Operationalizing Team Science at the Academic Cancer Center Network to Unveil the Structure and Function of the Gut Microbiome
Abstract
:1. Introduction
2. States of the Gut Microbiome in Health and Disease
3. The Impact of Gut Microbiome Dysbiosis on Cancer Predisposition, Treatment Toxicity, and Therapeutic Efficacy
4. Towards a More Complete Accounting of the Gut Microbiome: Genomic and Functional Advances
4.1. The Promise of Metagenomics
4.2. Deciphering the Molecular Metabolome of the Gut Microbiome
4.3. Towards a More Complete Accounting of the Gut Microbiome Molecular Inventory
4.4. Understanding the Microbiome from a Systems Biology Perspective
4.5. Leveraging AI to Discover Gut Microbiome Structure and Function
4.5.1. AI Insights into the Temporal/Spatial Structure of the Gut Microbiome
4.5.2. AI Methods, Models, and Algorithms to Investigate the Function and Operations of the Gut Microbiome
5. Gut Microbiome Therapeutic Intervention
5.1. Genetic Engineering and Synthetic Biology
5.1.1. Genetic Engineering
5.1.2. Synthetic Biology
5.2. Advances in Gut Microbiome PreBiotics, ProBiotics and Fecal Transplantation
6. Pathways to Accelerating Microbiome-Related Discovery and Clinical Translation
6.1. Team Collaborative Science Accelerates Gut Microbiome Research
6.2. The COH Model of Microbiome Collaborative Team Science
6.2.1. The Academic/Community Oncology Alliance at the COH
6.2.2. The COH Clinical Network
6.2.3. COH Resources Available for Microbiome Research
7. TGen, IMC, and QIIME 2
7.1. TGen
7.2. IMC
7.3. QIIME 2
8. The COH CQM
9. COH AAI/DS
10. Microbiome Advances at COH
11. Meeting the Challenges of Gut Microbiome Research at COH
12. Summary and Conclusions
Funding
Conflicts of Interest
References
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Elinav, E.; Garrett, W.S.; Trinchieri, G.; Wargo, J. The cancer microbiome. Nat. Rev. Cancer 2019, 19, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.; Bork, P.; Fraser, C.; Knight, R.; Wang, J. The microbiome explored: Recent insights and future challenges. Nat. Rev. Microbiol. 2013, 11, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Reynoso-García, J.; Miranda-Santiago, A.E.; Meléndez-Vázquez, N.M.; Acosta-Pagán, K.; Sánchez-Rosado, M.; Díaz-Rivera, J.; Rosado-Quiñones, A.M.; Acevedo-Márquez, L.; Cruz-Roldán, L.; Tosado-Rodríguez, E.L. A complete guide to human microbiomes: Body niches, transmission, development, dysbiosis, and restoration. Front. Syst. Biol. 2022, 2, 951403. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Afzaal, M.; Saeed, F.; Shah, Y.A.; Hussain, M.; Rabail, R.; Socol, C.T.; Hassoun, A.; Pateiro, M.; Lorenzo, J.M.; Rusu, A.V. Human gut microbiota in health and disease: Unveiling the relationship. Front. Microbiol. 2022, 13, 999001. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Kordahi, M.C.; Stanaway, I.B.; Avril, M.; Chac, D.; Blanc, M.P.; Ross, B.; Diener, C.; Jain, S.; McCleary, P.; Parker, A.; et al. Genomic and functional characterization of a mucosal symbiont involved in early-stage colorectal cancer. Cell Host Microbe 2021, 29, 1589–1598 e1586. [Google Scholar] [CrossRef]
- Guo, S.; Chen, J.; Chen, F.; Zeng, Q.; Liu, W.L.; Zhang, G. Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut 2021, 70, 1507–1519. [Google Scholar] [CrossRef]
- Aggarwal, N.; Kitano, S.; Puah, G.R.Y.; Kittelmann, S.; Hwang, I.Y.; Chang, M.W. Microbiome and Human Health: Current Understanding, Engineering, and Enabling Technologies. Chem. Rev. 2023, 123, 31–72. [Google Scholar] [CrossRef] [PubMed]
- De Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- McCallum, G.; Tropini, C. The gut microbiota and its biogeography. Nat. Rev. Microbiol. 2024, 22, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Ağagündüz, D.; Cocozza, E.; Cemali, Ö.; Bayazıt, A.D.; Nanì, M.F.; Cerqua, I.; Morgillo, F.; Saygili, S.K.; Berni Canani, R.; Amero, P.; et al. Understanding the role of the gut microbiome in gastrointestinal cancer: A review. Front. Pharmacol. 2023, 14, 1130562. [Google Scholar] [CrossRef]
- Eloe-Fadrosh, E.A.; Rasko, D.A. The human microbiome: From symbiosis to pathogenesis. Annu. Rev. Med. 2013, 64, 145–163. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.Z.; Haque, M. The ecological community of commensal, symbiotic, and pathogenic gastrointestinal microorganisms—An appraisal. Clin. Exp. Gastroenterol. 2017, 10, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Mohajeri, M.H.; Brummer, R.J.M.; Rastall, R.A.; Weersma, R.K.; Harmsen, H.J.M.; Faas, M.; Eggersdorfer, M. The role of the microbiome for human health: From basic science to clinical applications. Eur. J. Nutr. 2018, 57, 1–14. [Google Scholar] [CrossRef]
- Murphy, K.; O’Donovan, A.N.; Caplice, N.M.; Ross, R.P.; Stanton, C. Exploring the Gut Microbiota and Cardiovascular Disease. Metabolites 2021, 11, 493. [Google Scholar] [CrossRef] [PubMed]
- Hitch, T.C.A.; Hall, L.J.; Walsh, S.K.; Leventhal, G.E.; Slack, E.; de Wouters, T.; Walter, J.; Clavel, T. Microbiome-based interventions to modulate gut ecology and the immune system. Mucosal Immunol. 2022, 15, 1095–1113. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Kumar, M.; Singh, P.; Murugesan, S.; Vetizou, M.; McCulloch, J.; Badger, J.H.; Trinchieri, G.; Al Khodor, S. Microbiome as an Immunological Modifier. Methods Mol. Biol. 2020, 2055, 595–638. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef]
- Barker-Tejeda, T.C.; Zubeldia-Varela, E.; Macías-Camero, A.; Alonso, L.; Martín-Antoniano, I.A.; Rey-Stolle, M.F.; Mera-Berriatua, L.; Bazire, R.; Cabrera-Freitag, P.; Shanmuganathan, M.; et al. Comparative characterization of the infant gut microbiome and their maternal lineage by a multi-omics approach. Nat. Commun. 2024, 15, 3004. [Google Scholar] [CrossRef]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe 2018, 24, 133–145.e5. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Praveen, P.; Jordan, F.; Priami, C.; Morine, M.J. The role of breast-feeding in infant immune system: A systems perspective on the intestinal microbiome. Microbiome 2015, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Catassi, G.; Aloi, M.; Giorgio, V.; Gasbarrini, A.; Cammarota, G.; Ianiro, G. The Role of Diet and Nutritional Interventions for the Infant Gut Microbiome. Nutrients 2024, 16, 400. [Google Scholar] [CrossRef] [PubMed]
- Laursen, M.F. Gut Microbiota Development: Influence of Diet from Infancy to Toddlerhood. Ann. Nutr. Metab. 2021, 77, 21–34. [Google Scholar] [CrossRef]
- Patangia, D.V.; Anthony Ryan, C.; Dempsey, E.; Paul Ross, R.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. Microbiologyopen 2022, 11, e1260. [Google Scholar] [CrossRef]
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef] [PubMed]
- Mullish, B.H.; Williams, H.R. Clostridium difficile infection and antibiotic-associated diarrhoea. Clin. Med. 2018, 18, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Young, V.B. Interactions Between the Gastrointestinal Microbiome and Clostridium difficile. Annu. Rev. Microbiol. 2015, 69, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Seekatz, A.M.; Young, V.B. Clostridium difficile and the microbiota. J. Clin. Investig. 2014, 124, 4182–4189. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.; Taminiau, B.; Rodriguez, C.; Daube, G. Gut Microbiota Composition Associated with Clostridioides difficile Colonization and Infection. Pathogens 2022, 11, 781. [Google Scholar] [CrossRef]
- Brandt, L.J.; Borody, T.J.; Campbell, J. Endoscopic fecal microbiota transplantation: “first-line” treatment for severe clostridium difficile infection? J. Clin. Gastroenterol. 2011, 45, 655–657. [Google Scholar] [CrossRef]
- Valdés-Varela, L.; Gueimonde, M.; Ruas-Madiedo, P. Probiotics for Prevention and Treatment of Clostridium difficile Infection. In Updates on Clostridioides Difficile in Europe: Advances in Microbiology, Infectious Diseases and Public Health Volume 18; Springer: Cham, Switzerland, 2024; pp. 101–116. [Google Scholar]
- Sadrekarimi, H.; Gardanova, Z.R.; Bakhshesh, M.; Ebrahimzadeh, F.; Yaseri, A.F.; Thangavelu, L.; Hasanpoor, Z.; Zadeh, F.A.; Kahrizi, M.S. Emerging role of human microbiome in cancer development and response to therapy: Special focus on intestinal microflora. J. Transl. Med. 2022, 20, 301. [Google Scholar] [CrossRef]
- Maddern, A.S.; Coller, J.K.; Bowen, J.M.; Gibson, R.J. The Association between the Gut Microbiome and Development and Progression of Cancer Treatment Adverse Effects. Cancers 2023, 15, 4301. [Google Scholar] [CrossRef]
- Kunika; Frey, N.; Rangrez, A.Y. Exploring the Involvement of Gut Microbiota in Cancer Therapy-Induced Cardiotoxicity. Int. J. Mol. Sci. 2023, 24, 7261. [Google Scholar] [CrossRef]
- Sun, J.; Chen, F.; Wu, G. Potential effects of gut microbiota on host cancers: Focus on immunity, DNA damage, cellular pathways, and anticancer therapy. ISME J. 2023, 17, 1535–1551. [Google Scholar] [CrossRef]
- Ma, W.; Mao, Q.; Xia, W.; Dong, G.; Yu, C.; Jiang, F. Gut Microbiota Shapes the Efficiency of Cancer Therapy. Front. Microbiol. 2019, 10, 1050. [Google Scholar] [CrossRef] [PubMed]
- Al Bander, Z.; Nitert, M.D.; Mousa, A.; Naderpoor, N. The Gut Microbiota and Inflammation: An Overview. Int. J. Environ. Res. Public Health 2020, 17, 7618. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.; Sears, C.L. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: Contributions to colorectal cancer development. Genome Med. 2019, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstern, C.R.; Lamichhane-Khadka, R. A tale of two bacteria–Bacteroides fragilis, Escherichia coli, and colorectal cancer. Front. Bacteriol. 2023, 2, 1229077. [Google Scholar] [CrossRef]
- Ou, S.; Wang, H.; Tao, Y.; Luo, K.; Ye, J.; Ran, S.; Guan, Z.; Wang, Y.; Hu, H.; Huang, R. Fusobacterium nucleatum and colorectal cancer: From phenomenon to mechanism. Front. Cell Infect. Microbiol. 2022, 12, 1020583. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Dubinsky, V.; Dotan, I.; Gophna, U. Carriage of Colibactin-producing Bacteria and Colorectal Cancer Risk. Trends Microbiol. 2020, 28, 874–876. [Google Scholar] [CrossRef]
- Arthur, J.C. Microbiota and colorectal cancer: Colibactin makes its mark. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 317–318. [Google Scholar] [CrossRef]
- Lucafò, M.; Curci, D.; Franzin, M.; Decorti, G.; Stocco, G. Inflammatory Bowel Disease and Risk of Colorectal Cancer: An Overview From Pathophysiology to Pharmacological Prevention. Front. Pharmacol. 2021, 12, 772101. [Google Scholar] [CrossRef]
- Terlouw, D.; Boot, A.; Ducarmon, Q.R.; Nooij, S.; Suerink, M.; van Leerdam, M.E.; van Egmond, D.; Tops, C.M.; Zwittink, R.D.; Ruano, D. Enrichment of colibactin-associated mutational signatures in unexplained colorectal polyposis patients. BMC Cancer 2024, 24, 104. [Google Scholar] [CrossRef]
- White, M.T.; Sears, C.L. The microbial landscape of colorectal cancer. Nat. Rev. Microbiol. 2024, 22, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.; Parida, S.; Lingipilli, B.T.; Krishnan, R.; Podipireddy, D.R.; Muniraj, N. Role of Gut Microbiota in Breast Cancer and Drug Resistance. Pathogens 2023, 12, 468. [Google Scholar] [CrossRef]
- Arnone, A.A.; Cook, K.L. Gut and Breast Microbiota as Endocrine Regulators of Hormone Receptor-positive Breast Cancer Risk and Therapy Response. Endocrinology 2022, 164, bqac177. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, G.; Le Noci, V.; Di Modica, M.; Montanari, E.; Triulzi, T.; Pupa, S.M.; Tagliabue, E.; Sommariva, M.; Sfondrini, L. The Emerging Role of the Microbiota in Breast Cancer Progression. Cells 2023, 12, 1945. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Matsushita, M.; Banno, E.; De Velasco, M.A.; Hatano, K.; Nonomura, N.; Uemura, H. Gut microbiome and prostate cancer. Int. J. Urol. 2022, 29, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Matsushita, M.; De Velasco, M.A.; Hatano, K.; Minami, T.; Nonomura, N.; Uemura, H. The Gut-Prostate Axis: A New Perspective of Prostate Cancer Biology through the Gut Microbiome. Cancers 2023, 15, 1375. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, Y.; Li, S.; Peng, Z.; Liu, X.; Chen, J.; Zheng, X. Role of lung and gut microbiota on lung cancer pathogenesis. J. Cancer Res. Clin. Oncol. 2021, 147, 2177–2186. [Google Scholar] [CrossRef]
- Du, Y.; Wang, Q.; Zheng, Z.; Zhou, H.; Han, Y.; Qi, A.; Jiao, L.; Gong, Y. Gut microbiota influence on lung cancer risk through blood metabolite mediation: From a comprehensive Mendelian randomization analysis and genetic analysis. Front. Nutr. 2024, 11, 1425802. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, Y.; Zang, D.; Zhang, M.; Li, X.; Liu, D.; Gao, B.; Zhou, H.; Sun, J.; Han, X.; et al. The Role of Gut Microbiota in Lung Cancer: From Carcinogenesis to Immunotherapy. Front. Oncol. 2021, 11, 720842. [Google Scholar] [CrossRef]
