Deep Learning-Adjusted Monitoring of In-Hospital Mortality after Liver Transplantation
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Groups and Predictive Variables
2.2. Follow-Up
2.3. Statistical Analysis
3. Results
3.1. Patient Data
3.2. Transplantation and Donor Data
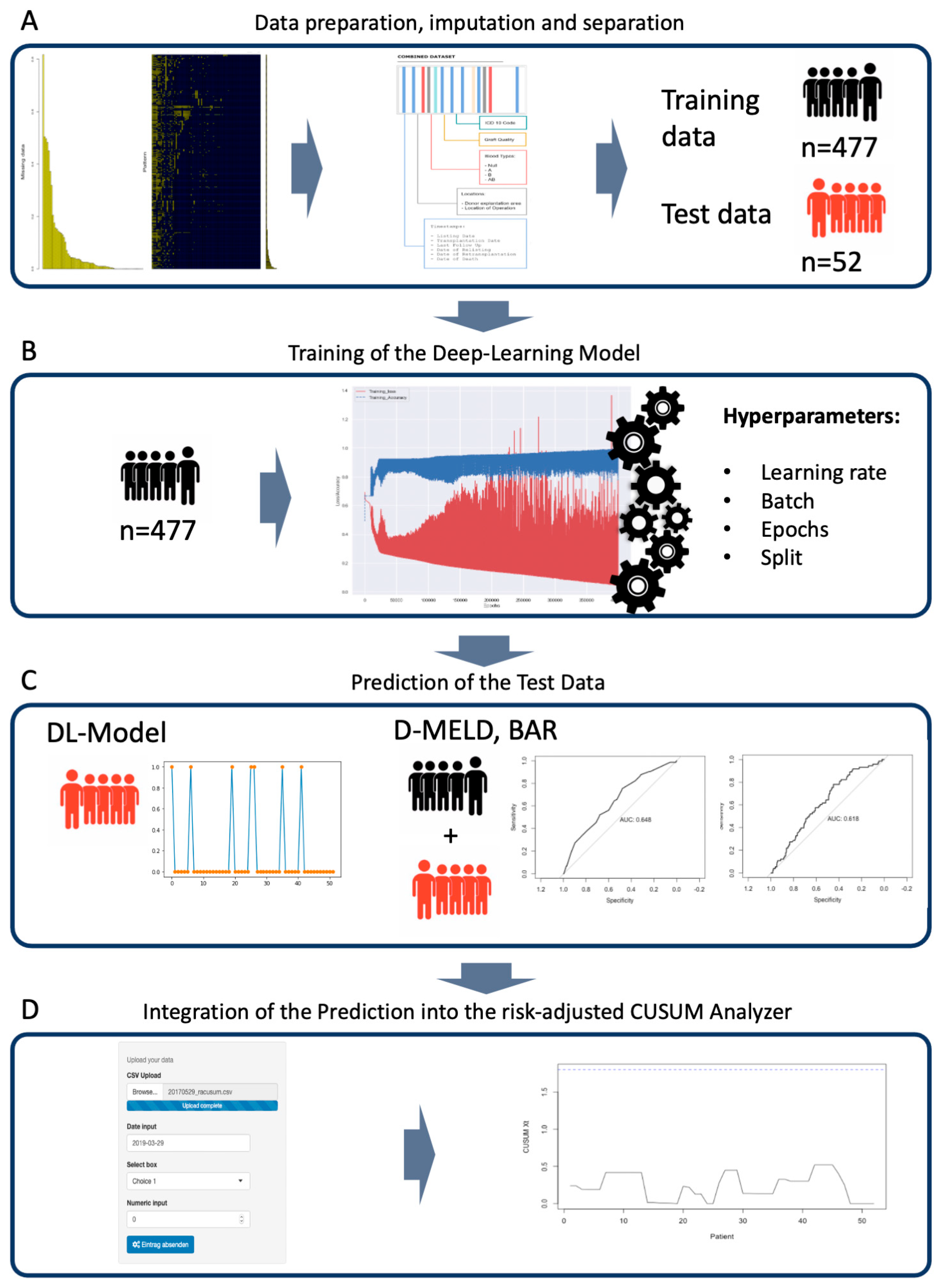
3.3. Separation of the Data Sets and Training of the Deep Learning Model
3.4. Predicting In-Hospital Mortality
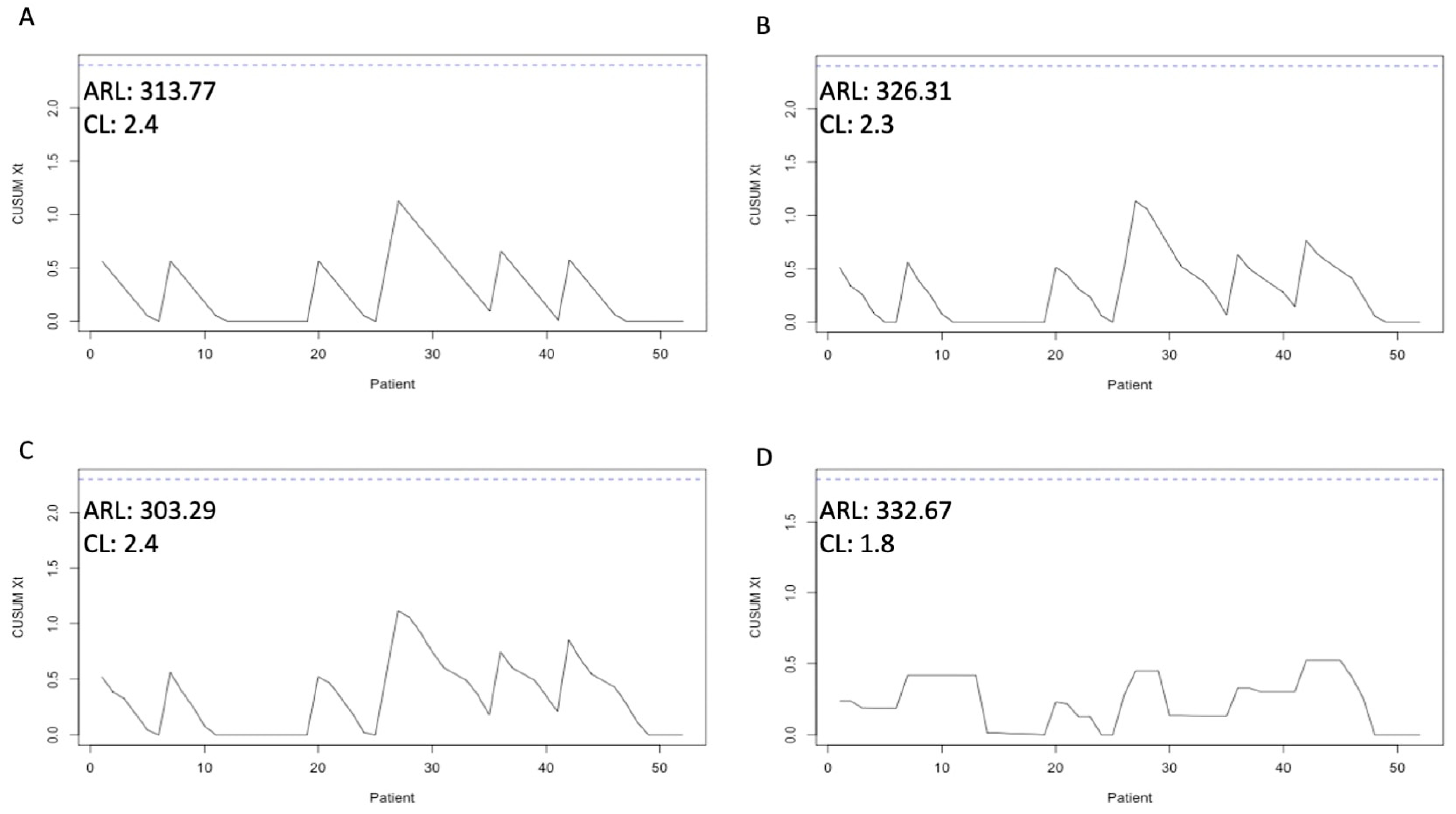
3.5. Merging of Risk Adjustment and CUSUM Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Craig, E.V.; Heller, M.T. Complications of liver transplant. Abdom. Radiol. 2021, 46, 43–67. [Google Scholar] [CrossRef] [PubMed]
- Mazzocco, K.; Petitti, D.B.; Fong, K.T.; Bonacum, D.; Brookey, J.; Graham, S.; Lasky, R.E.; Sexton, J.B.; Thomas, E.J. Surgical team behaviors and patient outcomes. Am. J. Surg. 2009, 197, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Suliburk, J.W.; Buck, Q.M.; Pirko, C.J.; Massarweh, N.N.; Barshes, N.R.; Singh, H.; Rosengart, T.K. Analysis of Human Performance Deficiencies Associated With Surgical Adverse Events. JAMA Netw. Open 2019, 2, e198067. [Google Scholar] [CrossRef] [PubMed]
- Glaysher, M.A.; Cresswell, A.B. Management of common surgical complications. Surgery 2014, 32, 121–125. [Google Scholar]
- Haynes, A.B.; Weiser, T.G.; Berry, W.R.; Lipsitz, S.R.; Breizat, A.-H.S.; Dellinger, E.P.; Herbosa, T.; Joseph, S.; Kibatala, P.L.; Lapitan, M.C.M.; et al. A Surgical Safety Checklist to Reduce Morbidity and Mortality in a Global Population. N. Engl. J. Med. 2009, 360, 491–499. [Google Scholar] [CrossRef]
- Kessels-Habraken, M.; De Jonge, J.; Van der Schaaf, T.; Rutte, C. Prospective risk analysis prior to retrospective incident reporting and analysis as a means to enhance incident reporting behaviour: A quasi-experimental field study. Soc. Sci. Med. 2010, 70, 1309–1316. [Google Scholar] [CrossRef]
- Mullen, M.G.; Michaels, A.D.; Mehaffey, J.H.; Guidry, C.A.; Turrentine, F.E.; Hedrick, T.L.; Friel, C.M. Risk Associated With Complications and Mortality After Urgent Surgery vs. Elective and Emergency Surgery: Implications for Defining “Quality” and Reporting Outcomes for Urgent Surgery. JAMA Surg. 2017, 152, 768–774. [Google Scholar] [CrossRef]
