Prospective and External Validation of Machine Learning Models for Short- and Long-Term Mortality in Acutely Admitted Patients Using Blood Tests
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Settings
2.2. Medical Records
2.3. Biomarkers
2.4. Outcomes
2.5. Data Preparation
2.6. Model Construction
2.7. Algorithm Selection and Performance Measures
2.8. Calibration
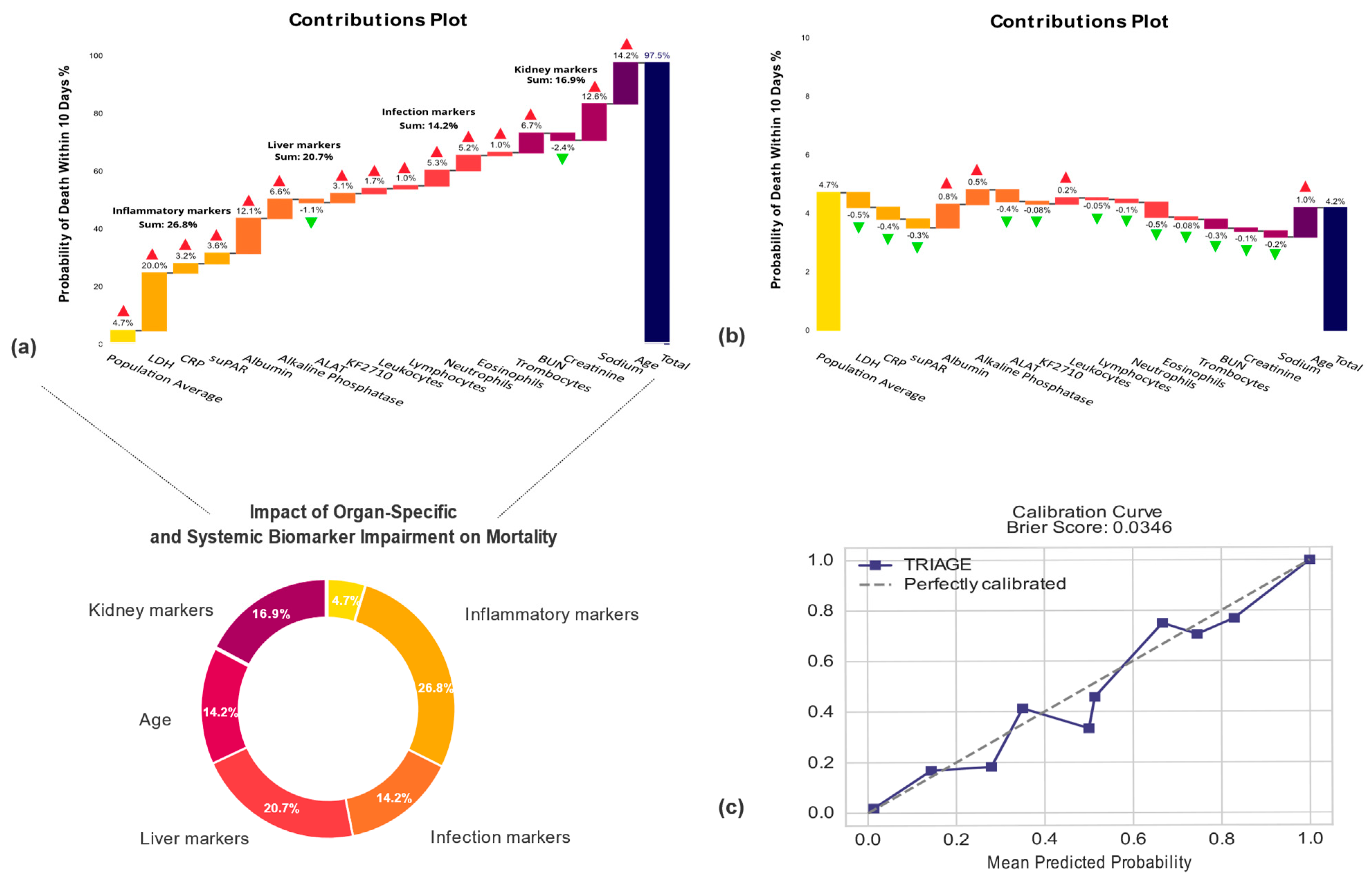
2.9. Explaining Model Predictions
2.10. Statistical Analysis
3. Results
3.1. Description of the Cohorts Used in the Study
3.2. Model Performance
3.3. Machine Learning Prediction Application
4. Discussion
4.1. Limitations
4.2. Future Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ONU. World Population, Ageing; ST/ESA/SER.A/390; Department of Economic and Social Affairs, Population Division: New York, NY, USA, 2015. [Google Scholar]
- Veser, A.; Sieber, F.; Groß, S.; Prückner, S. The demographic impact on the demand for emergency medical services in the urban and rural regions of bavaria, 2012–2032. J. Public Health 2015, 23, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Businger, A.P.; Kaderli, R.; Burghardt, L.R.; Osinga, R.; Liesch, M.; Furrer, M. Demographic Changes and Their Implications in a Nonacademic Emergency Department in Switzerland: An 11-Year Trend Analysis (2000–2010) of 104,510 Patients. ISRN Emerg. Med. 2012, 2012, 1–6. [Google Scholar] [CrossRef][Green Version]
- Lemeshow, S.; Gehlbach, S.H.; Klar, J.; Avrunin, J.S.; Teres, D.; Rapoport, J. Mortality Probability Models (MPM II) Based on an International Cohort of Intensive Care Unit Patients. JAMA J. Am. Med. Assoc. 1993, 270, 2478–2486. [Google Scholar] [CrossRef]
- Toma, T.; Abu-Hanna, A.; Bosman, R.J. Discovery and inclusion of SOFA score episodes in mortality prediction. J. Biomed. Inform. 2007, 40, 649–660. [Google Scholar] [CrossRef]
- Burch, V.C.; Tarr, G.; Morroni, C. Modified early warning score predicts the need for hospital admission and inhospital mortality. Emerg. Med. J. 2008, 25, 674–678. [Google Scholar] [CrossRef] [PubMed]
- HKlausen, H.H.; Petersen, J.; Bandholm, T.; Juul-Larsen, H.G.; Tavenier, J.; Eugen-Olsen, J.; Andersen, O. Association between routine laboratory tests and long-term mortality among acutely admitted older medical patients: A cohort study. BMC Geriatr. 2017, 17, 62. [Google Scholar] [CrossRef]
- Phungoen, P.; Khemtong, S.; Apiratwarakul, K.; Ienghong, K.; Kotruchin, P. Emergency Severity Index as a predictor of in-hospital mortality in suspected sepsis patients in the emergency department. Am. J. Emerg. Med. 2020, 38, 1854–1859. [Google Scholar] [CrossRef]