- Oh, B.; Boyle, F.; Pavlakis, N.; Clarke, S.; Guminski, A.; Eade, T.; Lamoury, G.; Carroll, S.; Morgia, M.; Kneebone, A.; et al. Emerging Evidence of the Gut Microbiome in Chemotherapy: A Clinical Review. Front. Oncol. 2021, 11, 706331. [Google Scholar] [CrossRef]
- Ervin, S.M.; Ramanan, S.V.; Bhatt, A.P. Relationship Between the Gut Microbiome and Systemic Chemotherapy. Dig. Dis. Sci. 2020, 65, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Bilenduke, E.; Sterrett, J.D.; Ranby, K.W.; Borges, V.F.; Grigsby, J.; Carr, A.L.; Kilbourn, K.; Lowry, C.A. Impacts of breast cancer and chemotherapy on gut microbiome, cognitive functioning, and mood relative to healthy controls. Sci. Rep. 2022, 12, 19547. [Google Scholar] [CrossRef]
- Chrysostomou, D.; Roberts, L.A.; Marchesi, J.R.; Kinross, J.M. Gut microbiota modulation of efficacy and toxicity of cancer chemotherapy and immunotherapy. Gastroenterology 2023, 164, 198–213. [Google Scholar] [CrossRef]
- Li, S.; Zhu, S.; Yu, J. The role of gut microbiota and metabolites in cancer chemotherapy. J. Adv. Res. 2024, 64, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Fei, Z.; Lijuan, Y.; Xi, Y.; Wei, W.; Jing, Z.; Miao, D.; Shuwen, H. Gut microbiome associated with chemotherapy-induced diarrhea from the CapeOX regimen as adjuvant chemotherapy in resected stage III colorectal cancer. Gut Pathog. 2019, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Roggiani, S.; Mengoli, M.; Conti, G.; Fabbrini, M.; Brigidi, P.; Barone, M.; D’Amico, F.; Turroni, S. Gut microbiota resilience and recovery after anticancer chemotherapy. Microbiome Res. Rep. 2023, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Touchefeu, Y.; Montassier, E.; Nieman, K.; Gastinne, T.; Potel, G.; Bruley des Varannes, S.; Le Vacon, F.; de La Cochetière, M.F. Systematic review: The role of the gut microbiota in chemotherapy- or radiation-induced gastrointestinal mucositis—Current evidence and potential clinical applications. Aliment. Pharmacol. Ther. 2014, 40, 409–421. [Google Scholar] [CrossRef]
- Wei, L.; Wen, X.S.; Xian, C.J. Chemotherapy-Induced Intestinal Microbiota Dysbiosis Impairs Mucosal Homeostasis by Modulating Toll-like Receptor Signaling Pathways. Int. J. Mol. Sci. 2021, 22, 9474. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, H.; Zhu, R.; Shang, G.; Wei, J.; Shang, H.; Tian, P.; Chen, T.; Wei, H. Combination of thalidomide and Clostridium butyricum relieves chemotherapy-induced nausea and vomiting via gut microbiota and vagus nerve activity modulation. Front. Immunol. 2023, 14, 1220165. [Google Scholar] [CrossRef]
- Deleemans, J.M.; Chleilat, F.; Reimer, R.A.; Baydoun, M.; Piedalue, K.A.; Lowry, D.E.; Henning, J.W.; Carlson, L.E. The Chemo-Gut Pilot Study: Associations between Gut Microbiota, Gastrointestinal Symptoms, and Psychosocial Health Outcomes in a Cross-Sectional Sample of Young Adult Cancer Survivors. Curr. Oncol. 2022, 29, 2973–2994. [Google Scholar] [CrossRef]
- Montassier, E.; Gastinne, T.; Vangay, P.; Al-Ghalith, G.; Bruley des Varannes, S.; Massart, S.; Moreau, P.; Potel, G.; de La Cochetière, M.; Batard, E. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment. Pharmacol. Ther. 2015, 42, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Chamseddine, A.N.; Ducreux, M.; Armand, J.P.; Paoletti, X.; Satar, T.; Paci, A.; Mir, O. Intestinal bacterial β-glucuronidase as a possible predictive biomarker of irinotecan-induced diarrhea severity. Pharmacol. Ther. 2019, 199, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Roberts, A.B.; Pollet, R.M.; Ingle, J.D.; Biernat, K.A.; Pellock, S.J.; Venkatesh, M.K.; Guthrie, L.; O’Neal, S.K.; Robinson, S.J.; et al. Structure and Inhibition of Microbiome β-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem. Biol. 2015, 22, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Hu, B.; Li, P.; Zhao, Y.; Xu, Y.; Wu, D. Roles of the intestinal microbiota and microbial metabolites in acute GVHD. Exp. Hematol. Oncol. 2021, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Häcker, G. GVHD prediction based on the microbiome. Blood 2022, 140, 2313–2314. [Google Scholar] [CrossRef] [PubMed]
- Fredricks, D.N. The gut microbiota and graft-versus-host disease. J. Clin. Investig. 2019, 129, 1808–1817. [Google Scholar] [CrossRef]
- Burgos da Silva, M.; Ponce, D.M.; Dai, A.; Devlin, S.M.; Gomes, A.L.C.; Moore, G.; Slingerland, J.; Shouval, R.; Armijo, G.K.; DeWolf, S.; et al. Preservation of the fecal microbiome is associated with reduced severity of graft-versus-host disease. Blood 2022, 140, 2385–2397. [Google Scholar] [CrossRef] [PubMed]
- Knisely, A.; Seo, Y.D.; Wargo, J.A.; Chelvanambi, M. Monitoring and Modulating Diet and Gut Microbes to Enhance Response and Reduce Toxicity to Cancer Treatment. Cancers 2023, 15, 777. [Google Scholar] [CrossRef]
- Kouidhi, S.; Zidi, O.; Belkhiria, Z.; Rais, H.; Ayadi, A.; Ben Ayed, F.; Mosbah, A.; Cherif, A.; El Gaaied, A.B.A. Gut microbiota, an emergent target to shape the efficiency of cancer therapy. Explor. Target. Antitumor Ther. 2023, 4, 240–265. [Google Scholar] [CrossRef]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef]
- Gori, S.; Inno, A.; Belluomini, L.; Bocus, P.; Bisoffi, Z.; Russo, A.; Arcaro, G. Gut microbiota and cancer: How gut microbiota modulates activity, efficacy and toxicity of antitumoral therapy. Crit. Rev. Oncol. Hematol. 2019, 143, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Deng, Y.; Chu, Q.; Zhang, P. Gut microbiome and cancer immunotherapy. Cancer Lett. 2019, 447, 41–47. [Google Scholar] [CrossRef]
- Pouncey, A.L.; Scott, A.J.; Alexander, J.L.; Marchesi, J.; Kinross, J. Gut microbiota, chemotherapy and the host: The influence of the gut microbiota on cancer treatment. Ecancermedicalscience 2018, 12, 868. [Google Scholar] [CrossRef]
- Yan, A.; Culp, E.; Perry, J.; Lau, J.T.; MacNeil, L.T.; Surette, M.G.; Wright, G.D. Transformation of the Anticancer Drug Doxorubicin in the Human Gut Microbiome. ACS Infect. Dis. 2018, 4, 68–76. [Google Scholar] [CrossRef]
- Gonçalves-Nobre, J.G.; Gaspar, I.; Alpuim Costa, D. Anthracyclines and trastuzumab associated cardiotoxicity: Is the gut microbiota a friend or foe?–a mini-review. Front. Microbiomes 2023, 2, 1217820. [Google Scholar] [CrossRef]
- Bawaneh, A.; Wilson, A.S.; Levi, N.; Howard-McNatt, M.M.; Chiba, A.; Soto-Pantoja, D.R.; Cook, K.L. Intestinal microbiota influence doxorubicin responsiveness in triple-negative breast cancer. Cancers 2022, 14, 4849. [Google Scholar] [CrossRef] [PubMed]
- Westman, E.L.; Canova, M.J.; Radhi, I.J.; Koteva, K.; Kireeva, I.; Waglechner, N.; Wright, G.D. Bacterial Inactivation of the Anticancer Drug Doxorubicin. Chem. Biol. 2012, 19, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Le Ngoc, K.; Pham, T.T.H.; Nguyen, T.K.; Huong, P.T. Pharmacomicrobiomics in precision cancer therapy: Bench to bedside. Front. Immunol. 2024, 15, 1428420. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, Y.; Huang, W.; Zhou, H.; Zhang, W. Drug-microbiota interactions: An emerging priority for precision medicine. Signal Transduct. Target. Ther. 2023, 8, 386. [Google Scholar] [CrossRef]
- Di Modica, M.; Gargari, G.; Regondi, V.; Bonizzi, A.; Arioli, S.; Belmonte, B.; De Cecco, L.; Fasano, E.; Bianchi, F.; Bertolotti, A. Gut microbiota condition the therapeutic efficacy of trastuzumab in HER2-positive breast cancer. Cancer Res. 2021, 81, 2195–2206. [Google Scholar] [CrossRef]
- Martini, G.; Ciardiello, D.; Dallio, M.; Famiglietti, V.; Esposito, L.; Corte, C.M.D.; Napolitano, S.; Fasano, M.; Gravina, A.G.; Romano, M.; et al. Gut microbiota correlates with antitumor activity in patients with mCRC and NSCLC treated with cetuximab plus avelumab. Int. J. Cancer 2022, 151, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Ryu, T.Y.; Kim, K.; Han, T.-S.; Lee, M.-O.; Lee, J.; Choi, J.; Jung, K.B.; Jeong, E.-J.; An, D.M.; Jung, C.-R.; et al. Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. ISME J. 2022, 16, 1205–1221. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Lau, H.C.-H.; Yu, J. Modulating gut microbiome in cancer immunotherapy: Harnessing microbes to enhance treatment efficacy. Cell Rep. Med. 2024, 5, 101478. [Google Scholar] [CrossRef] [PubMed]
- Aghamajidi, A.; Maleki Vareki, S. The Effect of the Gut Microbiota on Systemic and Anti-Tumor Immunity and Response to Systemic Therapy against Cancer. Cancers 2022, 14, 3563. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Zhu, X.; Wang, Y.; Lu, S. The gut microbiota improves the efficacy of immune-checkpoint inhibitor immunotherapy against tumors: From association to cause and effect. Cancer Lett. 2024, 598, 217123. [Google Scholar] [CrossRef]
- Lu, Y.; Yuan, X.; Wang, M.; He, Z.; Li, H.; Wang, J.; Li, Q. Gut microbiota influence immunotherapy responses: Mechanisms and therapeutic strategies. J. Hematol. Oncol. 2022, 15, 47. [Google Scholar] [CrossRef]
- Yousefi, Y.; Baines, K.J.; Maleki Vareki, S. Microbiome bacterial influencers of host immunity and response to immunotherapy. Cell Rep. Med. 2024, 5, 101487. [Google Scholar] [CrossRef]
- Pezo, R.C.; Wong, M.; Martin, A. Impact of the gut microbiota on immune checkpoint inhibitor-associated toxicities. Ther. Adv. Gastroenterol. 2019, 12, 1756284819870911. [Google Scholar] [CrossRef]
- Asokan, S.; Cullin, N.; Stein-Thoeringer, C.K.; Elinav, E. CAR-T Cell Therapy and the Gut Microbiota. Cancers 2023, 15, 794. [Google Scholar] [CrossRef]
- Smith, M.; Dai, A.; Ghilardi, G.; Amelsberg, K.V.; Devlin, S.M.; Pajarillo, R.; Slingerland, J.B.; Beghi, S.; Herrera, P.S.; Giardina, P. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 2022, 28, 713–723. [Google Scholar] [CrossRef]
- Gabrielli, G.; Shouval, R.; Ghilardi, G.; van den Brink, M.; Ruella, M. Harnessing the Gut Microbiota to Potentiate the Efficacy of CAR T Cell Therapy. Hemasphere 2023, 7, e950. [Google Scholar] [CrossRef]
- eBioMedicine. Emerging role of microbes in cancer development and anti-cancer therapy. eBioMedicine 2024, 101. [Google Scholar] [CrossRef]
- Hirayama, M.; Nishiwaki, H.; Hamaguchi, T.; Ito, M.; Ueyama, J.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ohno, K. Intestinal Collinsella may mitigate infection and exacerbation of COVID-19 by producing ursodeoxycholate. PLoS ONE 2021, 16, e0260451. [Google Scholar] [CrossRef]
- Hu, Y.; Li, J.; Ni, F.; Yang, Z.; Gui, X.; Bao, Z.; Zhao, H.; Wei, G.; Wang, Y.; Zhang, M.; et al. CAR-T cell therapy-related cytokine release syndrome and therapeutic response is modulated by the gut microbiome in hematologic malignancies. Nat. Commun. 2022, 13, 5313. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Smidt, H.; de Vos, W.M.; Belzer, C. The function of our microbiota: Who is out there and what do they do? Front. Cell. Infect. Microbiol. 2012, 2, 104. [Google Scholar] [CrossRef]
- Blaut, M. Composition and Function of the Gut Microbiome. In The Gut Microbiome in Health and Disease, Haller, D., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 5–30. [Google Scholar] [CrossRef]
- Ezzamouri, B.; Shoaie, S.; Ledesma-Amaro, R. Synergies of Systems Biology and Synthetic Biology in Human Microbiome Studies. Front. Microbiol. 2021, 12, 681982. [Google Scholar] [CrossRef] [PubMed]
- Altay, O.; Nielsen, J.; Uhlen, M.; Boren, J.; Mardinoglu, A. Systems biology perspective for studying the gut microbiota in human physiology and liver diseases. EBioMedicine 2019, 49, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Hernández Medina, R.; Kutuzova, S.; Nielsen, K.N.; Johansen, J.; Hansen, L.H.; Nielsen, M.; Rasmussen, S. Machine learning and deep learning applications in microbiome research. ISME Commun. 2022, 2, 98. [Google Scholar] [CrossRef]
- Sun, T.; Niu, X.; He, Q.; Chen, F.; Qi, R.-Q. Artificial Intelligence in microbiomes analysis: A review of applications in dermatology. Front. Microbiol. 2023, 14, 1112010. [Google Scholar] [CrossRef]
- Wang, W.L.; Xu, S.Y.; Ren, Z.G.; Tao, L.; Jiang, J.W.; Zheng, S.S. Application of metagenomics in the human gut microbiome. World J. Gastroenterol. 2015, 21, 803–814. [Google Scholar] [CrossRef]
- Martín, R.; Miquel, S.; Langella, P.; Bermúdez-Humarán, L.G. The role of metagenomics in understanding the human microbiome in health and disease. Virulence 2014, 5, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, F.; Zeng, Z.; Xu, M.; Sun, F.; Yang, L.; Bi, X.; Lin, Y.; Gao, Y.; Hao, H.; et al. Advances in Metagenomics and Its Application in Environmental Microorganisms. Front. Microbiol. 2021, 12, 766364. [Google Scholar] [CrossRef]
- Kim, N.; Ma, J.; Kim, W.; Kim, J.; Belenky, P.; Lee, I. Genome-resolved metagenomics: A game changer for microbiome medicine. Exp. Mol. Med. 2024, 56, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Yen, S.; Johnson, J.S. Metagenomics: A path to understanding the gut microbiome. Mamm. Genome 2021, 32, 282–296. [Google Scholar] [CrossRef]
