Differences in Grey Matter Concentrations and Functional Connectivity between Young Carriers and Non-Carriers of the APOE ε4 Genotype
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants, Study Design, and Ethics
2.2. Data Collection
2.2.1. Genotyping
2.2.2. Brain Imaging
2.3. Data Analysis
2.3.1. Demographic Data
2.3.2. Structural Brain Imaging Analysis
2.3.3. Functional Brain Imaging Analysis
3. Results
3.1. Demographic Characterization of the Sample
3.2. Structural Brain Differences between ε4(+) and ε4(−)
3.3. Functional Brain Differences between ε4(+) and ε4(−)
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prince, M.; Wimo, A.; Guerchet, M.; Ali, G.; Wu, Y.; Prina, M. The Global Impact of Dementia: An Analysis of Prevalence, Incidence, Cost and Trends; World Alzheimer Report. 2015. Available online: https://www.alzint.org/u/WorldAlzheimerReport2015.pdf (accessed on 17 July 2024).
- United Nations. World Population Prospects 2019: Highlights. Statistical Papers—United Nations (Ser. A), Population and Vital Statistics Report; United Nations: New York, NY, USA, 2019; p. 43. [Google Scholar]
- World Health Organization and Alzheimer’s Disease International. Dementia: A Public Health Priority; World Health Organization: Geneva, Switzerland, 2012; 1 Online Resource (112); Available online: https://www.who.int/publications/i/item/dementia-a-public-health-priority (accessed on 17 July 2024).
- Fiest, K.M.; Roberts, J.I.; Maxwell, C.J.; Hogan, D.B.; Smith, E.E.; Frolkis, A.; Cohen, A.; Kirk, A.; Pearson, D.; Pringsheim, T.; et al. The Prevalence and Incidence of Dementia Due to Alzheimer’s Disease: A Systematic Review and Meta-Analysis. Can. J. Neurol. Sci. 2016, 43 (Suppl 1), S51–S82. [Google Scholar] [CrossRef]
- McMurtray, A.; Clark, D.G.; Christine, D.; Mendez, M.F. Early-onset dementia: Frequency and causes compared to late-onset dementia. Dement. Geriatr. Cogn. Disord. 2006, 21, 59–64. [Google Scholar] [CrossRef]
- Barnes, J.; Dickerson, B.C.; Frost, C.; Jiskoot, L.C.; Wolk, D.; van der Flier, W.M. Alzheimer’s disease first symptoms are age dependent: Evidence from the NACC dataset. Alzheimers Dement. 2015, 11, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Bature, F.; Guinn, B.-A.; Pang, D.; Pappas, Y. Signs and symptoms preceding the diagnosis of Alzheimer’s disease: A systematic scoping review of literature from 1937 to 2016. BMJ Open 2017, 7, e015746. [Google Scholar] [CrossRef]
- Aisen, P.S.; Cummings, J.; Jack, C.R., Jr.; Morris, J.C.; Sperling, R.; Frölich, L.; Jones, R.W.; Dowsett, S.A.; Matthews, B.R.; Raskin, J.; et al. On the path to 2025: Understanding the Alzheimer’s disease continuum. Alzheimers Res. Ther. 2017, 9, 60. [Google Scholar] [CrossRef]
- Apostolova, L.G. Alzheimer Disease. Contin. Lifelong Learn. Neurol. 2016, 22, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, K.; Carriere, I.; Su, L.; O’Brien, J.T.; Lovestone, S.; Wells, K.; Ritchie, C.W. The midlife cognitive profiles of adults at high risk of late-onset Alzheimer’s disease: The PREVENT study. Alzheimers Dement. 2017, 13, 1089–1097. [Google Scholar] [CrossRef]