- Mahmoodpoor, A.; Sanaie, S.; Saghaleini, S.H.; Ostadi, Z.; Hosseini, M.-S.; Sheshgelani, N.; Vahedian-Azimi, A.; Samim, A.; Rahimi-Bashar, F. Prognostic value of National Early Warning Score and Modified Early Warning Score on intensive care unit readmission and mortality: A prospective observational study. Front. Med. 2022, 9, 938005. [Google Scholar] [CrossRef] [PubMed]
- Knaus, W.A. APACHE 1978-2001: The development of a quality assurance system based on prognosis: Milestones and personal reflections. Arch. Surg. 2002, 137, 37–41. [Google Scholar] [CrossRef]
- Sherazi, S.W.A.; Jeong, Y.J.; Jae, M.H.; Bae, J.-W.; Lee, J.Y. A machine learning-based 1-year mortality prediction model after hospital discharge for clinical patients with acute coronary syndrome. Health Inform. J. 2020, 26, 1289–1304. [Google Scholar] [CrossRef]
- Yadaw, A.S.; Li, Y.-C.; Bose, S.; Iyengar, R.; Bunyavanich, S.; Pandey, G. Clinical features of COVID-19 mortality: Development and validation of a clinical prediction model. Lancet Digit. Health 2020, 2, e516–e525. [Google Scholar] [CrossRef] [PubMed]
- Bonde, A.; Bonde, M.; Troelsen, A.; Sillesen, M. Assessing the utility of a sliding-windows deep neural network approach for risk prediction of trauma patients. Sci. Rep. 2023, 13, 5176. [Google Scholar] [CrossRef] [PubMed]
- Kijpaisalratana, N.; Sanglertsinlapachai, D.; Techaratsami, S.; Musikatavorn, K.; Saoraya, J. Machine learning algorithms for early sepsis detection in the emergency department: A retrospective study. Int. J. Med. Inform. 2022, 160, 104689. [Google Scholar] [CrossRef]
- HThorsen-Meyer, H.-C.; Nielsen, A.B.; Kaas-Hansen, B.S.; Toft, P.; Schierbeck, J.; Strøm, T.; Chmura, P.J.; Heimann, M.; Dybdahl, L.; Spangsege, L.; et al. Dynamic and explainable machine learning prediction of mortality in patients in the intensive care unit: A retrospective study of high-frequency data in electronic patient records. Lancet Digit. Health 2020, 2, e179–e191. [Google Scholar] [CrossRef]
- Kirk, J.W.; Nilsen, P. Implementing evidence-based practices in an emergency department: Contradictions exposed when prioritising a flow culture. J. Clin. Nurs. 2016, 25, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Li, C.; He, P.; Xiao, H.; Ping, Y.; Feng, P.; Tian, S.; Chen, H.; Mercaldo, F.; Santone, A.; et al. MamlFormer: Priori-experience guiding transformer network via manifold adversarial multi-modal learning for laryngeal histopathological grading. Inf. Fusion 2024, 108, 102333. [Google Scholar] [CrossRef]