- Zhang, W.; An, Y.; Qin, X.; Wu, X.; Wang, X.; Hou, H.; Song, X.; Liu, T.; Wang, B.; Huang, X.; et al. Gut Microbiota-Derived Metabolites in Colorectal Cancer: The Bad and the Challenges. Front. Oncol. 2021, 11, 739648. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.T.; Mao, Y.Q. The impact of the microbiome in cancer: Targeting metabolism of cancer cells and host. Front. Oncol. 2022, 12, 1029033. [Google Scholar] [CrossRef]
- Coker, O.O.; Liu, C.; Wu, W.K.K.; Wong, S.H.; Jia, W.; Sung, J.J.Y.; Yu, J. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome 2022, 10, 35. [Google Scholar] [CrossRef]
- He, Y.; Fu, L.; Li, Y.; Wang, W.; Gong, M.; Zhang, J.; Dong, X.; Huang, J.; Wang, Q.; Mackay, C.R.; et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metab. 2021, 33, 988–1000.e1007. [Google Scholar] [CrossRef]
- Bauermeister, A.; Mannochio-Russo, H.; Costa-Lotufo, L.V.; Jarmusch, A.K.; Dorrestein, P.C. Mass spectrometry-based metabolomics in microbiome investigations. Nat. Rev. Microbiol. 2022, 20, 143–160. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, D.; Zheng, S.; Ouyang, R.; Wang, Y.; Xu, G. Gut microbiota-related metabolome analysis based on chromatography-mass spectrometry. TrAC Trends Anal. Chem. 2021, 143, 116375. [Google Scholar] [CrossRef]
- Han, S.; Guiberson, E.R.; Li, Y.; Sonnenburg, J.L. High-throughput identification of gut microbiome-dependent metabolites. Nat. Protoc. 2024, 19, 2180–2205. [Google Scholar] [CrossRef]
- Awad, H.; Khamis, M.M.; El-Aneed, A. Mass spectrometry, review of the basics: Ionization. Appl. Spectrosc. Rev. 2015, 50, 158–175. [Google Scholar] [CrossRef]
- El-Aneed, A.; Cohen, A.; Banoub, J. Mass spectrometry, review of the basics: Electrospray, MALDI, and commonly used mass analyzers. Appl. Spectrosc. Rev. 2009, 44, 210–230. [Google Scholar] [CrossRef]
- Fang, A.S.; Miao, X.; Tidswell, P.W.; Towle, M.H.; Goetzinger, W.K.; Kyranos, J.N. Mass spectrometry analysis of new chemical entities for pharmaceutical discovery. Mass. Spectrom. Rev. 2008, 27, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.A.; Jones, O.A.; Beale, D.J.; Boughton, B.A.; Benheim, D.; Kouremenos, K.A.; Wolfender, J.-L.; Wishart, D.S. Current and future perspectives on the structural identification of small molecules in biological systems. Metabolites 2016, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.C.; Ridgeway, M.E.; Park, M.A.; Bleiholder, C. Tandem-trapped ion mobility spectrometry/mass spectrometry (t TIMS/MS): A promising analytical method for investigating heterogenous samples. Analyst 2022, 147, 2317–2337. [Google Scholar] [CrossRef]
- Zhao, Z.-X.; Wang, H.-Y.; Guo, Y.-L. Studies of heterogeneous/homogeneous ion-molecule reactions by ambient ionization mass spectrometry. Curr. Org. Chem. 2011, 15, 3734–3749. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Mas, S.; Åkesson, M.; Smedsgaard, J.; Nielsen, J. Mass spectrometry in metabolome analysis. Mass. Spectrom. Rev. 2005, 24, 613–646. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass. Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Magnúsdóttir, S.; Thiele, I. Modeling metabolism of the human gut microbiome. Curr. Opin. Biotechnol. 2018, 51, 90–96. [Google Scholar] [CrossRef]
- Sharon, G.; Garg, N.; Debelius, J.; Knight, R.; Dorrestein, P.C.; Mazmanian, S.K. Specialized metabolites from the microbiome in health and disease. Cell Metab. 2014, 20, 719–730. [Google Scholar] [CrossRef]
- Tajik, M.; Baharfar, M.; Donald, W.A. Single-cell mass spectrometry. Trends Biotechnol. 2022, 40, 1374–1392. [Google Scholar] [CrossRef]
- Zhang, L.; Vertes, A. Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angew. Chem. Int. Ed. 2018, 57, 4466–4477. [Google Scholar] [CrossRef] [PubMed]
- Slavov, N. Single-cell protein analysis by mass spectrometry. Curr. Opin. Chem. Biol. 2021, 60, 1–9. [Google Scholar] [CrossRef]
- Taylor, M.J.; Lukowski, J.K.; Anderton, C.R. Spatially resolved mass spectrometry at the single cell: Recent innovations in proteomics and metabolomics. J. Am. Soc. Mass. Spectrom. 2021, 32, 872–894. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Leng, Y.; Li, X.; Meng, Y.; Yin, Z.; Hang, W. High spatial resolution mass spectrometry imaging for spatial metabolomics: Advances, challenges, and future perspectives. TrAC Trends Anal. Chem. 2023, 159, 116902. [Google Scholar] [CrossRef]
- Matros, A.; Mock, H.-P. Mass spectrometry based imaging techniques for spatially resolved analysis of molecules. Front. Plant Sci. 2013, 4, 89. [Google Scholar] [CrossRef] [PubMed]
- Aretz, I.; Meierhofer, D. Advantages and pitfalls of mass spectrometry based metabolome profiling in systems biology. Int. J. Mol. Sci. 2016, 17, 632. [Google Scholar] [CrossRef]
- Theodoridis, G.; Gika, H.G.; Wilson, I.D. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass. Spectrom. Rev. 2011, 30, 884–906. [Google Scholar] [CrossRef]
- O’Reilly, F.J.; Rappsilber, J. Cross-linking mass spectrometry: Methods and applications in structural, molecular and systems biology. Nat. Struct. Mol. Biol. 2018, 25, 1000–1008. [Google Scholar] [CrossRef]
- Milman, B.L. General principles of identification by mass spectrometry. TrAC Trends Anal. Chem. 2015, 69, 24–33. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Wang, R.; Hermjakob, H.; Müller, M.; Vesada, V.; Vizcaíno, J.A. Open source libraries and frameworks for mass spectrometry based proteomics: A developer’s perspective. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2014, 1844, 63–76. [Google Scholar] [CrossRef]
- Knock, B.; Smith, I.; Wright, D.; Ridley, R.; Kelly, W. Compound identification by computer matching of low resolution mass spectra. Anal. Chem. 1970, 42, 1516–1520. [Google Scholar] [CrossRef]
- McLafferty, F.W.; Zhang, M.-Y.; Stauffer, D.B.; Loh, S.Y. Comparison of algorithms and databases for matching unknown mass spectra. J. Am. Soc. Mass. Spectrom. 1998, 9, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Hall, Z.; Politis, A.; Robinson, C.V. Structural modeling of heteromeric protein complexes from disassembly pathways and ion mobility-mass spectrometry. Structure 2012, 20, 1596–1609. [Google Scholar] [CrossRef]
- Biehn, S.E.; Lindert, S. Protein structure prediction with mass spectrometry data. Annu. Rev. Phys. Chem. 2022, 73, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Craig, R.; Cortens, J.; Fenyo, D.; Beavis, R.C. Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res. 2006, 5, 1843–1849. [Google Scholar] [CrossRef]
- Frewen, B.E.; Merrihew, G.E.; Wu, C.C.; Noble, W.S.; MacCoss, M.J. Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 2006, 78, 5678–5684. [Google Scholar] [CrossRef]
- Youngquist, R.S.; Fuentes, G.R.; Lacey, M.P.; Keough, T. Generation and screening of combinatorial peptide libraries designed for rapid sequencing by mass spectrometry. J. Am. Chem. Soc. 1995, 117, 3900–3906. [Google Scholar] [CrossRef]
- Parizadeh, M.; Arrieta, M.-C. The global human gut microbiome: Genes, lifestyles, and diet. Trends Mol. Med. 2023, 29, 789–801. [Google Scholar] [CrossRef]
- Martinez, J.E.; Kahana, D.D.; Ghuman, S.; Wilson, H.P.; Wilson, J.; Kim, S.C.; Lagishetty, V.; Jacobs, J.P.; Sinha-Hikim, A.P.; Friedman, T.C. Unhealthy lifestyle and gut dysbiosis: A better understanding of the effects of poor diet and nicotine on the intestinal microbiome. Front. Endocrinol. 2021, 12, 667066. [Google Scholar] [CrossRef] [PubMed]
- Do, N.M. From Leaky Gut to Leaky Skin: A Clinical Review of Lifestyle Influences on the Microbiome. Am. J. Lifestyle Med. 2024, 15598276241292605. [Google Scholar] [CrossRef]
- Lin, L.; Zhang, J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017, 18, 1–25. [Google Scholar] [CrossRef]
- Li, Z.; Quan, G.; Jiang, X.; Yang, Y.; Ding, X.; Zhang, D.; Wang, X.; Hardwidge, P.R.; Ren, W.; Zhu, G. Effects of metabolites derived from gut microbiota and hosts on pathogens. Front. Cell. Infect. Microbiol. 2018, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Zechner, E.L.; Kienesberger, S. Microbiota-derived small molecule genotoxins: Host interactions and ecological impact in the gut ecosystem. Gut Microbes 2024, 16, 2430423. [Google Scholar] [CrossRef]
- Hartl, K.; Sigal, M. Microbe-driven genotoxicity in gastrointestinal carcinogenesis. Int. J. Mol. Sci. 2020, 21, 7439. [Google Scholar] [CrossRef] [PubMed]
- Healy, A.R.; Herzon, S.B. Molecular basis of gut microbiome-associated colorectal cancer: A synthetic perspective. J. Am. Chem. Soc. 2017, 139, 14817–14824. [Google Scholar] [CrossRef]
- De Carvalho, C.C.; Caramujo, M.J. The various roles of fatty acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef]
- Das, U.N. Essential fatty acids-a review. Curr. Pharm. Biotechnol. 2006, 7, 467–482. [Google Scholar] [CrossRef]
- Sellin. Short chain fatty acids in health and disease. Aliment. Pharmacol. Ther. 1998, 12, 499–507. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [PubMed]
- Scheppach, W.; Bartram, H.; Richter, F. Role of short-chain fatty acids in the prevention of colorectal cancer. Eur. J. Cancer 1995, 31, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Afaghi, A.; Babakhani, S.; Sohrabi, M.R.; Hosseini-Fard, S.R.; Babolhavaeji, K.; Akbari, S.K.A.; Yousefimashouf, R.; Karampoor, S. Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed. Pharmacother. 2021, 139, 111619. [Google Scholar] [CrossRef]
- Feitelson, M.A.; Arzumanyan, A.; Medhat, A.; Spector, I. Short-chain fatty acids in cancer pathogenesis. Cancer Metastasis Rev. 2023, 42, 677–698. [Google Scholar] [CrossRef] [PubMed]
- Fusco, W.; Lorenzo, M.B.; Cintoni, M.; Porcari, S.; Rinninella, E.; Kaitsas, F.; Lener, E.; Mele, M.C.; Gasbarrini, A.; Collado, M.C.; et al. Short-Chain Fatty-Acid-Producing Bacteria: Key Components of the Human Gut Microbiota. Nutrients 2023, 15, 2211. [Google Scholar] [CrossRef] [PubMed]
- Deleu, S.; Machiels, K.; Raes, J.; Verbeke, K.; Vermeire, S. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? eBioMedicine 2021, 66, 103293. [Google Scholar] [CrossRef]
- Zeb, F.; Naqeeb, H.; Osaili, T.; Faris, M.E.; Ismail, L.C.; Obaid, R.S.; Naja, F.; Radwan, H.; Hasan, H.; Hashim, M.; et al. Molecular crosstalk between polyphenols and gut microbiota in cancer prevention. Nutr. Res. 2024, 124, 21–42. [Google Scholar] [CrossRef]
- Zhou, Y.; Zheng, J.; Li, Y.; Xu, D.P.; Li, S.; Chen, Y.M.; Li, H.B. Natural Polyphenols for Prevention and Treatment of Cancer. Nutrients 2016, 8, 515. [Google Scholar] [CrossRef]
- Rasouli, H.; Farzaei, M.H.; Khodarahmi, R. Polyphenols and their benefits: A review. Int. J. Food Prop. 2017, 20, 1700–1741. [Google Scholar] [CrossRef]
- Abbas, M.; Saeed, F.; Anjum, F.M.; Afzaal, M.; Tufail, T.; Bashir, M.S.; Ishtiaq, A.; Hussain, S.; Suleria, H.A.R. Natural polyphenols: An overview. Int. J. Food Prop. 2017, 20, 1689–1699. [Google Scholar] [CrossRef]
- Guo, J.; Wang, P.; Cui, Y.; Hu, X.; Chen, F.; Ma, C. Protective Effects of Hydroxyphenyl Propionic Acids on Lipid Metabolism and Gut Microbiota in Mice Fed a High-Fat Diet. Nutrients 2023, 15, 1043. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, R.; Zhao, W.; Zhao, Y.; Wang, D.; Zhao, S.; Ge, Z.; Ma, Y.; Zhao, X. Gut microbiota-derived 4-hydroxyphenylacetic acid from resveratrol supplementation prevents obesity through SIRT1 signaling activation. Gut Microbes 2025, 17, 2446391. [Google Scholar] [CrossRef] [PubMed]
- García-Villalba, R.; Giménez-Bastida, J.A.; Cortés-Martín, A.; Ávila-Gálvez, M.; Tomás-Barberán, F.A.; Selma, M.V.; Espín, J.C.; González-Sarrías, A. Urolithins: A Comprehensive Update on their Metabolism, Bioactivity, and Associated Gut Microbiota. Mol. Nutr. Food Res. 2022, 66, e2101019. [Google Scholar] [CrossRef] [PubMed]
- Hervert-Hernández, D.; Goñi, I. Dietary polyphenols and human gut microbiota: A review. Food Rev. Int. 2011, 27, 154–169. [Google Scholar] [CrossRef]
- Wang, X.; Qi, Y.; Zheng, H. Dietary polyphenol, gut microbiota, and health benefits. Antioxidants 2022, 11, 1212. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Lonardo, M.S.; Cacciapuoti, N.; Muscariello, E.; Guida, B.; Faraonio, R.; Santillo, M.; Damiano, S. Dietary polyphenols, microbiome, and multiple sclerosis: From molecular anti-inflammatory and neuroprotective mechanisms to clinical evidence. Int. J. Mol. Sci. 2023, 24, 7247. [Google Scholar] [CrossRef]
- Li, H.; Christman, L.M.; Li, R.; Gu, L. Synergic interactions between polyphenols and gut microbiota in mitigating inflammatory bowel diseases. Food Funct. 2020, 11, 4878–4891. [Google Scholar] [CrossRef]
- Sánchez-Alcoholado, L.; Ramos-Molina, B.; Otero, A.; Laborda-Illanes, A.; Ordóñez, R.; Medina, J.A.; Gómez-Millán, J.; Queipo-Ortuño, M.I. The role of the gut microbiome in colorectal cancer development and therapy response. Cancers 2020, 12, 1406. [Google Scholar] [CrossRef]
- Piekarska-Radzik, L.; Klewicka, E. Mutual influence of polyphenols and Lactobacillus spp. bacteria in food: A review. Eur. Food Res. Technol. 2021, 247, 9–24. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, W.; Zhang, L.; Wang, M.; Chang, W. The Interaction of Polyphenols and the Gut Microbiota in Neurodegenerative Diseases. Nutrients 2022, 14, 5373. [Google Scholar] [CrossRef]