- Ritchie, K.; Ritchie, C.W.; Yaffe, K.; Skoog, I.; Scarmeas, N. Is late-onset Alzheimer’s disease really a disease of midlife? Alzheimers Dement. 2015, 1, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.; Crean, S.; Mercaldi, C.J.; Collins, J.M.; Boyd, D.; Cook, M.N.; Arrighi, H.M. Prevalence of apolipoprotein E4 genotype and homozygotes (APOE e4/4) among patients diagnosed with Alzheimer’s disease: A systematic review and meta-analysis. Neuroepidemiology 2012, 38, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Crean, S.; Ward, A.; Mercaldi, C.J.; Collins, J.M.; Cook, M.N.; Baker, N.L.; Arrighi, H.M. Apolipoprotein E epsilon4 prevalence in Alzheimer’s disease patients varies across global populations: A systematic literature review and meta-analysis. Dement. Geriatr. Cogn. Disord. 2011, 31, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Suri, S.; Heise, V.; Trachtenberg, A.J.; Mackay, C.E. The forgotten APOE allele: A review of the evidence and suggested mechanisms for the protective effect of APOE ɛ2. Neurosci. Biobehav. Rev. 2013, 37 Pt 2, 2878–2886. [Google Scholar] [CrossRef]
- De-Almada, B.; De-Almeida, L.; Camporez, D.; De-Moraes, M.; Morelato, R.; Perrone, A.; Belcavello, L.; Louro, I.; De-Paula, F. Protective effect of the APOE-e3 allele in Alzheimer‘s’ disease. Braz. J. Med. Biol. Res. 2012, 45, 8–12. [Google Scholar] [CrossRef]
- Piers, R.J. Structural brain volume differences between cognitively intact ApoE4 carriers and non-carriers across the lifespan. Neural Regen. Res. 2018, 13, 1309–1312. [Google Scholar] [CrossRef]
- O’Dawyer, L.; Lamberton, F.; Matura, S.; Tanner, C.; Scheibe, M.; Miller, J.; Rujescu, D.; Prvulovic, D.; Hampel, H. Reduced hippocampal volume in healthy young ApoE4 carriers: An MRI study. PLoS ONE 2012, 7, e48895. [Google Scholar] [CrossRef] [PubMed]
- Stening, E.; Persson, J.; Eriksson, E.; Wahlund, L.-O.; Zetterberg, H.; Söderlund, H. Apolipoprotein E ϵ4 is positively related to spatial performance but unrelated to hippocampal volume in healthy young adults. Behav. Brain Res. 2016, 299, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Dounavi, M.-E.; Mak, E.; Wells, K.; Ritchie, K.; Ritchie, C.W.; Su, L.; Brien, J.T.O. Volumetric alterations in the hippocampal subfields of subjects at increased risk of dementia. Neurobiol. Aging 2020, 91, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Burggren, A.; Zeineh, M.; Ekstrom, A.; Braskie, M.; Thompson, P.; Small, G.; Bookheimer, S. Reduced cortical thickness in hippocampal subregions among cognitively normal apolipoprotein E e4 carriers. Neuroimage 2008, 41, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Risacher, S.L.; Apostolova, L.G. Neuroimaging in Dementia. Contin. Lifelong Learn. Neurol. 2023, 29, 219–254. [Google Scholar] [CrossRef]
- Shen, J.; Qin, W.; Xu, Q.; Xu, L.; Xu, J.; Zhang, P.; Liu, H.; Liu, B.; Jiang, T.; Yu, C. Modulation of APOE and SORL1 genes on hippocampal functional connectivity in healthy young adults. Brain Struct. Funct. 2017, 222, 2877–2889. [Google Scholar] [CrossRef]
- Li, W.; Antuono, P.G.; Xie, C.; Chen, G.; Jones, J.L.; Ward, B.D.; Singh, S.P.; Franczak, M.B.; Goveas, J.S.; Li, S.J. Aberrant functional connectivity in Papez circuit correlates with memory performance in cognitively intact middle-aged APOE4 carriers. Cortex 2014, 57, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Sheline, Y.I.; Morris, J.C.; Snyder, A.Z.; Price, J.L.; Yan, Z.; D’Angelo, G.; Liu, C.; Dixit, S.; Benzinger, T.; Fagan, A.; et al. APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF Aβ42. J. Neurosci. 2010, 30, 17035–17040. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Seeley, W.W. Network dysfunction in Alzheimer’s disease and frontotemporal dementia: Implications for psychiatry. Biol. Psychiatry 2014, 75, 565–573. [Google Scholar] [CrossRef]