- Huang, P.; Xiao, H.; He, P.; Li, C.; Guo, X.; Tian, S.; Feng, P.; Chen, H.; Sun, Y.; Mercaldo, F.; et al. LA-ViT: A Network With Transformers Constrained by Learned-Parameter-Free Attention for Interpretable Grading in a New Laryngeal Histopathology Image Dataset. IEEE J. Biomed. Health Inform. 2024, 28, 3557–3570. [Google Scholar] [CrossRef]
- Singh, P.; Chahat, R.K.; Kr, S.; Anup, M.; Yadav, L.; Singla, S. Comparative Analysis of Tree-Based Models and Deep Learning Architectures for Tabular Data: Performance Disparities and Underlying Factors. In Proceedings of the 2023 International Conference on Advanced Computing & Communication Technologies (ICACCTech), Banur, India, 23–24 December 2023; pp. 224–231. Available online: https://api.semanticscholar.org/CorpusID:268045915 (accessed on 28 August 2024).
- Frasca, M.; La Torre, D.; Pravettoni, G.; Cutica, I. Explainable and interpretable artificial intelligence in medicine: A systematic bibliometric review. Discov. Artif. Intell. 2024, 4, 15. [Google Scholar] [CrossRef]
- Jawad, B.N.; Shaker, S.M.; Altintas, I.; Eugen-Olsen, J.; Nehlin, J.O.; Andersen, O.; Kallemose, T. Development and validation of prognostic machine learning models for short- and long-term mortality among acutely admitted patients based on blood tests. Sci. Rep. 2024, 14, 5942. [Google Scholar] [CrossRef]
- Plesner, L.L.; Iversen, A.K.S.; Langkjær, S.; Nielsen, T.L.; Østervig, R.; Warming, P.E.; Salam, I.A.; Kristensen, M.; Schou, M.; Eugen-Olsen, J.; et al. The formation and design of the TRIAGE study—Baseline data on 6005 consecutive patients admitted to hospital from the emergency department. Scand. J. Trauma. Resusc. Emerg. Med. 2015, 23, 106. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD Statement. Eur. Urol. 2015, 67, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Arendt, J.F.H.; Hansen, A.T.; Ladefoged, S.A.; Sørensen, H.T.; Pedersen, L.; Adelborg, K. Existing data sources in clinical epidemiology: Laboratory information system databases in Denmark. Clin. Epidemiol. 2020, 12, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Lemaître, G.; Nogueira, F.; Aridas, C.K. Imbalanced-learn: A python toolbox to tackle the curse of imbalanced datasets in machine learning. J. Mach. Learn. Res. 2017, 18, 1–5. [Google Scholar]
- Yeo, I.; Johnson, R.A. A new family of power transformations to improve normality or symmetry. Biometrika 2000, 87, 954–959. [Google Scholar] [CrossRef]
- Moez, A. PyCaret: An Open Source, Low-Code Machine Learning Library in Python. 2020. Available online: https://www.pycaret.org (accessed on 8 March 2023).