- Corrêa, T.A.F.; Rogero, M.M.; Hassimotto, N.M.A.; Lajolo, F.M. The Two-Way Polyphenols-Microbiota Interactions and Their Effects on Obesity and Related Metabolic Diseases. Front. Nutr. 2019, 6, 188. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Halder, C.V.; Faria, A.V.d.S.; Andrade, S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best. Pract. Res. Clin. Gastroenterol. 2017, 31, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Vlahcevic, R.Z.; Heuman, D.M.; Hylemon, P.B. Regulation of bile acid synthesis. Hepatology 1991, 13, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Barth, C. Regulation and interaction of cholesterol, bile salt and lipoprotein synthesis in liver. Klin. Wochenschr. 1983, 61, 1163–1170. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.; Jaeschke, H.; Copple, B.L. Bile acids induce inflammatory genes in hepatocytes: A novel mechanism of inflammation during obstructive cholestasis. Am. J. Pathol. 2011, 178, 175–186. [Google Scholar] [CrossRef]
- Pavlidis, P.; Powell, N.; Vincent, R.; Ehrlich, D.; Bjarnason, I.; Hayee, B. Systematic review: Bile acids and intestinal inflammation-luminal aggressors or regulators of mucosal defence? Aliment. Pharmacol. Ther. 2015, 42, 802–817. [Google Scholar] [CrossRef]
- Li, M.; Cai, S.-Y.; Boyer, J.L. Mechanisms of bile acid mediated inflammation in the liver. Mol. Asp. Med. 2017, 56, 45–53. [Google Scholar] [CrossRef]
- Kandell, R.L.; Bernstein, C. Bile salt/acid induction of DNA damage in bacterial and mammalian cells: Implications for colon cancer. Nutr. Cancer 1991, 16, 227–238. [Google Scholar] [CrossRef]
- Dvorak, K.; Payne, C.M.; Chavarria, M.; Ramsey, L.; Dvorakova, B.; Bernstein, H.; Holubec, H.; Sampliner, R.E.; Guy, N.; Condon, A. Bile acids in combination with low pH induce oxidative stress and oxidative DNA damage: Relevance to the pathogenesis of Barrett’s oesophagus. Gut 2007, 56, 763–771. [Google Scholar] [CrossRef]
- Deuk Kim, N.; Im, E.; Hyun Yoo, Y.; Hyun Choi, Y. Modulation of the cell cycle and induction of apoptosis in human cancer cells by synthetic bile acids. Curr. Cancer Drug Targets 2006, 6, 681–689. [Google Scholar] [CrossRef]
- Gándola, Y.B.; Fontana, C.; Bojorge, M.A.; Luschnat, T.T.; Moretton, M.A.; Chiapetta, D.A.; Verstraeten, S.V.; González, L. Concentration-dependent effects of sodium cholate and deoxycholate bile salts on breast cancer cells proliferation and survival. Mol. Biol. Rep. 2020, 47, 3521–3539. [Google Scholar] [CrossRef]
- Baptissart, M.; Vega, A.; Maqdasy, S.; Caira, F.; Baron, S.; Lobaccaro, J.-M.A.; Volle, D.H. Bile acids: From digestion to cancers. Biochimie 2013, 95, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Nagengast, F.; Grubben, M.; Van Munster, I. Role of bile acids in colorectal carcinogenesis. Eur. J. Cancer 1995, 31, 1067–1070. [Google Scholar] [CrossRef]
- Fu, J.; Yu, M.; Xu, W.; Yu, S. Research progress of bile acids in cancer. Front. Oncol. 2022, 11, 778258. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Li, F.; Yang, R. The Roles of Gut Microbiota Metabolites in the Occurrence and Development of Colorectal Cancer: Multiple Insights for Potential Clinical Applications. Gastro Hep Adv. 2024, 3, 855–870. [Google Scholar] [CrossRef]
- Song, X.; An, Y.; Chen, D.; Zhang, W.; Wu, X.; Li, C.; Wang, S.; Dong, W.; Wang, B.; Liu, T.; et al. Microbial metabolite deoxycholic acid promotes vasculogenic mimicry formation in intestinal carcinogenesis. Cancer Sci. 2022, 113, 459–477. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Ung, T.T.; Li, S.; Sah, D.K.; Park, S.Y.; Lian, S.; Jung, Y.D. Lithocholic Acid Induces miR21, Promoting PTEN Inhibition via STAT3 and ERK-1/2 Signaling in Colorectal Cancer Cells. Int. J. Mol. Sci. 2021, 22, 10209. [Google Scholar] [CrossRef]
- Lin, H.; Yu, Y.; Zhu, L.; Lai, N.; Zhang, L.; Guo, Y.; Lin, X.; Yang, D.; Ren, N.; Zhu, Z.; et al. Implications of hydrogen sulfide in colorectal cancer: Mechanistic insights and diagnostic and therapeutic strategies. Redox Biol. 2023, 59, 102601. [Google Scholar] [CrossRef]
- Li, W.; Chen, H.; Tang, J. Interplay between Bile Acids and Intestinal Microbiota: Regulatory Mechanisms and Therapeutic Potential for Infections. Pathogens 2024, 13, 702. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef]
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Peters, J. Tryptophan nutrition and metabolism: An overview. In Kynurenine and Serotonin Pathways. Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 1991; pp. 345–358. [Google Scholar]
- Leathwood, P.D. Tryptophan availability and serotonin synthesis. Proc. Nutr. Soc. 1987, 46, 143–156. [Google Scholar] [CrossRef]
- Sainio, E.-L.; Pulkki, K.; Young, S. L-Tryptophan: Biochemical, nutritional and pharmacological aspects. Amino Acids 1996, 10, 21–47. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Zhang, X.; Song, Z.; Dai, Z.; Luo, K.; Chen, B.; Zhou, Z.; Cui, Y.; Feng, B.; Zhu, Z.; et al. Targeting gut microbiota–derived kynurenine to predict and protect the remodeling of the pressure-overloaded young heart. Sci. Adv. 2023, 9, eadg7417. [Google Scholar] [CrossRef] [PubMed]
- Jaglin, M.; Rhimi, M.; Philippe, C.; Pons, N.; Bruneau, A.; Goustard, B.; Daugé, V.; Maguin, E.; Naudon, L.; Rabot, S. Indole, a Signaling Molecule Produced by the Gut Microbiota, Negatively Impacts Emotional Behaviors in Rats. Front. Neurosci. 2018, 12, 216. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.D.; Wargo, J.A. From bugs to drugs: Bacterial 3-IAA enhances efficacy of chemotherapy in pancreatic cancer. Cell Rep. Med. 2023, 4, 101039. [Google Scholar] [CrossRef] [PubMed]
- Ward, F.W. The fate of indolepropionic acid in the animal organism. Biochem. J. 1923, 17, 907. [Google Scholar] [CrossRef]
- Stone, T.W.; Williams, R.O. Modulation of T cells by tryptophan metabolites in the kynurenine pathway. Trends Pharmacol. Sci. 2023, 44, 442–456. [Google Scholar] [CrossRef]
- Fiore, A.; Murray, P.J. Tryptophan and indole metabolism in immune regulation. Curr. Opin. Immunol. 2021, 70, 7–14. [Google Scholar] [CrossRef]
- Mándi, Y.; Vécsei, L. The kynurenine system and immunoregulation. J. Neural Transm. (Vienna) 2012, 119, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Osuch, B.; Misztal, T.; Pałatyńska, K.; Tomaszewska-Zaremba, D. Implications of Kynurenine Pathway Metabolism for the Immune System, Hypothalamic–Pituitary–Adrenal Axis, and Neurotransmission in Alcohol Use Disorder. Int. J. Mol. Sci. 2024, 25, 4845. [Google Scholar] [CrossRef]
- Ye, X.; Li, H.; Anjum, K.; Zhong, X.; Miao, S.; Zheng, G.; Liu, W.; Li, L. Dual Role of Indoles Derived From Intestinal Microbiota on Human Health. Front. Immunol. 2022, 13, 903526. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Pei, Z.; Pan, T.; Wang, H.; Chen, W.; Lu, W. Indole metabolites and colorectal cancer: Gut microbial tryptophan metabolism, host gut microbiome biomarkers, and potential intervention mechanisms. Microbiol. Res. 2023, 272, 127392. [Google Scholar] [CrossRef] [PubMed]
- Cover, C.M.; Hsieh, S.J.; Tran, S.H.; Hallden, G.; Kim, G.S.; Bjeldanes, L.F.; Firestone, G.L. Indole-3-carbinol inhibits the expression of cyclin-dependent kinase-6 and induces a G1 cell cycle arrest of human breast cancer cells independent of estrogen receptor signaling. J. Biol. Chem. 1998, 273, 3838–3847. [Google Scholar] [CrossRef] [PubMed]
- Gouasmi, R.; Ferraro-Peyret, C.; Nancey, S.; Coste, I.; Renno, T.; Chaveroux, C.; Aznar, N.; Ansieau, S. The Kynurenine Pathway and Cancer: Why Keep It Simple When You Can Make It Complicated. Cancers 2022, 14, 2793. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.-Y.; Hofree, M.; Ideker, T. A decade of systems biology. Annu. Rev. Cell Dev. Biol. 2010, 26, 721–744. [Google Scholar] [CrossRef]
- Casotti, M.C.; Meira, D.D.; Zetum, A.S.S.; Campanharo, C.V.; da Silva, D.R.C.; Giacinti, G.M.; da Silva, I.M.; Moura, J.A.D.; Barbosa, K.R.M.; Altoé, L.S.C. Integrating frontiers: A holistic, quantum and evolutionary approach to conquering cancer through systems biology and multidisciplinary synergy. Front. Oncol. 2024, 14, 1419599. [Google Scholar] [CrossRef]
- Dutta, B.; Lahiri, D.; Nag, M.; Sarkar, N.; Ray, R.R.; Bhattacharya, D. Systems Biology Approaches for Cancer Biology. In Systems Biology Approaches: Prevention, Diagnosis, and Understanding Mechanisms of Complex Diseases; Springer: Singapore, 2024; pp. 537–559. [Google Scholar]
- Passi, A.; Tibocha-Bonilla, J.D.; Kumar, M.; Tec-Campos, D.; Zengler, K.; Zuniga, C. Genome-Scale Metabolic Modeling Enables In-Depth Understanding of Big Data. Metabolites 2021, 12, 14. [Google Scholar] [CrossRef]
- Gu, C.; Kim, G.B.; Kim, W.J.; Kim, H.U.; Lee, S.Y. Current status and applications of genome-scale metabolic models. Genome Biol. 2019, 20, 121. [Google Scholar] [CrossRef]
- Tarzi, C.; Zampieri, G.; Sullivan, N.; Angione, C. Emerging methods for genome-scale metabolic modeling of microbial communities. Trends Endocrinol. Metab. 2024, 35, 533–548. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.J.; Monk, J.M.; Palsson, B.O. Using Genome-scale Models to Predict Biological Capabilities. Cell 2015, 161, 971–987. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.L.; Constantinidou, C.; Alam, M.T. Applications of genome-scale metabolic models to investigate microbial metabolic adaptations in response to genetic or environmental perturbations. Brief. Bioinform. 2023, 25, bbad439. [Google Scholar] [CrossRef]
- Proffitt, C.; Bidkhori, G.; Lee, S.; Tebani, A.; Mardinoglu, A.; Uhlen, M.; Moyes, D.L.; Shoaie, S. Genome-scale metabolic modelling of the human gut microbiome reveals changes in the glyoxylate and dicarboxylate metabolism in metabolic disorders. iScience 2022, 25, 104513. [Google Scholar] [CrossRef]
- Diener, C.; Gibbons Sean, M.; Resendis-Antonio, O. MICOM: Metagenome-Scale Modeling To Infer Metabolic Interactions in the Gut Microbiota. mSystems 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Esvap, E.; Ulgen, K.O. Advances in Genome-Scale Metabolic Modeling toward Microbial Community Analysis of the Human Microbiome. ACS Synth. Biol. 2021, 10, 2121–2137. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, A.; Nielsen, J. Genome scale metabolic modeling of cancer. Metab. Eng. 2017, 43, 103–112. [Google Scholar] [CrossRef]
- Lee, G.; Lee, S.M.; Lee, S.; Jeong, C.W.; Song, H.; Lee, S.Y.; Yun, H.; Koh, Y.; Kim, H.U. Prediction of metabolites associated with somatic mutations in cancers by using genome-scale metabolic models and mutation data. Genome Biol. 2024, 25, 66. [Google Scholar] [CrossRef]
- Gilbert, J.A. Our unique microbial identity. Genome Biol. 2015, 16, 97. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Huang, K.; Meadow, J.F.; Gevers, D.; Lemon, K.P.; Bohannan, B.J.; Huttenhower, C. Identifying personal microbiomes using metagenomic codes. Proc. Natl. Acad. Sci. USA 2015, 112, E2930–E2938. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Godoy-Vitorino, F.; Knight, R.; Blaser, M.J. Role of the microbiome in human development. Gut 2019, 68, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Shahab, M.; Shahab, N. Coevolution of the Human Host and Gut Microbiome: Metagenomics of Microbiota. Cureus 2022, 14, e26310. [Google Scholar] [CrossRef] [PubMed]
- Groussin, M.; Mazel, F.; Alm, E.J. Co-evolution and Co-speciation of Host-Gut Bacteria Systems. Cell Host Microbe 2020, 28, 12–22. [Google Scholar] [CrossRef]
- Stolfi, C.; Maresca, C.; Monteleone, G.; Laudisi, F. Implication of Intestinal Barrier Dysfunction in Gut Dysbiosis and Diseases. Biomedicines 2022, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Takiishi, T.; Fenero, C.I.M.; Câmara, N.O.S. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers 2017, 5, e1373208. [Google Scholar] [CrossRef] [PubMed]
- Akbar, N.; Khan, N.A.; Muhammad, J.S.; Siddiqui, R. The role of gut microbiome in cancer genesis and cancer prevention. Health Sci. Rev. 2022, 2, 100010. [Google Scholar] [CrossRef]
- Roy, R.; Singh, S.K. The Microbiome Modulates the Immune System to Influence Cancer Therapy. Cancers 2024, 16, 779. [Google Scholar] [CrossRef]
- Schluter, J.; Peled, J.U.; Taylor, B.P.; Markey, K.A.; Smith, M.; Taur, Y.; Niehus, R.; Staffas, A.; Dai, A.; Fontana, E.; et al. The gut microbiota is associated with immune cell dynamics in humans. Nature 2020, 588, 303–307. [Google Scholar] [CrossRef]
- Luchini, C.; Pea, A.; Scarpa, A. Artificial intelligence in oncology: Current applications and future perspectives. Br. J. Cancer 2022, 126, 4–9. [Google Scholar] [CrossRef]
- Graham, D.B.; Xavier, R.J. Conditioning of the immune system by the microbiome. Trends Immunol. 2023, 44, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Wu, H.; Wang, L.; Zhao, Y.; Huang, H. Altered intestinal microbiome and epithelial damage aggravate intestinal graft-versus-host disease. Gut Microbes 2023, 15, 2221821. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Palaniyandi, S.; Strattan, E.; Hildebrandt, G.C. Microbiome: An emerging new frontier in graft-versus-host disease. Inflamm. Res. 2020, 70, 1–5. [Google Scholar] [CrossRef]