- Kucikova, L.; Goerdten, J.; Dounavi, M.E.; Mak, E.; Su, L.; Waldman, A.D.; Danso, S.; Muniz-Terrera, G.; Ritchie, C.W. Resting-state brain connectivity in healthy young and middle-aged adults at risk of progressive Alzheimer’s disease. Neurosci. Biobehav. Rev. 2021, 129, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Foo, H.; Mather, K.A.; Jiang, J.; Thalamuthu, A.; Wen, W.; Sachdev, P.S. Genetic influence on ageing-related changes in resting-state brain functional networks in healthy adults: A systematic review. Neurosci. Biobehav. Rev. 2020, 113, 98–110. [Google Scholar] [CrossRef]
- Su, Y.Y.; Liang, X.; Schoepf, U.J.; Varga-Szemes, A.; West, H.C.; Qi, R.; Kong, X.; Chen, H.J.; Lu, G.M.; Zhang, L.J. APOE Polymorphism Affects Brain Default Mode Network in Healthy Young Adults: A STROBE Article. Medicine 2015, 94, e1734. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.Y.; Zhang, X.D.; Schoepf, U.J.; Varga-Szemes, A.; Stubenrauch, A.; Liang, X.; Zheng, L.J.; Zheng, G.; Kong, X.; Xu, Q.; et al. Lower functional connectivity of default mode network in cognitively normal young adults with mutation of APP, presenilins and APOE ε4. Brain Imaging Behav. 2017, 11, 818–828. [Google Scholar] [CrossRef]
- Trachtenberg, A.J.; Filippini, N.; Ebmeier, K.P.; Smith, S.M.; Karpe, F.; Mackay, C.E. The effects of APOE on the functional architecture of the resting brain. Neuroimage 2012, 59, 565–572. [Google Scholar] [CrossRef]
- JASP Team. JASP, (Version 0.18.3); Windows: Redmond, WA, USA, 2024.
- Friston, K.J.; Ashburner, J.; Kiebel, S.; Nichols, T.; Penny, W. Statistical Parametric Mapping: The Analysis of Funtional Brain Images; Elsevier: Amsterdam The Netherland; Academic Press: Boston, MA, USA, 2007. [Google Scholar]
- Ashburner, J.; Friston, K.J. Voxel-based morphometry—The methods. Neuroimage 2000, 11 Pt 1, 805–821. [Google Scholar] [CrossRef] [PubMed]
- Gaser, C.; Dahnke, R. CAT—A Computational Anatomy Toolbox for the Analysis of Structural MRI Data, in HBM. 2016. Available online: https://neuro-jena.github.io/cat12-help/ (accessed on 17 July 2024).
- Gaser, C.; Dahnke, R.; Thompson, P.M.; Kurth, F.; Luders, E.; the Alzheimer’s Disease Neuroimaging Initiative. CAT: A computational anatomy toolbox for the analysis of structural MRI data. GigaScience 2024, 13, 1–13. Available online: https://jasp-stats.org/ (accessed on 17 July 2024). [CrossRef]
- Whitfield-Gabrieli, S.; Nieto-Castanon, A. Conn: A functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2012, 2, 125–141. [Google Scholar] [CrossRef]
- Nieto-Castanon, A. Handbook of Functional Connectivity Magnetic Resonance Imaging Methods in CONN; Hilbert Press: Boston, MA, USA, 2020; Available online: https://www.hilbertpress.org/link-nieto-castanon2020 (accessed on 17 July 2024).
- Nieto-Castanon, A. Citing CONN. Available online: https://web.conn-toolbox.org/resources/citing-conn (accessed on 8 January 2024).