- Grigg, O.A.; Farewell, V.T.; Spiegelhalter, D.J. Use of risk-adjusted CUSUM and RSPRT charts for monitoring in medical contexts. Stat. Methods Med. Res. 2003, 12, 147–170. [Google Scholar] [CrossRef]
- Steiner, S.H.; Cook, R.J.; Farewell, V.T.; Treasure, T. Monitoring surgical performance using risk-adjusted cumulative sum charts. Biostatistics 2000, 1, 441–452. [Google Scholar] [CrossRef]
- Axelrod, D.A.; Guidinger, M.K.; Metzger, R.A.; Wiesner, R.H.; Webb, R.L.; Merion, R.M. Transplant center quality assessment using a continuously updatable, risk-adjusted technique (CUSUM). Am. J. Transplant. 2006, 6, 313–323. [Google Scholar] [CrossRef]
- Snyder, J.J.; Salkowski, N.; Zaun, D.; Leppke, S.N.; Leighton, T.; Israni, A.K.; Kasiske, B.L. New quality monitoring tools provided by the Scientific Registry of Transplant Recipients: CUSUM. Am. J. Transplant. 2014, 14, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Ngiam, K.Y.; Khor, I.W. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019, 20, e262–e273. [Google Scholar] [CrossRef] [PubMed]
- Parisi, G.I.; Kemker, R.; Part, J.L.; Kanan, C.; Wermter, S. Continual lifelong learning with neural networks: A review. Neural Netw. 2019, 113, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Schoenberg, M.B.; Ehmer, U.; Umgelter, A.; Bucher, J.N.; Koch, D.T.; Börner, N.; Nieß, H.; Denk, G.; De Toni, E.N.; Seidensticker, M.; et al. Liver transplantation versus watchful waiting in hepatocellular carcinoma patients with complete response to bridging therapy—A retrospective observational study. Transpl. Int. 2021, 34, 465–473. [Google Scholar] [CrossRef]
- Herzer, K.; Strassburg, C.P.; Braun, F.; Engelmann, C.; Guba, M.; Lehner, F.; Nadalin, S.; Pascher, A.; Scherer, M.N.; Schnitzbauer, A.A.; et al. Selection and use of immunosuppressive therapies after liver transplantation: Current German practice. Clin. Transplant. 2016, 30, 487–501. [Google Scholar] [CrossRef]
- Bolondi, G.; Mocchegiani, F.; Montalti, R.; Nicolini, D.; Vivarelli, M.; De Pietri, L. Predictive factors of short term outcome after liver transplantation: A review. World J. Gastroenterol. 2016, 22, 5936–5949. [Google Scholar] [CrossRef] [PubMed]