- Gyriki, D.; Nikolaidis, C.; Stavropoulou, E.; Bezirtzoglou, I.; Tsigalou, C.; Vradelis, S.; Bezirtzoglou, E. Exploring the Gut Microbiome’s Role in Inflammatory Bowel Disease: Insights and Interventions. J. Pers. Med. 2024, 14, 507. [Google Scholar] [CrossRef] [PubMed]
- La Flamme, A.C.; Milling, S.W.F. Immunological partners: The gut microbiome in homeostasis and disease. Immunology 2020, 161, 1. [Google Scholar] [CrossRef]
- Tepekule, B.; Lim, A.I.; Jessica, C.; Metcalf, E. The ontogeny of immune tolerance: A model of the early-life gut microbiome and adaptive immunity. bioRxiv 2024. [Google Scholar] [CrossRef]
- Sharma, P.; Jain, T.; Sethi, V.; Iyer, S.; Dudeja, V. Gut Microbiome: The Third Musketeer in the Cancer-Immune System Cross-Talk. J. Pancreatol. 2020, 3, 181–187. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.K. Potential Role of the Gut Microbiome In Colorectal Cancer Progression. Front. Immunol. 2022, 12, 807648. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Shishir, M.A.; Oyshe, I.I.; Amin, S.M.T.; Hossain, A.; Sarna, I.J.; Jerin, N.; Mitra, D.K. Microbial Architects of Malignancy: Exploring the Gut Microbiome’s Influence in Cancer Initiation and Progression. Cancer Plus 2023, 5, 397. [Google Scholar] [CrossRef]
- Anderson, S.M.; Sears, C.L. The Role of the Gut Microbiome in Cancer: A Review, With Special Focus on Colorectal Neoplasia and Clostridioides difficile. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2023, 77 (Suppl. 6), S471–S478. [Google Scholar] [CrossRef]
- Kann, B.H.; Hosny, A.; Aerts, H. Artificial intelligence for clinical oncology. Cancer Cell 2021, 39, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Kurian, M.; Adashek, J.J.; West, H. Cancer Care in the Era of Artificial Intelligence. JAMA Oncol. 2024, 10, 683. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, K.J. Leveraging the Academic Artificial Intelligence Silecosystem to Advance the Community Oncology Enterprise. J. Clin. Med. 2023, 12, 4830. [Google Scholar] [CrossRef]
- Kumar, Y.; Koul, A.; Singla, R.; Ijaz, M.F. Artificial intelligence in disease diagnosis: A systematic literature review, synthesizing framework and future research agenda. J. Ambient. Intell. Humaniz. Comput. 2023, 14, 8459–8486. [Google Scholar] [CrossRef]
- Kulkarni, P.A.; Singh, H. Artificial Intelligence in Clinical Diagnosis: Opportunities, Challenges, and Hype. JAMA 2023, 330, 317–318. [Google Scholar] [CrossRef] [PubMed]
- Peris-Bondia, F.; Latorre, A.; Artacho, A.; Moya, A.; D’Auria, G. The active human gut microbiota differs from the total microbiota. PLoS ONE 2011, 6, e22448. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed]
- D’Elia, D.; Truu, J.; Lahti, L.; Berland, M.; Papoutsoglou, G.; Ceci, M.; Zomer, A.; Lopes, M.B.; Ibrahimi, E.; Gruca, A.; et al. Advancing microbiome research with machine learning: Key findings from the ML4Microbiome COST action. Front. Microbiol. 2023, 14, 1257002. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Luo, H.; Ji, B.; Nielsen, J. Machine learning for data integration in human gut microbiome. Microb. Cell Factories 2022, 21, 241. [Google Scholar] [CrossRef]
- Abavisani, M.; Khoshrou, A.; Foroushan, S.K.; Ebadpour, N.; Sahebkar, A. Deciphering the gut microbiome: The revolution of artificial intelligence in microbiota analysis and intervention. Curr. Res. Biotechnol. 2024, 7, 100211. [Google Scholar] [CrossRef]
- Manandhar, I.; Alimadadi, A.; Aryal, S.; Munroe, P.B.; Joe, B.; Cheng, X. Gut microbiome-based supervised machine learning for clinical diagnosis of inflammatory bowel diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G328–G337. [Google Scholar] [CrossRef]
- Dudek, N.K.; Chakhvadze, M.; Kobakhidze, S.; Kantidze, O.; Gankin, Y. Supervised machine learning for microbiomics: Bridging the gap between current and best practices. Mach. Learn. Appl. 2024, 18, 100607. [Google Scholar] [CrossRef]
- Liang, H.; Jo, J.-H.; Zhang, Z.; MacGibeny, M.A.; Han, J.; Proctor, D.M.; Taylor, M.E.; Che, Y.; Juneau, P.; Apolo, A.B. Predicting cancer immunotherapy response from gut microbiomes using machine learning models. Oncotarget 2022, 13, 876. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, L.; Peterson, C.B.; Do, K.-A.; Jenq, R.R. Performance determinants of unsupervised clustering methods for microbiome data. Microbiome 2022, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Papoutsoglou, G.; Tarazona, S.; Lopes, M.B.; Klammsteiner, T.; Ibrahimi, E.; Eckenberger, J.; Novielli, P.; Tonda, A.; Simeon, A.; Shigdel, R. Machine learning approaches in microbiome research: Challenges and best practices. Front. Microbiol. 2023, 14, 1261889. [Google Scholar] [CrossRef] [PubMed]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Loncar Turukalo, T.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K.; et al. Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment. Front. Microbiol. 2021, 12, 634511. [Google Scholar] [CrossRef]
- Liu, W.; Fang, X.; Zhou, Y.; Dou, L.; Dou, T. Machine learning-based investigation of the relationship between gut microbiome and obesity status. Microbes Infect. 2022, 24, 104892. [Google Scholar] [CrossRef]
- Thompson, J.; Johansen, R.; Dunbar, J.; Munsky, B. Machine learning to predict microbial community functions: An analysis of dissolved organic carbon from litter decomposition. PLoS ONE 2019, 14, e0215502. [Google Scholar] [CrossRef]
- Chen, T.F.; Chen, R.M.; Tsai, J.J.P.; Hu, R.M. Fine Classification of Human Gut Microbiota by Using Hierarchical Clustering Approach. In Proceedings of the 2016 IEEE 16th International Conference on Bioinformatics and Bioengineering (BIBE), Taichung, Taiwan, 31 October–2 November 2016; pp. 109–112. [Google Scholar]
- Yang, D.; Xu, W. Clustering on Human Microbiome Sequencing Data: A Distance-Based Unsupervised Learning Model. Microorganisms 2020, 8, 1612. [Google Scholar] [CrossRef]
- Leong, C.; Haszard, J.J.; Heath, A.-L.M.; Tannock, G.W.; Lawley, B.; Cameron, S.L.; Szymlek-Gay, E.A.; Gray, A.R.; Taylor, B.J.; Galland, B.C.; et al. Using compositional principal component analysis to describe children’s gut microbiota in relation to diet and body composition. Am. J. Clin. Nutr. 2020, 111, 70–78. [Google Scholar] [CrossRef]
- Novielli, P.; Romano, D.; Magarelli, M.; Bitonto, P.D.; Diacono, D.; Chiatante, A.; Lopalco, G.; Sabella, D.; Venerito, V.; Filannino, P.; et al. Explainable artificial intelligence for microbiome data analysis in colorectal cancer biomarker identification. Front. Microbiol. 2024, 15, 1348974. [Google Scholar] [CrossRef] [PubMed]
- Giuffrè, M.; Moretti, R.; Tiribelli, C. Gut Microbes Meet Machine Learning: The Next Step towards Advancing Our Understanding of the Gut Microbiome in Health and Disease. Int. J. Mol. Sci. 2023, 24, 5229. [Google Scholar] [CrossRef] [PubMed]
- Wickramaratne, D.N.; Wijesinghe, C.R.; Weerasinghe, A.R. A Deep Learning Approach to Predict Health Status Using Microbiome Profiling. In Proceedings of the 2022 22nd International Conference on Advances in ICT for Emerging Regions (ICTer), Colombo, Sri Lanka, 30 November–1 December 2022; pp. 075–079. [Google Scholar]
- Bai, X.; Huang, Z.; Duraj-Thatte, A.M.; Ebert, M.P.; Zhang, F.; Burgermeister, E.; Liu, X.; Scott, B.M.; Li, G.; Zuo, T. Engineering the gut microbiome. Nat. Rev. Bioeng. 2023, 1, 665–679. [Google Scholar] [CrossRef]
- Arnold, J.; Glazier, J.; Mimee, M. Genetic engineering of resident bacteria in the gut microbiome. J. Bacteriol. 2023, 205, e00127-23. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.-B.; Li, T.-T.; Huo, D.; Qu, S.; Li, X.V.; Arifuzzaman, M.; Lima, S.F.; Shi, H.-Q.; Wang, A.; Putzel, G.G.; et al. Genetic manipulation of gut microbes enables single-gene interrogation in a complex microbiome. Cell 2022, 185, 547–562.e522. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Zheng, C.; Jia, J.; Zhao, H.; Feng, Q.; Zhang, H.; Wang, L.; Zhang, Z.; Zhang, Y. A Probiotic Spore-Based Oral Autonomous Nanoparticles Generator for Cancer Therapy. Adv. Mater. 2019, 31, e1903793. [Google Scholar] [CrossRef]
- Molla, K.A.; Yang, Y. CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef]
- Hess, G.T.; Tycko, J.; Yao, D.; Bassik, M.C. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol. Cell 2017, 68, 26–43. [Google Scholar] [CrossRef] [PubMed]
- Porto, E.M.; Komor, A.C.; Slaymaker, I.M.; Yeo, G.W. Base editing: Advances and therapeutic opportunities. Nat. Rev. Drug Discov. 2020, 19, 839–859. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Saber Sichani, A.; Ranjbar, M.; Baneshi, M.; Torabi Zadeh, F.; Fallahi, J. A review on advanced CRISPR-based genome-editing tools: Base editing and prime editing. Mol. Biotechnol. 2023, 65, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Brodel, A.K.; Charpenay, L.H.; Galtier, M.; Fuche, F.J.; Terrasse, R.; Poquet, C.; Havranek, J.; Pignotti, S.; Krawczyk, A.; Arraou, M.; et al. In situ targeted base editing of bacteria in the mouse gut. Nature 2024, 632, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Ronda, C.; Chen, S.P.; Cabral, V.; Yaung, S.J.; Wang, H.H. Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods 2019, 16, 167–170. [Google Scholar] [CrossRef]
- Zheng, L.; Shen, J.; Chen, R.; Hu, Y.; Zhao, W.; Leung, E.L.-H.; Dai, L. Genome engineering of the human gut microbiome. J. Genet. Genom. 2024, 51, 479–491. [Google Scholar] [CrossRef]
- Benner, S.A.; Sismour, A.M. Synthetic biology. Nat. Rev. Genet. 2005, 6, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Hanczyc, M.M. Engineering Life: A Review of Synthetic Biology. Artif. Life 2020, 26, 260–273. [Google Scholar] [CrossRef]
- Garner, K.L. Principles of synthetic biology. Essays Biochem. 2021, 65, 791–811. [Google Scholar] [CrossRef] [PubMed]
- Tanna, T.; Ramachanderan, R.; Platt, R.J. Engineered bacteria to report gut function: Technologies and implementation. Curr. Opin. Microbiol. 2021, 59, 24–33. [Google Scholar] [CrossRef]
- Woo, S.-G.; Moon, S.-J.; Kim, S.K.; Kim, T.H.; Lim, H.S.; Yeon, G.-H.; Sung, B.H.; Lee, C.-H.; Lee, S.-G.; Hwang, J.H.; et al. A designed whole-cell biosensor for live diagnosis of gut inflammation through nitrate sensing. Biosens. Bioelectron. 2020, 168, 112523. [Google Scholar] [CrossRef]
- Ayati, M.H.; Araj-Khodaei, M.; Haghgouei, T.; Ahmadalipour, A.; Mobed, A.; Sanaie, S. Biosensors: The nanomaterial-based method in detection of human gut microbiota. Mater. Chem. Phys. 2023, 307, 127854. [Google Scholar] [CrossRef]
- Dunham, K.E.; Venton, B.J. Electrochemical and biosensor techniques to monitor neurotransmitter changes with depression. Anal. Bioanal. Chem. 2024, 416, 2301–2318. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, H.; Wang, G.; Wang, Q.; Wang, Y.; Li, Q.; Bi, S.; Qi, Q.; Wang, Q. Engineering Chimeric Chemoreceptors and Two-Component Systems for Orthogonal and Leakless Biosensing of Extracellular γ-Aminobutyric Acid. J. Agric. Food Chem. 2024, 72, 14216–14228. [Google Scholar] [CrossRef] [PubMed]
- Baumann, L.; Rajkumar, A.S.; Morrissey, J.P.; Boles, E.; Oreb, M. A Yeast-Based Biosensor for Screening of Short- and Medium-Chain Fatty Acid Production. ACS Synth. Biol. 2018, 7, 2640–2646. [Google Scholar] [CrossRef] [PubMed]
- Wasiewska, L.A.; Uhlig, F.; Barry, F.; Teixeira, S.; Clarke, G.; Schellekens, H. Developments in sensors for rapid detection of short-chain fatty acids (SCFAs): Research and limitation in their applications in gut health and the microbiota-gut-brain axis. TrAC Trends Anal. Chem. 2024, 184, 118118. [Google Scholar] [CrossRef]
- van der Lelie, D.; Oka, A.; Taghavi, S.; Umeno, J.; Fan, T.J.; Merrell, K.E.; Watson, S.D.; Ouellette, L.; Liu, B.; Awoniyi, M.; et al. Rationally designed bacterial consortia to treat chronic immune-mediated colitis and restore intestinal homeostasis. Nat. Commun. 2021, 12, 3105. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wu, J.; Tang, H.; Liu, D.; Jeon, B.H.; Jin, W.; Wang, Y.; Zheng, Y.; Khan, A.; Han, H.; et al. Enhancing tumor-specific recognition of programmable synthetic bacterial consortium for precision therapy of colorectal cancer. NPJ Biofilms Microbiomes 2024, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, P.T.; Brul, S.; Zhang, J.; Wortel, M.T. Synthetic microbial communities (SynComs) of the human gut: Design, assembly, and applications. FEMS Microbiol. Rev. 2023, 47, fuad012. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.; Ntemiri, A.; Tan, H.; Harris, H.M.B.; Roager, H.M.; Ribiere, C.; O’Toole, P.W. A synthetic consortium of 100 gut commensals modulates the composition and function in a colon model of the microbiome of elderly subjects. Gut Microbes 2021, 13, 1–19. [Google Scholar] [CrossRef]