- DiBattista, A.M.; Stevens, B.W.; Rebeck, G.W.; Green, A.E. Two Alzheimer’s disease risk genes increase entorhinal cortex volume in young adults. Front. Hum. Neurosci. 2014, 8, 779. [Google Scholar] [CrossRef]
- Walhovd, K.B.; Fjell, A.M.; Sørensen, Ø.; Mowinckel, A.M.; Reinbold, C.S.; Idland, A.-V.; Watne, L.O.; Franke, A.; Dobricic, V.; Kilpert, F.; et al. Genetic risk for Alzheimer disease predicts hippocampal volume through the human lifespan. Neurol. Genet. 2020, 6, e506. [Google Scholar] [CrossRef]
- Ramusino, M.C.; Altomare, D.; Bacchin, R.; Ingala, S.; Bnà, C.; Bonetti, M.; Costa, A.; Barkhof, F.; Nicolosi, V.; Festari, C.; et al. Medial temporal lobe atrophy and posterior atrophy scales normative values. Neuroimage Clin. 2019, 24, 101936. [Google Scholar] [CrossRef]
- Konishi, K.; Bhat, V.; Banner, H.; Poirier, J.; Joober, R.; Bohbot, V.D. APOE2 Is Associated with Spatial Navigational Strategies and Increased Gray Matter in the Hippocampus. Front. Hum. Neurosci. 2016, 10, 349. [Google Scholar] [CrossRef] [PubMed]
- McKeever, A.; Paris, A.F.; Cullen, J.; Hayes, L.; Ritchie, C.W.; Ritchie, K.; Waldman, A.D.; Wells, K.; Busza, A.; Carriere, I.; et al. Hippocampal Subfield Volumes in Middle-Aged Adults at Risk of Dementia. J. Alzheimers Dis. 2020, 75, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.K.; Diniz, B.S.; Forlenza, O.V.; Busatto, G.F.; Zanetti, M.V. Neurostructural predictors of Alzheimer’s disease: A meta-analysis of VBM studies. Neurobiol. Aging 2011, 32, 1733–1741. [Google Scholar] [CrossRef]
- Park, H.Y.; Park, C.R.; Suh, C.H.; Shim, W.H.; Kim, S.J. Diagnostic performance of the medial temporal lobe atrophy scale in patients with Alzheimer’s disease: A systematic review and meta-analysis. Eur. Radiol. 2021, 31, 9060–9072. [Google Scholar] [CrossRef]
- Mak, E.; Gabel, S.; Mirette, H.; Su, L.; Williams, G.B.; Waldman, A.; Wells, K.; Ritchie, K.; Ritchie, C.; O’Brien, J.T. Structural neuroimaging in preclinical dementia: From microstructural deficits and grey matter atrophy to macroscale connectomic changes. Ageing Res. Rev. 2017, 35, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Kucikova, L.; Zeng, J.; Muñoz-Neira, C.; Muniz-Terrera, G.; Huang, W.; Gregory, S.; Ritchie, C.; O’Brien, J.T.; Su, L. Genetic risk factors of Alzheimer’s Disease disrupt resting-state functional connectivity in cognitively intact young individuals. J. Neurol. 2023, 270, 4949–4958. [Google Scholar] [CrossRef]