- Börner, N.; Schoenberg, M.B.; Pöschke, P.; Pöllmann, B.; Koch, D.; Drefs, M.; Koliogiannis, D.; Böhm, C.; Werner, J.; Guba, M. A custom build multidimensional medical combined imputation application for a transplantation dataset. Comput. Methods Programs Biomed. Updat. 2022, 2, 100083. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. BMJ 2015, 350, g7594. [Google Scholar] [CrossRef]
- London, A.J. Artificial Intelligence and Black-Box Medical Decisions: Accuracy versus Explainability. Hastings Cent. Rep. 2019, 49, 15–21. [Google Scholar] [CrossRef]
- Saito, T.; Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef]
- Halldorson, J.B.; Bakthavatsalam, R.; Fix, O.; Reyes, J.D.; Perkins, J.D. D-MELD, a simple predictor of post liver transplant mortality for optimization of donor/recipient matching. Am. J. Transplant. 2009, 9, 318–326. [Google Scholar] [PubMed]
- Dutkowski, P.; Oberkofler, C.E.; Slankamenac, K.; Puhan, M.A.; Schadde, E.; Müllhaupt, B.; Geier, A.; Clavien, P.A. Are there better guidelines for allocation in liver transplantation?: A novel score targeting justice and utility in the model for end-stage liver disease era. Ann. Surg. 2011, 254, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Schrem, H.; Volz, S.; Koch, H.-F.; Gwiasda, J.; Kürsch, P.; Goldis, A.; Pöhnert, D.; Winny, M.; Klempnauer, J.; Kaltenborn, A. Statistical approach to quality assessment in liver transplantation. Langenbeck’s Arch. Surg. 2018, 403, 61–71. [Google Scholar]
- Barjaktarevic, I.; Lopez, R.C.; Steadman, R.; Wray, C.; Qadir, N.; Chang, S.Y.; Wang, T. Perioperative Considerations in Liver Transplantation. Semin. Respir. Crit. Care Med. 2018, 39, 609–624. [Google Scholar] [PubMed]
- Neuberger, J.; Madden, S.; Collett, D. Review of methods for measuring and comparing center performance after organ transplantation. Liver Transplant. 2010, 16, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, Y.; Li, Y.; Sun, S.; Gao, X. A self-adaptive deep learning algorithm for accelerating multi-component flash calculation. Comput. Methods Appl. Mech. Eng. 2020, 369, 113207. [Google Scholar]
- Raveh, Y.; Livingstone, J.; Mahan, J.; Tekin, A.; Selvaggi, G.; Bowdon-Romero, M.; Nicolau-Raducu, R. Comprehensive Frailty Severity Index for End-Stage Liver Disease Predicts Early Outcomes After Liver Transplantation. JPEN J. Parenter. Enter. Nutr. 2020, 44, 1079–1088. [Google Scholar]