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- Bartol, T. Thoughtful use of diagnostic testing: Making practical sense of sensitivity, specificity, and predictive value. Nurse Pract. 2015, 40, 10–12. [Google Scholar] [CrossRef]
- Galvin, R.; Gilleit, Y.; Wallace, E.; Cousins, G.; Bolmer, M.; Rainer, T.; Smith, S.M.; Fahey, T. Adverse outcomes in older adults attending emergency departments: A systematic review and meta-analysis of the Identification of Seniors At Risk (ISAR) screening tool. Age Ageing 2017, 46, 179–186. [Google Scholar] [CrossRef]
- Trentino, K.M.; Schwarzbauer, K.M.; Mitterecker, A.M.; Hofmann, A.D.R.M.; Lloyd, A.M.; Leahy, M.F.M.; Tschoellitsch, T.; Böck, C.M.; Hochreiter, S.; Meier, J. Machine Learning-Based Mortality Prediction of Patients at Risk During Hospital Admission. J. Patient Saf. 2022, 18, 494–498. [Google Scholar] [CrossRef]
- Xie, F.; Ong, M.E.H.; Liew, J.N.M.H.; Tan, K.B.K.; Ho, A.F.W.; Nadarajan, G.D.; Low, L.L.; Kwan, Y.H.; Goldstein, B.A.; Matchar, D.B.; et al. Development and Assessment of an Interpretable Machine Learning Triage Tool for Estimating Mortality after Emergency Admissions. JAMA Netw. Open 2021, 4, e2118467. [Google Scholar] [CrossRef]
- Yu, J.Y.; Xie, F.; Nan, L.; Yoon, S.; Ong, M.E.H.; Ng, Y.Y.; Cha, W.C. An external validation study of the Score for Emergency Risk Prediction (SERP), an interpretable machine learning-based triage score for the emergency department. Sci. Rep. 2022, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Brajer, N.; Cozzi, B.; Gao, M.; Nichols, M.; Revoir, M.; Balu, S.; Futoma, J.; Bae, J.; Setji, N.; Hernandez, A.; et al. Prospective and External Evaluation of a Machine Learning Model to Predict In-Hospital Mortality of Adults at Time of Admission. JAMA Netw. Open 2020, 3, e1920733. [Google Scholar] [CrossRef] [PubMed]
- Sahni, N.; Simon, G.; Arora, R. Development and Validation of Machine Learning Models for Prediction of 1-Year Mortality Utilizing Electronic Medical Record Data Available at the End of Hospitalization in Multicondition Patients: A Proof-of-Concept Study. J. Gen. Intern. Med. 2018, 33, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, T.B.; Kristensen, K.; Uffelmann, J.; Brandslund, I. Erroneous data: The Achilles’ heel of AI and personalized medicine. Front. Digit. Health 2022, 4, 862095. [Google Scholar] [CrossRef]


| Variable | Retrospective | Prospective | Prospective |
|---|---|---|---|
| Cohort | AHH (29K) | NZH (Triage) | AHH (RESPOND-COVID) |
| Year | 2013–2017 | 2013 | 2020–2022 |
| N of unique patients | 28,671 | 6124 | 8451 |
| N of admissions | 48,841 | 6150 | 10,493 |
| Demographics | |||
| Age | 65.6 (48.2–78.5) | 63.0 (46.0–76.0) | 66.0 (49.1–78.2) |