- Clark, R.L.; Connors, B.M.; Stevenson, D.M.; Hromada, S.E.; Hamilton, J.J.; Amador-Noguez, D.; Venturelli, O.S. Design of synthetic human gut microbiome assembly and butyrate production. Nat. Commun. 2021, 12, 3254. [Google Scholar] [CrossRef]
- Nazir, A.; Hussain, F.H.N.; Raza, A. Advancing microbiota therapeutics: The role of synthetic biology in engineering microbial communities for precision medicine. Front. Bioeng. Biotechnol. 2024, 12, 1511149. [Google Scholar] [CrossRef]
- Gonzalez-Garcia, R.A.; McCubbin, T.; Navone, L.; Stowers, C.; Nielsen, L.K.; Marcellin, E. Microbial Propionic Acid Production. Fermentation 2017, 3, 21. [Google Scholar] [CrossRef]
- McCarty, N.S.; Ledesma-Amaro, R. Synthetic Biology Tools to Engineer Microbial Communities for Biotechnology. Trends Biotechnol. 2019, 37, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhou, Y.; Dai, L.; Yang, A.; Qiao, J. Assembly of functional microbial ecosystems: From molecular circuits to communities. FEMS Microbiol. Rev. 2024, 48, fuae026. [Google Scholar] [CrossRef]
- El Hage, R.; Hernandez-Sanabria, E.; Calatayud Arroyo, M.; Props, R.; Van de Wiele, T. Propionate-producing consortium restores antibiotic-induced dysbiosis in a dynamic in vitro model of the human intestinal microbial ecosystem. Front. Microbiol. 2019, 10, 1206. [Google Scholar] [CrossRef] [PubMed]
- Raghu, A.K.; Palanikumar, I.; Raman, K. Designing function-specific minimal microbiomes from large microbial communities. npj Syst. Biol. Appl. 2024, 10, 46. [Google Scholar] [CrossRef]
- Lee, H.L.; Shen, H.; Hwang, I.Y.; Ling, H.; Yew, W.S.; Lee, Y.S.; Chang, M.W. Targeted Approaches for In Situ Gut Microbiome Manipulation. Genes 2018, 9, 351. [Google Scholar] [CrossRef] [PubMed]
- Nogal, A.; Louca, P.; Zhang, X.; Wells, P.M.; Steves, C.J.; Spector, T.D.; Falchi, M.; Valdes, A.M.; Menni, C. Circulating levels of the short-chain fatty acid acetate mediate the effect of the gut microbiome on visceral fat. Front. Microbiol. 2021, 12, 711359. [Google Scholar] [CrossRef]
- Hosmer, J.; McEwan, A.G.; Kappler, U. Bacterial acetate metabolism and its influence on human epithelia. Emerg. Top. Life Sci. 2024, 8, 1–13. [Google Scholar] [CrossRef]
- Li, H.Y.; Zhou, D.D.; Gan, R.Y.; Huang, S.Y.; Zhao, C.N.; Shang, A.; Xu, X.Y.; Li, H.B. Effects and Mechanisms of Probiotics, Prebiotics, Synbiotics, and Postbiotics on Metabolic Diseases Targeting Gut Microbiota: A Narrative Review. Nutrients 2021, 13, 3211. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Holscher, H.D. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, A.; Bakhsh Javaid, A.; Irshad, S.; Hoseinifar, S.H.; Xiong, H. The functionality of prebiotics as immunostimulant: Evidences from trials on terrestrial and aquatic animals. Fish. Shellfish. Immunol. 2018, 76, 272–278. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef]
- Birkeland, E.; Gharagozlian, S.; Birkeland, K.I.; Valeur, J.; Måge, I.; Rud, I.; Aas, A.M. Prebiotic effect of inulin-type fructans on faecal microbiota and short-chain fatty acids in type 2 diabetes: A randomised controlled trial. Eur. J. Nutr. 2020, 59, 3325–3338. [Google Scholar] [CrossRef]
- Riva, A.; Rasoulimehrabani, H.; Cruz-Rubio, J.M.; Schnorr, S.L.; von Baeckmann, C.; Inan, D.; Nikolov, G.; Herbold, C.W.; Hausmann, B.; Pjevac, P.; et al. Identification of inulin-responsive bacteria in the gut microbiota via multi-modal activity-based sorting. Nat. Commun. 2023, 14, 8210. [Google Scholar] [CrossRef] [PubMed]
- Le Bastard, Q.; Chapelet, G.; Javaudin, F.; Lepelletier, D.; Batard, E.; Montassier, E. The effects of inulin on gut microbial composition: A systematic review of evidence from human studies. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 403–413. [Google Scholar] [CrossRef]
- Becerril-Alarcón, Y.; Campos-Gómez, S.; Valdez-Andrade, J.J.; Campos-Gómez, K.A.; Reyes-Barretero, D.Y.; Benítez-Arciniega, A.D.; Valdés-Ramos, R.; Soto-Piña, A.E. Inulin Supplementation Reduces Systolic Blood Pressure in Women with Breast Cancer Undergoing Neoadjuvant Chemotherapy. Cardiovasc. Ther. 2019, 2019, 5707150. [Google Scholar] [CrossRef] [PubMed]
- Rafter, J.; Bennett, M.; Caderni, G.; Clune, Y.; Hughes, R.; Karlsson, P.C.; Klinder, A.; O’Riordan, M.; O’Sullivan, G.C.; Pool-Zobel, B.; et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am. J. Clin. Nutr. 2007, 85, 488–496. [Google Scholar] [CrossRef]
- Garcia-Peris, P.; Velasco, C.; Hernandez, M.; Lozano, M.A.; Paron, L.; de la Cuerda, C.; Breton, I.; Camblor, M.; Guarner, F. Effect of inulin and fructo-oligosaccharide on the prevention of acute radiation enteritis in patients with gynecological cancer and impact on quality-of-life: A randomized, double-blind, placebo-controlled trial. Eur. J. Clin. Nutr. 2016, 70, 170–174. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Suez, J.; Zmora, N.; Segal, E.; Elinav, E. The pros, cons, and many unknowns of probiotics. Nat. Med. 2019, 25, 716–729. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Guarner, F.; Guerrant, R.; Holt, P.R.; Quigley, E.M.; Sartor, R.B.; Sherman, P.M.; Mayer, E.A. An update on the use and investigation of probiotics in health and disease. Gut 2013, 62, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Kiepś, J.; Dembczyński, R. Current Trends in the Production of Probiotic Formulations. Foods 2022, 11, 2330. [Google Scholar] [CrossRef] [PubMed]
- Fenster, K.; Freeburg, B.; Hollard, C.; Wong, C.; Rønhave Laursen, R.; Ouwehand, A.C. The Production and Delivery of Probiotics: A Review of a Practical Approach. Microorganisms 2019, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, T.; Sequoia, J. Probiotics for Gastrointestinal Conditions: A Summary of the Evidence. Am. Fam. Physician 2017, 96, 170–178. [Google Scholar] [PubMed]
- Zhou, Z.; Chen, X.; Sheng, H.; Shen, X.; Sun, X.; Yan, Y.; Wang, J.; Yuan, Q. Engineering probiotics as living diagnostics and therapeutics for improving human health. Microb. Cell Fact. 2020, 19, 56. [Google Scholar] [CrossRef]
- Bober, J.R.; Beisel, C.L.; Nair, N.U. Synthetic Biology Approaches to Engineer Probiotics and Members of the Human Microbiota for Biomedical Applications. Annu. Rev. Biomed. Eng. 2018, 20, 277–300. [Google Scholar] [CrossRef]
- Mays, Z.J.; Nair, N.U. Synthetic biology in probiotic lactic acid bacteria: At the frontier of living therapeutics. Curr. Opin. Biotechnol. 2018, 53, 224–231. [Google Scholar] [CrossRef]
- Romero-Luna, H.E.; Hernández-Mendoza, A.; González-Córdova, A.F.; Peredo-Lovillo, A. Bioactive peptides produced by engineered probiotics and other food-grade bacteria: A review. Food Chem. X 2022, 13, 100196. [Google Scholar] [CrossRef]
- Wang, G.; Chen, Y.; Xia, Y.; Song, X.; Ai, L. Characteristics of Probiotic Preparations and Their Applications. Foods 2022, 11, 2472. [Google Scholar] [CrossRef] [PubMed]
- Ciorba, M.A. A gastroenterologist’s guide to probiotics. Clin. Gastroenterol. Hepatol. 2012, 10, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Su, G.L.; Ko, C.W.; Bercik, P.; Falck-Ytter, Y.; Sultan, S.; Weizman, A.V.; Morgan, R.L. AGA Clinical Practice Guidelines on the Role of Probiotics in the Management of Gastrointestinal Disorders. Gastroenterology 2020, 159, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Śliżewska, K.; Markowiak-Kopeć, P.; Śliżewska, W. The Role of Probiotics in Cancer Prevention. Cancers 2020, 13, 20. [Google Scholar] [CrossRef]
- Naeem, H.; Hassan, H.U.; Shahbaz, M.; Imran, M.; Memon, A.G.; Hasnain, A.; Murtaza, S.; Alsagaby, S.A.; Al Abdulmonem, W.; Hussain, M.; et al. Role of Probiotics against Human Cancers, Inflammatory Diseases, and Other Complex Malignancies. J. Food Biochem. 2024, 2024, 6632209. [Google Scholar] [CrossRef]
- Jiang, S.; Ma, W.; Ma, C.; Zhang, Z.; Zhang, W.; Zhang, J. An emerging strategy: Probiotics enhance the effectiveness of tumor immunotherapy via mediating the gut microbiome. Gut Microbes 2024, 16, 2341717. [Google Scholar] [CrossRef] [PubMed]
- Marinelli, L.; Tenore, G.C.; Novellino, E. Probiotic species in the modulation of the anticancer immune response. Semin. Cancer Biol. 2017, 46, 182–190. [Google Scholar] [CrossRef]
- Redman, M.G.; Ward, E.J.; Phillips, R.S. The efficacy and safety of probiotics in people with cancer: A systematic review. Ann. Oncol. 2014, 25, 1919–1929. [Google Scholar] [CrossRef]
- Lu, Y.; Luo, X.; Yang, D.; Li, Y.; Gong, T.; Li, B.; Cheng, J.; Chen, R.; Guo, X.; Yuan, W. Effects of probiotic supplementation on related side effects after chemoradiotherapy in cancer patients. Front. Oncol. 2022, 12, 1032145. [Google Scholar] [CrossRef]
- Zaharuddin, L.; Mokhtar, N.M.; Muhammad Nawawi, K.N.; Raja Ali, R.A. A randomized double-blind placebo-controlled trial of probiotics in post-surgical colorectal cancer. BMC Gastroenterol. 2019, 19, 131. [Google Scholar] [CrossRef]
- Xia, C.; Jiang, C.; Li, W.; Wei, J.; Hong, H.; Li, J.; Feng, L.; Wei, H.; Xin, H.; Chen, T. A Phase II Randomized Clinical Trial and Mechanistic Studies Using Improved Probiotics to Prevent Oral Mucositis Induced by Concurrent Radiotherapy and Chemotherapy in Nasopharyngeal Carcinoma. Front. Immunol. 2021, 12, 618150. [Google Scholar] [CrossRef] [PubMed]
- Dizman, N.; Meza, L.; Bergerot, P.; Alcantara, M.; Dorff, T.; Lyou, Y.; Frankel, P.; Cui, Y.; Mira, V.; Llamas, M.; et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: A randomized phase 1 trial. Nat. Med. 2022, 28, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Lythgoe, M.P.; Ghani, R.; Mullish, B.H.; Marchesi, J.R.; Krell, J. The potential of fecal microbiota transplantation in oncology. Trends Microbiol. 2022, 30, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Cao, C.; Ren, Y.; Weng, S.; Liu, L.; Guo, C.; Wang, L.; Han, X.; Ren, J.; Liu, Z. Antitumor effects of fecal microbiota transplantation: Implications for microbiome modulation in cancer treatment. Front. Immunol. 2022, 13, 949490. [Google Scholar] [CrossRef]
- Drew, L. Faecal transplants can treat some cancers—But probably won’t ever be widely used. Nature 2024. [Google Scholar] [CrossRef] [PubMed]
- Yadegar, A.; Bar-Yoseph, H.; Monaghan, T.M.; Pakpour, S.; Severino, A.; Kuijper, E.J.; Smits, W.K.; Terveer, E.M.; Neupane, S.; Nabavi-Rad, A.; et al. Fecal microbiota transplantation: Current challenges and future landscapes. Clin. Microbiol. Rev. 2024, 37, e0006022. [Google Scholar] [CrossRef]
- Vindigni, S.M.; Surawicz, C.M. Fecal Microbiota Transplantation. Gastroenterol. Clin. N. Am. 2017, 46, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Luo, Y.; Walsh, S.; Grinspan, A. Oral Fecal Microbiota Transplant Capsules Are Safe and Effective for Recurrent Clostridioides difficile Infection: A Systematic Review and Meta-Analysis. J. Clin. Gastroenterol. 2021, 55, 300–308. [Google Scholar] [CrossRef]
- Kao, D.; Roach, B.; Silva, M.; Beck, P.; Rioux, K.; Kaplan, G.G.; Chang, H.J.; Coward, S.; Goodman, K.J.; Xu, H.; et al. Effect of Oral Capsule- vs Colonoscopy-Delivered Fecal Microbiota Transplantation on Recurrent Clostridium difficile Infection: A Randomized Clinical Trial. Jama 2017, 318, 1985–1993. [Google Scholar] [CrossRef]
- Cho, J.M.; Pestana, L.; Pardi, R.; Pardi, D.S.; Khanna, S. Fecal microbiota transplant via colonoscopy may be preferred due to intraprocedure findings. Intest. Res. 2019, 17, 434–437. [Google Scholar] [CrossRef]
- Allegretti, J.R.; Korzenik, J.R.; Hamilton, M.J. Fecal microbiota transplantation via colonoscopy for recurrent C. difficile Infection. J. Vis. Exp. 2014. [Google Scholar] [CrossRef]
- Skjevling, L.K.; Hanssen, H.M.; Valle, P.C.; Goll, R.; Juul, F.E.; Arlov, Ø.; Johnsen, P.H. Colonic distribution of FMT by different enema procedures compared to colonoscopy—Proof of concept study using contrast fluid. BMC Gastroenterol. 2023, 23, 363. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Kuo, C.-H.; Kuo, F.-C.; Wang, Y.-K.; Hsu, W.-H.; Yu, F.-J.; Hu, H.-M.; Hsu, P.-I.; Wang, J.-Y.; Wu, D.-C. Fecal microbiota transplantation: Review and update. J. Formos. Med. Assoc. 2019, 118, S23–S31. [Google Scholar] [CrossRef]
- Nicco, C.; Paule, A.; Konturek, P.; Edeas, M. From Donor to Patient: Collection, Preparation and Cryopreservation of Fecal Samples for Fecal Microbiota Transplantation. Diseases 2020, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zaman, A.; Ramakrishna, B.; Olesen, S.W. Stool Banking for Fecal Microbiota Transplantation: Methods and Operations at a Large Stool Bank. Front. Cell Infect. Microbiol. 2021, 11, 622949. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, X.X.; Han, X.; Chen, B.X.; Zhang, X.H.; Gao, S.; Xu, D.Q.; Wang, Y.; Gao, Z.K.; Yu, L.; et al. Fecal microbiota transplantation inhibits colorectal cancer progression: Reversing intestinal microbial dysbiosis to enhance anti-cancer immune responses. Front. Microbiol. 2023, 14, 1126808. [Google Scholar] [CrossRef]