- Calvani, R.; Picca, A.; Marini, F.; Biancolillo, A.; Gervasoni, J.; Persichilli, S.; Primiano, A.; Coelho-Junior, H.J.; Cesari, M.; Bossola, M.; et al. Identification of biomarkers for physical frailty and sarcopenia through a new multi-marker approach: Results from the BIOSPHERE study. GeroScience 2020, 43, 727–740. [Google Scholar]
- Corbett-Davies, S.; Goel, S. The Measure and Mismeasure of Fairness: A Critical Review of Fair Machine Learning. arXiv 2023, arXiv:1808.00023. [Google Scholar]
| Characteristics | Study Cohort | Training Data | Test Data | Training vs. Test |
|---|---|---|---|---|
| n = 529 | n = 477 | n = 52 | p-Value | |
| Demographics | ||||
| Age at operation in years, mean ± SD | 50.28 ± 12.29 | 50.06 ± 12.46 | 52.31 ± 10.58 | 0.2113 |
| Male/female | 357/172 | 318/159 | 39/13 | 0.2755 |
| Height (m), mean ± SD | 1.73 ± 0.10 | 1.73 ± 0.10 | 1.73 ± 0.09 | 0.9754 |
| Weight (kg), mean ± SD | 77.57 ± 16.39 | 77.79 ± 16.36 | 75.58 ± 16.66 | 0.3543 |
| BMI, mean ± SD | 25.67 ± 4.59 | 25.74 ± 4.57 | 25.03 ± 4.44 | 0.2903 |
| Liver disease features | ||||
| Ascites, Y/N | 332/197 | 301/176 | 31/21 | 0.6518 |
| Encephalopathy, Y/N | 216/313 | 194/283 | 22/30 | 0.8822 |
| Dialysis, Y/N | 77/452 | 72/407 | 5/47 | 0.2921 |
| MELD, mean ± SD | 23.79 ± 11.08 | 23.86 ± 11.16 | 23.17 ± 10.50 | 0.6710 |
| Allocation MELD, mean ± SD | 27.75 ± 8.55 | 27.83 ± 8.66 | 27.15 ± 7.64 | 0.5912 |
| Laboratory values | ||||
| Na mmol/L, mean ± SD | 135.98 ± 5.42 | 135.98 ± 5.43 | 135.98 ± 5.38 | 0.9983 |
| K mmol/L, mean ± SD | 4.10 ± 0.50 | 4.11 ± 0.49 | 3.95 ± 0.55 | 0.0268 |
| Bilirubin mg/dL, mean ± SD | 12.12 ± 13.56 | 12.02 ± 13.32 | 12.97 ± 15.83 | 0.6296 |
| Albumin g/L, mean ± SD | 3.15 ± 0.67 | 3.15 ± 0.68 | 3.16 ± 0.60 | 0.8627 |
| ALT U/L, mean ± SD | 328.94 ± 876.02 | 306 ± 829.33 | 421.81 ± 1023.03 | 0.0967 |
| AST U/L, mean ± SD | 454.85 ± 1318.16 | 389.63 ± 1125.18 | 684.92 ± 1854.65 | 0.3536 |
| GGT U/L, mean ± SD | 141.45 ± 186.77 | 140.23 ± 186.29 | 144.37 ± 189.98 | 0.8796 |
| AP U/L, mean ± SD | 231.38 ± 252.37 | 225.67 ± 251.54 | 246.48 ± 237.75 | 0.5693 |
| Hemoglobin g/dL, mean ± SD | 10.58 ± 2.50 | 10.60 ± 2.50 | 10.43 ± 2.47 | 0.6348 |