- Zheng, L.J.; Su, Y.Y.; Wang, Y.F.; Schoepf, U.J.; Varga-Szemes, A.; Pannell, J.; Liang, X.; Zheng, G.; Lu, G.M.; Yang, G.F.; et al. Different Hippocampus Functional Connectivity Patterns in Healthy Young Adults with Mutations of APP/Presenilin-1/2 and APOEepsilon4. Mol. Neurobiol. 2018, 55, 3439–3450. [Google Scholar] [CrossRef]
- Zheng, L.J.; Lin, L.; Schoepf, U.J.; Varga-Szemes, A.; Savage, R.H.; Zhang, H.; Wang, Y.F.; Zhang, X.Y.; Luo, S.; Liu, Y.; et al. Different posterior hippocampus and default mode network modulation in young APOE epsilon4 carriers: A functional connectome-informed phenotype longitudinal study. Mol. Neurobiol. 2021, 58, 2757–2769. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.C.; Huang, C.C.; Liu, M.E.; Liou, Y.J.; Hong, C.J.; Lo, M.T.; Huang, N.E.; Peng, C.K.; Lin, C.P.; Tsai, S.J. The APOE epsilon4 allele affects complexity and functional connectivity of resting brain activity in healthy adults. Hum. Brain Mapp. 2014, 35, 3238–3248. [Google Scholar] [CrossRef] [PubMed]
- Korthauer, L.E.; Zhan, L.; Ajilore, O.; Leow, A.; Driscoll, I. Disrupted topology of the resting state structural connectome in middle-aged APOE epsilon4 carriers. Neuroimage 2018, 178, 295–305. [Google Scholar] [CrossRef]
- Berron, D.; van Westen, D.; Ossenkoppele, R.; Strandberg, O.; Hansson, O. Medial temporal lobe connectivity and its associations with cognition in early Alzheimer’s disease. Brain 2020, 143, 1233–1248. [Google Scholar] [CrossRef]
- Kenny, E.R.; Blamire, A.M.; Firbank, M.J.; O’Brien, J.T. Functional connectivity in cortical regions in dementia with Lewy bodies and Alzheimer’s disease. Brain 2012, 135 Pt 2, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, R.; Fleisher, A.S.; Reiman, E.M.; Guan, X.; Zhang, Y.; Chen, K.; Yao, L. Altered default mode network connectivity in Alzheimer’s disease—A resting functional MRI and Bayesian network study. Hum. Brain Mapp. 2011, 32, 1868–1881. [Google Scholar] [CrossRef]
- Regy, M.; Dugravot, A.; Sabia, S.; Fayosse, A.; Mangin, J.F.; Chupin, M.; Fischer, C.; Bouteloup, V.; Dufouil, C.; Chene, G.; et al. Association of APOE epsilon4 with cerebral gray matter volumes in non-demented older adults: The MEMENTO cohort study. Neuroimage 2022, 250, 118966. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Holland, D.; Dale, A.M.; Miller, M.I.; Alzheimer’s Disease Neuroimaging Initiative. APOE Affects the Volume and Shape of the Amygdala and the Hippocampus in Mild Cognitive Impairment and Alzheimer’s Disease: Age Matters. J. Alzheimers Dis. 2015, 47, 645–660. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, J.T.; Wang, H.F.; Han, P.R.; Tan, C.C.; Wang, C.; Meng, X.F.; Risacher, S.L.; Saykin, A.J.; Tan, L. APOE genotype and neuroimaging markers of Alzheimer’s disease: Systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 2015, 86, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Schuff, N.; Woerner, N.; Boreta, L.; Kornfield, T.; Shaw, L.M.; Trojanowski, J.Q.; Thompson, P.M.; Jack, C.R., Jr.; Weiner, M.W.; Alzheimer’s Disease Neuroimaging Initiative. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to APOE genotype and biomarkers. Brain 2009, 132 Pt 4, 1067–1077. [Google Scholar] [CrossRef]
- Lee, H.; Stirnberg, R.; Wu, S.; Wang, X.; Stöcker, T.; Jung, S.; Montag, C.; Axmacher, N. Genetic Alzheimer’s Disease Risk Affects the Neural Mechanisms of Pattern Separation in Hippocampal Subfields. Curr. Biol. 2020, 30, 4201–4212 e3. [Google Scholar] [CrossRef]
- Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Mahley, R.W.; Weisgraber, K.H.; Huang, Y. Apolipoprotein E4: A causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2006, 103, 5644–5651. [Google Scholar] [CrossRef] [PubMed]
- Han, S.D.; Bondi, M.W. Revision of the apolipoprotein E compensatory mechanism recruitment hypothesis. Alzheimers Dement. 2008, 4, 251–254. [Google Scholar] [CrossRef]
- Bondi, M.W.; Houston, W.S.; Eyler, L.T.; Brown, G.G. fMRI evidence of compensatory mechanisms in older adults at genetic risk for Alzheimer disease. Neurology 2005, 64, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Bookheimer, S.Y.; Strojwas, M.H.; Cohen, M.S.; Saunders, A.M.; Pericak-Vance, M.A.; Mazziotta, J.C.; Small, G.W. Patterns of brain activation in people at risk for Alzheimer’s disease. N. Engl. J. Med. 2000, 343, 450–456. [Google Scholar] [CrossRef]
- Tuminello, E.R.; Han, S.D. The apolipoprotein e antagonistic pleiotropy hypothesis: Review and recommendations. Int. J. Alzheimers Dis. 2011, 2011, 1. [Google Scholar] [CrossRef] [PubMed]
- Filippini, N.; Ebmeier, K.; MacIntosh, B.; Trachtenberg, A.; Frisoni, G.; Wilcock, G.; Beckmann, C.; Smith, S.; Matthews, P.; Mackay, C.E. Differential effects of the APOE genotype on brain function across the lifespan. Neuroimage 2011, 54, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Seitzman, B.A.; Snyder, A.Z.; Leuthardt, E.C.; Shimony, J.S. The State of Resting State Networks. Top. Magn. Reson. Imaging 2019, 28, 189–196. [Google Scholar] [CrossRef]

| Parameter | Descriptive Statistics per Group | Comparisons | |||
|---|---|---|---|---|---|
| Structural Neuroimaging Analysis (VBM) | Functional Neuroimaging Analysis (SBC) | ||||
| ε4(+) (n = 28) | ε4(−) (n = 123) | ε4(+) (n = 27) | ε4(−) (n = 102) | ||