- Khoruts, A. Can FMT Cause or Prevent CRC? Maybe, But There Is More to Consider. Gastroenterology 2021, 161, 1103–1105. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-W.; Lee, H.-C.; Li, L.-H.; Chiang Chiau, J.-S.; Wang, T.-E.; Chuang, W.-H.; Chen, M.-J.; Wang, H.-Y.; Shih, S.-C.; Liu, C.-Y.; et al. Fecal Microbiota Transplantation Prevents Intestinal Injury, Upregulation of Toll-Like Receptors, and 5-Fluorouracil/Oxaliplatin-Induced Toxicity in Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 386. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, K.; Shi, C.; Li, G. Cancer Immunotherapy: Fecal Microbiota Transplantation Brings Light. Curr. Treat. Options Oncol. 2022, 23, 1777–1792. [Google Scholar] [CrossRef]
- Vongsavath, T.; Rahmani, R.; Tun, K.M.; Manne, V. The Use of Fecal Microbiota Transplant in Overcoming and Modulating Resistance to Anti-PD-1 Therapy in Patients with Skin Cancer. Cancers 2024, 16, 499. [Google Scholar] [CrossRef]
- Chen, D.; Wu, J.; Jin, D.; Wang, B.; Cao, H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int. J. Cancer 2019, 145, 2021–2031. [Google Scholar] [CrossRef] [PubMed]
- Le Bastard, Q.; Ward, T.; Sidiropoulos, D.; Hillmann, B.M.; Chun, C.L.; Sadowsky, M.J.; Knights, D.; Montassier, E. Fecal microbiota transplantation reverses antibiotic and chemotherapy-induced gut dysbiosis in mice. Sci. Rep. 2018, 8, 6219. [Google Scholar] [CrossRef] [PubMed]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021, 371, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, Y.; Zheng, Q.; Li, L. The Efficacy of Probiotics, Prebiotics, Synbiotics, and Fecal Microbiota Transplantation in Irritable Bowel Syndrome: A Systematic Review and Network Meta-Analysis. Nutrients 2024, 16, 2114. [Google Scholar] [CrossRef] [PubMed]
- Kaźmierczak-Siedlecka, K.; Daca, A.; Fic, M.; van de Wetering, T.; Folwarski, M.; Makarewicz, W. Therapeutic methods of gut microbiota modification in colorectal cancer management—Fecal microbiota transplantation, prebiotics, probiotics, and synbiotics. Gut Microbes 2020, 11, 1518–1530. [Google Scholar] [CrossRef]
- Ciernikova, S.; Sevcikova, A.; Drgona, L.; Mego, M. Modulating the gut microbiota by probiotics, prebiotics, postbiotics, and fecal microbiota transplantation: An emerging trend in cancer patient care. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2023, 1878, 188990. [Google Scholar] [CrossRef] [PubMed]
- Oprita, R.; Bratu, M.; Oprita, B.; Diaconescu, B. Fecal transplantation—The new, inexpensive, safe, and rapidly effective approach in the treatment of gastrointestinal tract diseases. J. Med. Life 2016, 9, 160–162. [Google Scholar]
- Kelly, C.R.; Yen, E.F.; Grinspan, A.M.; Kahn, S.A.; Atreja, A.; Lewis, J.D.; Moore, T.A.; Rubin, D.T.; Kim, A.M.; Serra, S.; et al. Fecal Microbiota Transplantation Is Highly Effective in Real-World Practice: Initial Results From the FMT National Registry. Gastroenterology 2021, 160, 183–192.e183. [Google Scholar] [CrossRef]
- Khanna, S.; Assi, M.; Lee, C.; Yoho, D.; Louie, T.; Knapple, W.; Aguilar, H.; Garcia-Diaz, J.; Wang, G.P.; Berry, S.M.; et al. Efficacy and Safety of RBX2660 in PUNCH CD3, a Phase III, Randomized, Double-Blind, Placebo-Controlled Trial with a Bayesian Primary Analysis for the Prevention of Recurrent Clostridioides difficile Infection. Drugs 2022, 82, 1527–1538. [Google Scholar] [CrossRef]
- Dubberke, E.R.; Lee, C.H.; Orenstein, R.; Khanna, S.; Hecht, G.; Gerding, D.N. Results From a Randomized, Placebo-Controlled Clinical Trial of a RBX2660—A Microbiota-Based Drug for the Prevention of Recurrent Clostridium difficile Infection. Clin. Infect. Dis. 2018, 67, 1198–1204. [Google Scholar] [CrossRef]
- Mullard, A. FDA approves second microbiome-based C. difficile therapy. Nat. Rev. Drug Discov. 2023, 23, 436. [Google Scholar] [CrossRef] [PubMed]
- Feuerstadt, P.; Louie, T.J.; Lashner, B.; Wang, E.E.; Diao, L.; Bryant, J.A.; Sims, M.; Kraft, C.S.; Cohen, S.H.; Berenson, C.S. SER-109, an oral microbiome therapy for recurrent Clostridioides difficile infection. N. Engl. J. Med. 2022, 386, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Ramai, D.; Zakhia, K.; Ofosu, A.; Ofori, E.; Reddy, M. Fecal microbiota transplantation: Donor relation, fresh or frozen, delivery methods, cost-effectiveness. Ann. Gastroenterol. 2019, 32, 30–38. [Google Scholar] [CrossRef]
- Aung, Y.Y.M.; Wong, D.C.S.; Ting, D.S.W. The promise of artificial intelligence: A review of the opportunities and challenges of artificial intelligence in healthcare. Br. Med. Bull. 2021, 139, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef]
- Kurant, D.E. Opportunities and Challenges with Artificial Intelligence in Genomics. Clin. Lab. Med. 2023, 43, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Lombard, V.; Henrissat, B. Complex carbohydrate utilization by the healthy human microbiome. PLoS ONE 2012, 7, e28742. [Google Scholar] [CrossRef]
- Halfvarson, J.; Brislawn, C.J.; Lamendella, R.; Vázquez-Baeza, Y.; Walters, W.A.; Bramer, L.M.; D’Amato, M.; Bonfiglio, F.; McDonald, D.; Gonzalez, A.; et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2017, 2, 17004. [Google Scholar] [CrossRef] [PubMed]
- Liechty, Z.S.; Agans, R.T.; Barbato, R.A.; Colston, S.M.; Christian, M.R.; Hammamieh, R.; Kardish, M.R.; Karl, J.P.; Leary, D.H.; Mauzy, C.A.; et al. Meeting report of the seventh annual Tri-Service Microbiome Consortium Symposium. BMC Proc. 2024, 18, 25. [Google Scholar] [CrossRef]
- Rabesandratana, T. Microbiome conservancy stores global fecal samples. Science 2018, 362, 510–511. [Google Scholar] [CrossRef]
- Carthey, A.J.; Blumstein, D.T.; Gallagher, R.V.; Tetu, S.G.; Gillings, M.R. Conserving the holobiont. Funct. Ecol. 2020, 34, 764–776. [Google Scholar] [CrossRef]
- Zimmer, A. Collect, preserve, cultivate intestinal microbiotes. A rescue biology. Ecol. Polit. 2019, 58, 135–150. [Google Scholar]
- Jain, N.; Umar, T.P.; Fahner, A.-F.; Gibietis, V. Advancing therapeutics for recurrent clostridioides difficile infections: An overview of vowst’s FDA approval and implications. Gut Microbes 2023, 15, 2232137. [Google Scholar] [CrossRef]
- Groussin, M.; Poyet, M.; Sistiaga, A.; Kearney, S.M.; Moniz, K.; Noel, M.; Hooker, J.; Gibbons, S.M.; Segurel, L.; Froment, A.; et al. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell 2021, 184, 2053–2067.e2018. [Google Scholar] [CrossRef] [PubMed]
- Schaan, A.P.; Vidal, A.; Zhang, A.-N.; Poyet, M.; Alm, E.J.; Groussin, M.; Ribeiro-dos-Santos, Â. Temporal dynamics of gut microbiomes in non-industrialized urban Amazonia. mSystems 2024, 9, e00707-23. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; DeMott, M.S.; Byrne, S.R.; Flores, K.; Poyet, M.; Groussin, M.; Microbiome Conservancy, G.; Berdy, B.; Comstock, L.; Alm, E.J.; et al. Phosphorothioate DNA modification by BREX Type 4 systems in the human gut microbiome. bioRxiv 2024. [Google Scholar] [CrossRef]
- Lokmer, A.; Aflalo, S.; Amougou, N.; Lafosse, S.; Froment, A.; Tabe, F.E.; Poyet, M.; Groussin, M.; Said-Mohamed, R.; Ségurel, L. Response of the human gut and saliva microbiome to urbanization in Cameroon. Sci. Rep. 2020, 10, 2856. [Google Scholar] [CrossRef]
- Melas, M.; Subbiah, S.; Saadat, S.; Rajurkar, S.; McDonnell, K.J. The Community Oncology and Academic Medical Center Alliance in the Age of Precision Medicine: Cancer Genetics and Genomics Considerations. J. Clin. Med. 2020, 9, 2125. [Google Scholar] [CrossRef]
- Bosserman, L.D.; Cianfrocca, M.; Yuh, B.; Yeon, C.; Chen, H.; Sentovich, S.; Polverini, A.; Zachariah, F.; Deaville, D.; Lee, A.B.; et al. Integrating Academic and Community Cancer Care and Research through Multidisciplinary Oncology Pathways for Value-Based Care: A Review and the City of Hope Experience. J. Clin. Med. 2021, 10, 188. [Google Scholar] [CrossRef]
- Karimi, M.; Wang, C.; Bahadini, B.; Hajjar, G.; Fakih, M. Integrating Academic and Community Practices in the Management of Colorectal Cancer: The City of Hope Model. J. Clin. Med. 2020, 9, 1687. [Google Scholar] [CrossRef]
- Presant, C.A.; Ashing, K.; Raz, D.; Yeung, S.; Gascon, B.; Stewart, A.; Macalintal, J.; Sandoval, A.; Ehrunmwunsee, L.; Phillips, T.; et al. Overcoming Barriers to Tobacco Cessation and Lung Cancer Screening among Racial and Ethnic Minority Groups and Underserved Patients in Academic Centers and Community Network Sites: The City of Hope Experience. J. Clin. Med. 2023, 12, 1275. [Google Scholar] [CrossRef] [PubMed]
- Villalona-Calero, M.A.; Malhotra, J.; Chung, V.; Xing, Y.; Gray, S.W.; Hampel, H.; Gruber, S.; McDonnell, K. Integrating Early-Stage Drug Development with Clinical Networks; Challenges and Opportunities: The City of Hope Developing Experience. J. Clin. Med. 2023, 12, 4061. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; González, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vázquez-Baeza, Y. QIIME 2 enables comprehensive end-to-end analysis of diverse microbiome data and comparative studies with publicly available data. Curr. Protoc. Bioinform. 2020, 70, e100. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.W.; Yost, S.E.; Lee, J.S.; Gillece, J.D.; Folkerts, M.; Reining, L.; Highlander, S.K.; Eftekhari, Z.; Mortimer, J.; Yuan, Y. Analysis of Gut Microbiome Using Explainable Machine Learning Predicts Risk of Diarrhea Associated With Tyrosine Kinase Inhibitor Neratinib: A Pilot Study. Front. Oncol. 2021, 11, 604584. [Google Scholar] [CrossRef]
- Murad, J.P.; Christian, L.; Yamaguchi, Y.; Lopez, L.; Ouyang, C.; Sato, H.; Jarman, B.; Stromberg, S.; Forman, S.J.; Van Dien, S.; et al. Abstract 6676: Microbiome modification impacts PSCA directed chimeric antigen receptor (CAR) T cell therapy for prostate cancer. Cancer Res. 2024, 84, 6676. [Google Scholar] [CrossRef]
- Gong, J.; Dizman, N.; Poroyko, V.; Won, H.; Bergerot, C.D.; Bergerot, P.G.; Maia, M.C.; Hsu, J.; Frankel, P.H.; Jones, J.; et al. Gut microbiome composition and response to sunitinib in metastatic renal cell carcinoma (mRCC). J. Clin. Oncol. 2018, 36, 657. [Google Scholar] [CrossRef]
- Meza, L.A.; Choi, Y.; Govindarajan, A.; Dizman, N.; Zengin, Z.B.; Hsu, J.; Salgia, N.; Salgia, S.; Malhotra, J.; Chawla, N.S.; et al. Association of intra-tumoral microbiome and response to immune checkpoint inhibitors (ICIs) in patients with metastatic renal cell carcinoma (mRCC). J. Clin. Oncol. 2022, 40, 372. [Google Scholar] [CrossRef]
- Meza, L.A.; Dizman, N.; Bergerot, P.G.; Dorff, T.B.; Lyou, Y.; Frankel, P.H.; Mira, V.; Llamas, M.; Hsu, J.; Zengin, Z.B.; et al. First results of a randomized phase IB study comparing nivolumab/ipilimumab with or without CBM-588 in patients with metastatic renal cell carcinoma. J. Clin. Oncol. 2021, 39, 4513. [Google Scholar] [CrossRef]
- Wong, C.W.; Yost, S.E.; Lee, J.S.; Highlander, S.K.; Yuan, Y. Abstract 336: Gut microbiome predicts response to CDK4/6 inhibitor and immune check point inhibitor combination in patients with hormone receptor positive metastatic breast cancer. Cancer Res. 2021, 81, 336. [Google Scholar] [CrossRef]
- Yi, Y.; Shen, L.; Shi, W.; Xia, F.; Zhang, H.; Wang, Y.; Zhang, J.; Wang, Y.; Sun, X.; Zhang, Z.; et al. Gut Microbiome Components Predict Response to Neoadjuvant Chemoradiotherapy in Patients with Locally Advanced Rectal Cancer: A Prospective, Longitudinal Study. Clin. Cancer Res. 2021, 27, 1329–1340. [Google Scholar] [CrossRef] [PubMed]
- Moshayedi, N.; Yang, J.; Lagishetty, V.; Jacobs, J.; Placencio-Hickok, V.; Osipov, A.; Hendifar, A.E.; Gong, J. Fecal microbiome composition in pancreatic cancer cachexia and response to nutrition support. J. Clin. Oncol. 2021, 39, 4129. [Google Scholar] [CrossRef]
- Zengin, Z.B.; Malhotra, J.; Salgia, S.K.; Dizman, N.; Yost, S.; Chawla, N.; Govindarajan, A.; Hsu, J.; Salgia, N.; Bergerot, P.G.; et al. 1759MO Associations between sarcopenia and gut microbiota in patients (pts) with metastatic renal cell carcinoma (mRCC) and breast cancer (BC). Ann. Oncol. 2021, 32, S1211. [Google Scholar] [CrossRef]
- Paredes, J.; Dai, A.; Ramos, R.J.F.; Adintori, P.; Fei, T.; Nguyen, C.; Elias, H.; Victor, K.; Ghale, R.; Pohl, C.; et al. Abstract 2794: Consumption of dietary fiber correlates with a significant increase in a-diversity in the intestinal Microbiome of allo-HCT recipients and with lower GVHD-lethality in pre-clinical models. Cancer Res. 2024, 84, 2794. [Google Scholar] [CrossRef]
- Huang, N.; Hwang, A.; Lagishetty, V.; Epeldegui, M.; Yu, Y.; Buchanan, L.; Yu, G.; Nathwani, B.N.; Millstein, J.; Mack, T.M.; et al. Differences in Fecal Microbiota in Long-Term Adolscent/Young Adult Hodgkin Lymphoma Survivors and Their Unaffected Twins. Blood 2017, 130, 4084. [Google Scholar] [CrossRef]
- Cozen, W.; Hamilton, A.; Salam, M.; Deapen, D.; Nathwani, B.; Weiss, L.; Mack, T. Abstract #2146: Environmental exposures associated with increased Th2 cytokines may increase risk of young adult Hodgkin’s lymphoma. Cancer Res. 2009, 69, 2146. [Google Scholar]
- NIH Report. Available online: https://report.nih.gov (accessed on 20 July 2024).
