, H.; Seegert, D.; et al. Efficacy of Sterile Fecal Filtrate Transfer for Treating Patients with Clostridium difficile Infection. Gastroenterology 2017, 152, 799–811.e7. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Wong, S.H.; Lam, K.; Lui, R.; Cheung, K.; Tang, W.; Ching, J.Y.L.; Chan, P.K.S.; Chan, M.C.W.; Wu, J.C.Y.; et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 2018, 67, 634–643. [Google Scholar] [CrossRef]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef]
- Henn, M.R.; O’Brien, E.J.; Diao, L.; Feagan, B.G.; Sandborn, W.J.; Huttenhower, C.; Wortman, J.R.; McGovern, B.H.; Wang-Weigand, S.; Lichter, D.I.; et al. A Phase 1b Safety Study of SER-287, a Spore-Based Microbiome Therapeutic, for Active Mild to Moderate Ulcerative Colitis. Gastroenterology 2021, 160, 115–127.e30. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Miquel, S.; Benevides, L.; Bridonneau, C.; Robert, V.; Hudault, S.; Chain, F.; Berteau, O.; Azevedo, V.; Chatel, J.M.; et al. Functional characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: A step forward in the use of F. prausnitzii as a next-generation probiotic. Front. Microbiol. 2017, 8, 1226. [Google Scholar] [CrossRef]
- Clinical Trial: EXL01 in the Maintenance of Steroid-Induced Clinical Response/Remission in Participants with Mild to Moderate Crohn’s Disease (MAINTAIN). Available online: https://clinicaltrials.gov/study/NCT05542355 (accessed on 20 March 2023).
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia municiphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Png, C.W.; Lindén, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H.J. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Earley, H.; Lennon, G.; Balfe, Á.; Coffey, J.C.; Winter, D.C.; O’Connell, P.R. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci. Rep. 2019, 9, 15683. [Google Scholar] [CrossRef] [PubMed]
- Vigsnæs, L.K.; Brynskov, J.; Steenholdt, C.; Wilcks, A.; Licht, T.R. Gram-negative bacteria account for main differences between faecal microbiota from patients with ulcerative colitis and healthy controls. Benef. Microbes 2012, 3, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Enrich-Capó, N.; Aldeguer, X.; Sabat-Mir, M.; Duncan, S.H.; Garcia-Gil, J.; Martinez-Medina, M. Alterations in the Abundance and Co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the Colonic Mucosa of Inflammatory Bowel Disease Subjects. Front. Cell Infect. Microbiol. 2018, 8, 281. [Google Scholar] [CrossRef] [PubMed]
- EFSA (European Food Safety Authority). Safety of Pasteurised Akkermansia Muciniphila as a Novel Food Pursuant to Regulation (EU) 2015/2283. Available online: https://www.efsa.europa.eu/en/efsajournal/pub/6780 (accessed on 1 September 2021).
- Baxter, N.T.; Zackular, J.P.; Chen, G.Y.; Schloss, P.D. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2014, 2, 20. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, B.P.; Klopfleisch, R.; Loh, G.; Blaut, M. Commensal Akkermansia muciniphila Exacerbates Gut Inflammation in Salmonella Typhimurium-Infected Gnotobiotic Mice. PLoS ONE 2013, 8, e74963. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Michaudel, C.; Danne, C.; Agus, A.; Magniez, A.; Aucouturier, A.; Spatz, M.; Lefevre, A.; Kirchgesner, J.; Rolhion, N.; Wang, Y.; et al. Rewiring the altered tryptophan metabolism as a novel therapeutic strategy in inflammatory bowel diseases. Gut 2023, 72, 1296–1307. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, I.; Allaband, C.; Mannochio-Russo, H.; El Abiead, Y.; Hagey, L.R.; Knight, R.; Dorrestein, P.C. The changing metabolic landscape of bile acids—Keys to metabolism and immune regulation. Nat. Rev. Gastroenterol. Hepatol. 2024, 21, 493–516. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, W.; Lin, B.; Lei, Y.; Zhang, Y.; Chen, B.; Mao, Q.; Kim, J.J.; Cao, Q. Efficacy of probiotic supplementation and impact on fecal microbiota in patients with inflammatory bowel disease: A systematic review and meta-analysis of randomized controlled trials. Nutr. Rev. 2024, nuae022. [Google Scholar] [CrossRef]
- Carasso, S.; Zaatry, R.; Hajjo, H.; Kadosh-Kariti, D.; Ben-Assa, N.; Naddaf, R.; Mandelbaum, N.; Pressman, S.; Chowers, Y.; Gefen, T.; et al. Inflammation and bacteriophages affect DNA inversion states and functionality of the gut microbiota. Cell Host Microbe 2024, 32, 322–334.e9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, J.; Duan, L. The Role of Genetically Engineered Probiotics for Treatment of Inflammatory Bowel Disease: A Systematic Review. Nutrients 2023, 15, 1566. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jauregui-Amezaga, A.; Smet, A. The Microbiome in Inflammatory Bowel Disease. J. Clin. Med. 2024, 13, 4622. https://doi.org/10.3390/jcm13164622
Jauregui-Amezaga A, Smet A. The Microbiome in Inflammatory Bowel Disease. Journal of Clinical Medicine. 2024; 13(16):4622. https://doi.org/10.3390/jcm13164622
Chicago/Turabian StyleJauregui-Amezaga, Aranzazu, and Annemieke Smet. 2024. "The Microbiome in Inflammatory Bowel Disease" Journal of Clinical Medicine 13, no. 16: 4622. https://doi.org/10.3390/jcm13164622