| INR, mean ± SD | 1.76 ± 0.90 | 1.77 ± 0.94 | 1.62 ± 0.51 | 0.2541 |
| Creatinine mg/dL, mean ± SD | 1.66 ± 1.16 | 1.65 ± 1.14 | 1.83 ± 1.30 | 0.2843 |
| CRP mg/dL, mean ± SD | 2.51 ± 3.58 | 2.50 ± 3.64 | 2.60 ± 3.09 | 0.8481 |
| Leukocytes 106/L, mean ± SD | 8.15 ± 6.47 | 8.22 ± 6.66 | 7.50 ± 4.37 | 0.4426 |
| Platelets 106/L, mean ± SD | 100.27 ± 74.17 | 100.49 ± 75.54 | 98.17 ± 60.68 | 0.8305 |
| Characteristic | Study Cohort | Training Data | Test Data | Training vs. Test |
|---|---|---|---|---|
| n = 529 | n = 477 | n = 52 | p-Value | |
| Cold Ischemia Time (min) ± SD | 630.69 ± 156.61 | 634.28 ± 159.66 | 597.77 ± 121.49 | 0.1104 |
| Full/Split Liver ± SD | 499/30 | 447/30 | 52/0 | 0.0607 |
| Distance from Explanation to Transplantation (km) ± SD | 312.56 ± 210.99 | 328.52 ± 210.31 | 257.73 ± 208.38 | 0.0215 |
| Duration of Stay (Days) ± SD | 45.15 ± 39.87 | 44.79 ± 39.64 | 48.42 ± 42.13 | 0.5334 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Börner, N.; Schoenberg, M.B.; Pöllmann, B.; Pöschke, P.; Böhm, C.; Koch, D.; Drefs, M.; Koliogiannis, D.; Andrassy, J.; Werner, J.; et al. Deep Learning-Adjusted Monitoring of In-Hospital Mortality after Liver Transplantation. J. Clin. Med. 2024, 13, 6046. https://doi.org/10.3390/jcm13206046
Börner N, Schoenberg MB, Pöllmann B, Pöschke P, Böhm C, Koch D, Drefs M, Koliogiannis D, Andrassy J, Werner J, et al. Deep Learning-Adjusted Monitoring of In-Hospital Mortality after Liver Transplantation. Journal of Clinical Medicine. 2024; 13(20):6046. https://doi.org/10.3390/jcm13206046
Chicago/Turabian StyleBörner, Nikolaus, Markus B. Schoenberg, Benedikt Pöllmann, Philipp Pöschke, Christian Böhm, Dominik Koch, Moritz Drefs, Dionysios Koliogiannis, Joachim Andrassy, Jens Werner, and et al. 2024. "Deep Learning-Adjusted Monitoring of In-Hospital Mortality after Liver Transplantation" Journal of Clinical Medicine 13, no. 20: 6046. https://doi.org/10.3390/jcm13206046
APA StyleBörner, N., Schoenberg, M. B., Pöllmann, B., Pöschke, P., Böhm, C., Koch, D., Drefs, M., Koliogiannis, D., Andrassy, J., Werner, J., & Guba, M. O. (2024). Deep Learning-Adjusted Monitoring of In-Hospital Mortality after Liver Transplantation. Journal of Clinical Medicine, 13(20), 6046. https://doi.org/10.3390/jcm13206046