| Sex (female) 6 | 52.3% | 50.5% | 51.2% |
| Laboratory Biomarkers | |||
| ALAT 1 | 21.0 (15.0–33.0) | 20.8 (14.8–31.5) | 23.0 (16.0–35.0) |
| Albumin 2 | 34.0 (30.0–37.0) | 37.2 (33.5–39.8) | 34.0 (30.0–37.0) |
| Alkaline Phosphatase 1 | 75.6 (63.0–94.0) | 84.2 (69.3–105.9) | 79.0 (63.0–103.0) |
| Bilirubin 3 | 7.0 (5.0–10.1) | 7.9 (5.7–11.2) | 8.0 (5.0–11.0) |
| BUN 4 | 5.1 (3.8–7.2) | 5.2 (4.0–7.1) | 5.3 (3.9–7.7) |
| Creatinine 3 | 77.0 (62.0–97.0) | 71.0 (59.0–88.0) | 77.0 (62.0–98.0) |
| CRP 5 | 7.0 (2.0–39.0) | 5.2 (2.9–23.2) | 12.0 (2.6–54.0) |
| HB 4 | 8.1 (7.2–8.9) | 8.4 (7.6–9.0) | 8.2 (7.3–9.0) |
| INR | 1.0 (1.0–1.1) | 1.0 (0.9–1.1) | 1.0 (1.0–1.1) |
| Potassium 4 | 3.9 (3.6–4.2) | 4.0 (3.8–4.3) | 3.9 (3.6–4.2) |
| KF2710 | 0.9 (0.8–1.0) | 0.9 (0.8–1.1) | 0.8 (0.7–0.9) |
| LDH 1 | 186.0 (169.0–214.0) | 182.6 (157.6–217.3) | 214.0 (184.0–260.0) |
| Leukocytes 6 | 8.7 (6.9–11.3) | 8.2 (6.5–10.6) | 8.7 (6.6–11.8) |
| Lymphocytes 6 | 1.7 (1.1–2.3) | 1.6 (1.2–2.0) | 1.4 (0.9–2.1) |
| Monocytes 6 | 0.7 (0.5–0.9) | 0.6 (0.5–0.8) | 0.7 (0.5–0.9) |
| Neutrophils 6 | 5.8 (4.1–8.3) | 5.7 (4.0–7.9) | 6.0 (4.1–8.9) |
| suPAR (ng/mL) | 3.3 (2.3–5.0) | 4.5 (3.5–6.4) | 4.1 (2.9–5.9) |
| Thrombocytes 6 | 247.0 (201.0–302.0) | 238.0 (196.0–288.0) | 248.0 (196.0–310.0) |
| Eosinophils 6 | 0.1 (0.0–0.2) | 0.2 (0.1–0.4) | 0.1 (0.0–0.2) |
| eGFR (mL/min) | 80.0 (60.0–90.0) | 86.3 (67.6–90.0) | 77.0 (58.0–84.5) |
| Sodium 4 | 139.0 (136.0–141.0) | 139.0 (136.9–140.6) | 138.0 (135.0–140.0) |
| Mortality Rates | |||
| Mortality rate 10 days 6 | 4.4% (1252) | 2.9% (177) | 4.0% (341) |
| Mortality rate 30 days 6 | 8.2% (2338) | 4.6% (284) | 8.4% (712) |
| Mortality rate 90 days 6 | 11.8% (3394) | 7.8% (475) | 12.4% (1052) |
| Mortality rate 1 year 6 | 16.3% (4677) | 11.9% (729) | 18.5% (1560) |
| Test Data | N | AUC | Sensitivity | Specificity | PPV | NPV | MCC |
|---|---|---|---|---|---|---|---|
| 10-day Mortality | |||||||
| 29K | 7.327 (272) | 0.93 (0.92–0.94) | 0.90 (0.86–0.93) | 0.82 (0.81–0.83) | 0.12 (0.11–0.14) | 1.0 (1.0–1.0) | 0.30 (0.28–0.32) |
| RESPOND-COVID | 10.493 (341) | 0.88 (0.86–0.89) | 0.88 (0.84–0.91) | 0.70 (0.69–0.71) | 0.09 (0.08–0.10) | 0.99 (0.99–1.0) | 0.22 (0.22–0.24) |
| TRIAGE | 6.150 (177) | 0.87 (0.85–0.89) | 0.72 (0.65–0.79) | 0.84 (0.84–0.85) | 0.12 (0.10–0.14) | 0.99 (0.99–0.99) | 0.25 (0.22–0.29) |
| 30-day mortality | |||||||
| 29K | 7.327 (537) | 0.92 (0.90–0.92) | 0.89 (0.86–0.91) | 0.83 (0.82–0.83) | 0.23 (0.21–0.24) | 0.99 (0.99–0.99) | 0.40 (0.38–0.42) |
| RESPOND-COVID | 10.493 (712) | 0.88 (0.87–0.89) | 0.89 (0.86–0.91) | 0.68 (0.68–0.69) | 0.18 (0.17–0.19) | 0.99 (0.98–0.98) | 0.32 (0.30–0.33) |