| Age a | 19.53 ± 0.98 | 19.64 ± 0.87 | 19.57 ± 0.98 | 19.64 ± 0.87 | nsd for VBM and SBC |
| Years of Education a | 12.94 ± 0.54 | 13.11 ± 0.57 | 12.95 ± 0.55 | 13.14 ± 0.59 | nsd for VBM and SBC |
| Sex b | nsd for VBM and SBC | ||||
| %Female (n) | 57.14% (16) | 56.10% (69) | 57.14% (16) | 56.10% (69) | |
| %Male (n) | 42.86% (12) | 43.90% (54) | 42.86% (12) | 43.90% (54) | |
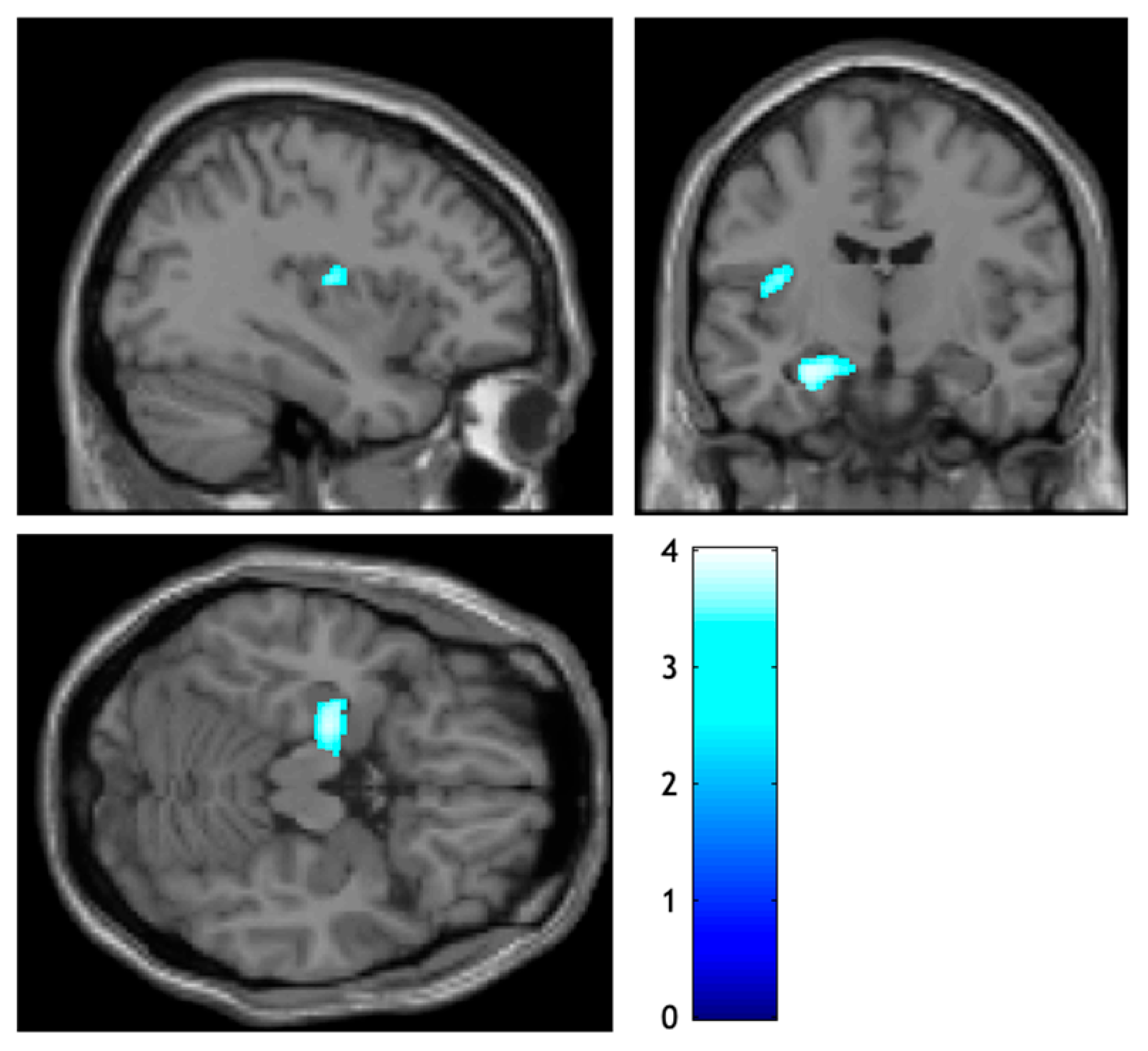
| VBM Analysis Contrast: APOE ε4 Carriers > APOE ε4 Non-Carriers puncorr < 0.001, 97.472 Expected Voxels per Cluster | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Structure (Hemisphere) | % | Types of Significance | MNI Coordinates | ||||||||
| Cluster-Level | Peak-Level | ||||||||||
| pFWEcorr | ke | puncorr | pFWEcorr | T | Ze | puncorr | mm | mm | mm | ||
| Hippocampus (Left) | 50.9 | 0.153 | 534 | 0.023 | 0.493 | 4.00 | 3.89 | 0.000 | −24 | −14 | −21 |
| Posterior Insula (Left) | 26.9 | 0.763 | 148 | 0.202 | 0.943 | 3.51 | 3.43 | 0.000 | −39 | −12 | 12 |
| SBC Analysis between the Hippocampus Left and Other Voxels across the Brain and the Posterior Insula Left and Other Voxels across the Brain | ||||
|---|---|---|---|---|
| Seed | Associated Brain Areas | T(123) | puncorr | pFDR |
| Hippocampus (Left) | 4.59 | 0.000011 | 0.000011 | |
| Middle Temporal Gyrus, posterior region (Left) | ||||
| Middle Temporal Gyrus, temporooccipital division (Left) | ||||
| Posterior Insula (Left) | None | na | na | >0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz-Neira, C.; Zeng, J.; Kucikova, L.; Huang, W.; Xiong, X.; Muniz-Terrera, G.; Ritchie, C.; O’Brien, J.T.; Su, L. Differences in Grey Matter Concentrations and Functional Connectivity between Young Carriers and Non-Carriers of the APOE ε4 Genotype. J. Clin. Med. 2024, 13, 5228. https://doi.org/10.3390/jcm13175228
Muñoz-Neira C, Zeng J, Kucikova L, Huang W, Xiong X, Muniz-Terrera G, Ritchie C, O’Brien JT, Su L. Differences in Grey Matter Concentrations and Functional Connectivity between Young Carriers and Non-Carriers of the APOE ε4 Genotype. Journal of Clinical Medicine. 2024; 13(17):5228. https://doi.org/10.3390/jcm13175228
Chicago/Turabian StyleMuñoz-Neira, Carlos, Jianmin Zeng, Ludmila Kucikova, Weijie Huang, Xiong Xiong, Graciela Muniz-Terrera, Craig Ritchie, John T. O’Brien, and Li Su. 2024. "Differences in Grey Matter Concentrations and Functional Connectivity between Young Carriers and Non-Carriers of the APOE ε4 Genotype" Journal of Clinical Medicine 13, no. 17: 5228. https://doi.org/10.3390/jcm13175228