| TRIAGE | 6.150 (284) | 0.88 (0.86–0.90) | 0.76 (0.71–0.81) | 0.84 (0.83–0.85) | 0.18 (0.16–0.21) | 0.99 (0.98–0.99) | 0.34 (0.30–0.37) |
| 90-day Mortality | |||||||
| 29K | 7.327 (982) | 0.91 (0.90–0.92) | 0.84 (0.82–0.86) | 0.85 (0.84–0.86) | 0.38 (0.36–0.40) | 0.98 (0.98–0.98) | 0.51 (0.49–0.53) |
| RESPOND-COVID | 10.493 (1052) | 0.87 (0.86–0.88) | 0.84 (0.82–0.86) | 0.73 (0.72–0.74) | 0.28 (0.26–0.29) | 0.97 (0.97–0.97) | 0.38 (0.36–0.40) |
| TRIAGE | 6.150 (475) | 0.88 (0.86–0.90) | 0.77 (0.73–0.81) | 0.84 (0.83–0.85) | 0.30 (0.27–0.32) | 0.98 (0.97–0.98) | 0.40 (0.37–0.43) |
| 365-day mortality | |||||||
| 29K | 7.327 (1812) | 0.91 (0.91–0.91) | 0.87 (0.86–0.88) | 0.79 (0.79–0.79) | 0.47 (0.46–0.48) | 0.97 (0.96–0.97) | 0.53 (0.51–0.55) |
| RESPOND-COVID | 10.493 (1569) | 0.88 (0.86–0.90) | 0.85 (0.83–0.87) | 0.78 (0.77–0.79) | 0.45 (0.44–0.47) | 0.96 (0.96–0.97) | 0.43 (0.42–0.45) |
| TRIAGE | 6.150 (729) | 0.90 (0.89–0.91) | 0.87 (0.84–0.89) | 0.75 (0.74–0.76) | 0.32 (0.30–0.34) | 0.98 (0.97–0.98) | 0.43 (0.41–0.45) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jawad, B.N.; Altintas, I.; Eugen-Olsen, J.; Niazi, S.; Mansouri, A.; Rasmussen, L.J.H.; Schultz, M.; Iversen, K.; Normann Holm, N.; Kallemose, T.; et al. Prospective and External Validation of Machine Learning Models for Short- and Long-Term Mortality in Acutely Admitted Patients Using Blood Tests. J. Clin. Med. 2024, 13, 6437. https://doi.org/10.3390/jcm13216437
Jawad BN, Altintas I, Eugen-Olsen J, Niazi S, Mansouri A, Rasmussen LJH, Schultz M, Iversen K, Normann Holm N, Kallemose T, et al. Prospective and External Validation of Machine Learning Models for Short- and Long-Term Mortality in Acutely Admitted Patients Using Blood Tests. Journal of Clinical Medicine. 2024; 13(21):6437. https://doi.org/10.3390/jcm13216437
Chicago/Turabian StyleJawad, Baker Nawfal, Izzet Altintas, Jesper Eugen-Olsen, Siar Niazi, Abdullah Mansouri, Line Jee Hartmann Rasmussen, Martin Schultz, Kasper Iversen, Nikolaj Normann Holm, Thomas Kallemose, and et al. 2024. "Prospective and External Validation of Machine Learning Models for Short- and Long-Term Mortality in Acutely Admitted Patients Using Blood Tests" Journal of Clinical Medicine 13, no. 21: 6437. https://doi.org/10.3390/jcm13216437
APA StyleJawad, B. N., Altintas, I., Eugen-Olsen, J., Niazi, S., Mansouri, A., Rasmussen, L. J. H., Schultz, M., Iversen, K., Normann Holm, N., Kallemose, T., Andersen, O., & Nehlin, J. O. (2024). Prospective and External Validation of Machine Learning Models for Short- and Long-Term Mortality in Acutely Admitted Patients Using Blood Tests. Journal of Clinical Medicine, 13(21), 6437. https://doi.org/10.3390/jcm13216437