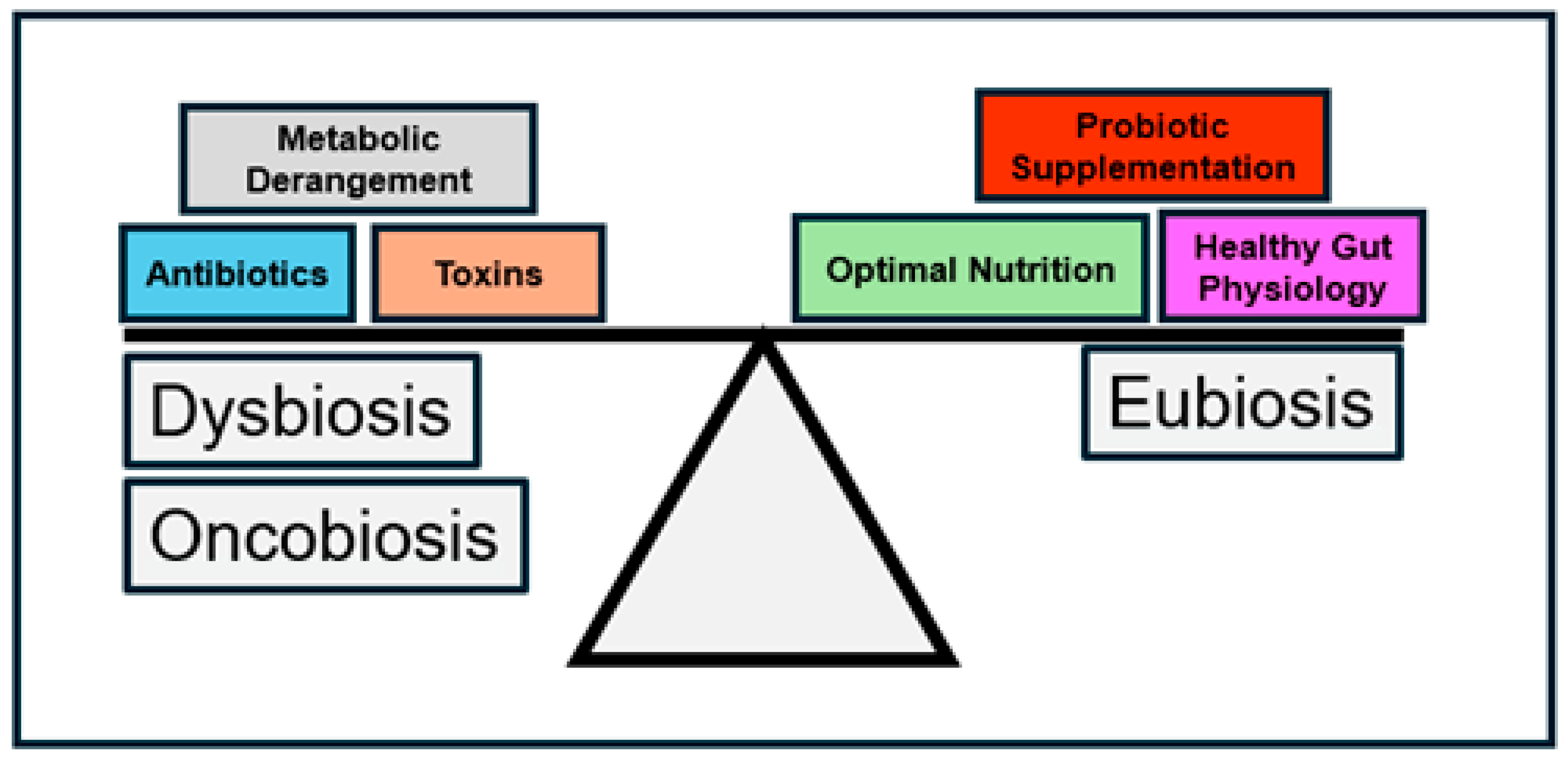
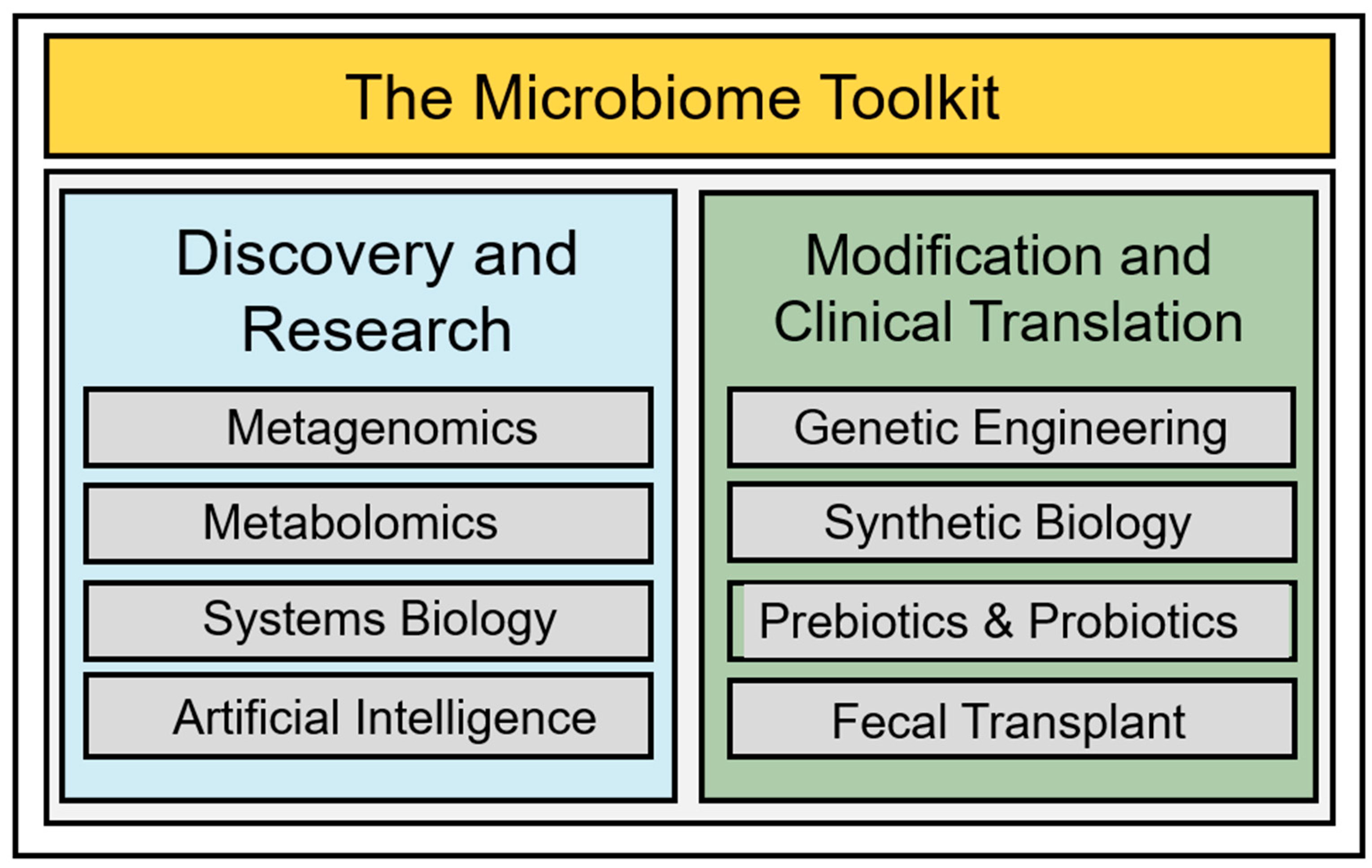

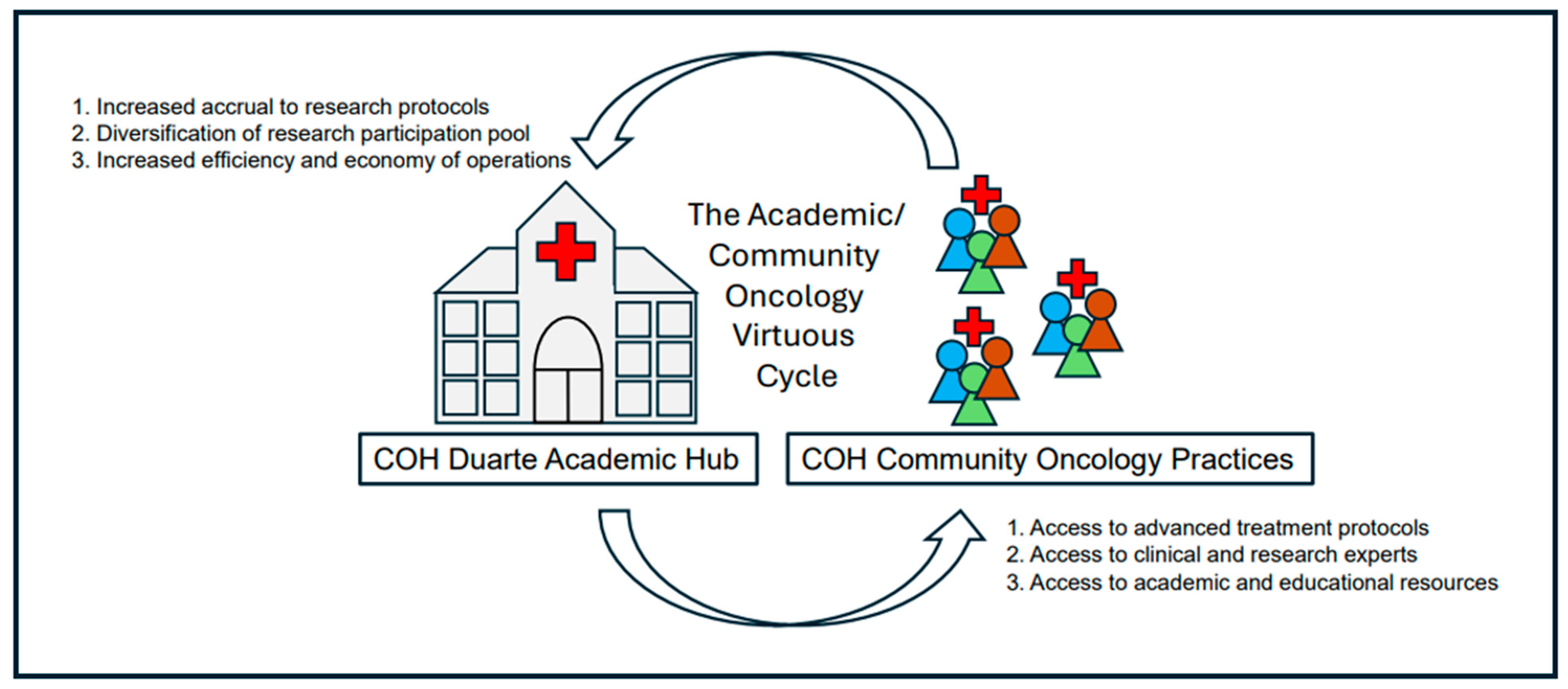
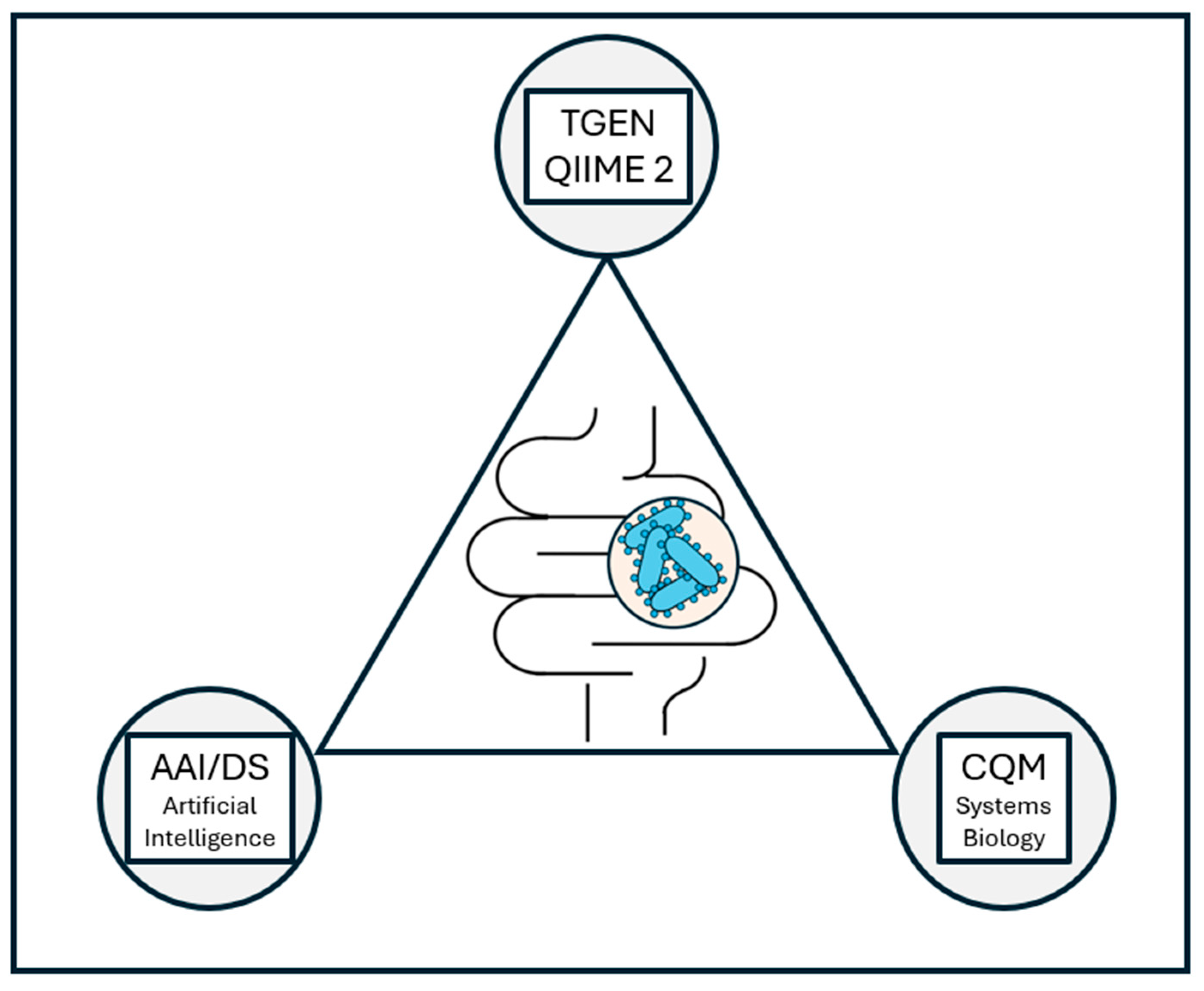
| Clinical Trial | NCT Identifier | Status |
|---|---|---|
| Study to Detect Changes in Urinary and Gut Microbiome During Androgen Deprivation Therapy and Radiation Therapy in Patients with Prostate Cancer | NCT04775355 | Recruiting |
| CBM588 in Combination with Nivolumab and Cabozantinib for the Treatment of Advanced or Metastatic Kidney Cancer | NCT05122546 | Active, not recruiting |
| TAC/MTX vs. TAC/MMF/PTCY for Prevention of Graft-versus-Host Disease and Microbiome and Immune Reconstitution Study (BMT CTN 1703/1801) | NCT03959241 | Completed |
| A Multiple Dose Study to Evaluate Safety, Tolerability, PK, and Efficacy of SER-155 in Adults Undergoing HSCT | NCT04995653 | Active, not recruiting |
| CBM588 Capsules in Combination with Nivolumab and Ipilimumab for the Treatment of Advanced Stage Kidney Cancer | NCT06399419 | Recruiting |
| Adding Itacitinib to Cyclophosphamide and Tacrolimus for the Prevention of Graft Versus Host Disease in Patients Undergoing Hematopoietic Stem Cell Transplants | NCT05364762 | Recruiting |
| CBM588 in Improving Clinical Outcomes in Patients Who Have Undergone Donor Hematopoietic Stem Cell Transplant | NCT03922035 | Active, not recruiting |
| Minnelide and Osimertinib for the Treatment of Advanced EGFR Mutated Non-Small Cell Lung Cancer | NCT05166616 | Recruiting |
| Atezolizumab, Guadecitabine, and CDX-1401 Vaccine in Treating Patients with Recurrent Ovarian, Fallopian Tube, or Primary Peritoneal Cancer | NCT03206047 | Active, not recruiting |
| Probiotic Yogurt Supplement in Reducing Diarrhea in Patients with Metastatic Kidney Cancer Being Treated With Vascular Endothelial Growth Factor-Tyrosine Kinase Inhibitor | NCT02944617 | Completed |
| CBM588, Nivolumab, and Ipilimumab in Treating Patients with Stage IV or Advanced Kidney Cancer | NCT03829111 | Completed |
| Pembrolizumab, Endocrine Therapy, and Palbociclib in Treating Postmenopausal Patients with Newly Diagnosed Metastatic Stage IV Estrogen Receptor Positive Breast Cancer | NCT02778685 | Recruiting |
| Study of IL-22 IgG2-Fc (F-652) for Subjects with Grade II–IV Lower GI aGVHD | NCT02406651 | Completed |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McDonnell, K.J. Operationalizing Team Science at the Academic Cancer Center Network to Unveil the Structure and Function of the Gut Microbiome. J. Clin. Med. 2025, 14, 2040. https://doi.org/10.3390/jcm14062040
McDonnell KJ. Operationalizing Team Science at the Academic Cancer Center Network to Unveil the Structure and Function of the Gut Microbiome. Journal of Clinical Medicine. 2025; 14(6):2040. https://doi.org/10.3390/jcm14062040
Chicago/Turabian StyleMcDonnell, Kevin J. 2025. "Operationalizing Team Science at the Academic Cancer Center Network to Unveil the Structure and Function of the Gut Microbiome" Journal of Clinical Medicine 14, no. 6: 2040. https://doi.org/10.3390/jcm14062040
APA StyleMcDonnell, K. J. (2025). Operationalizing Team Science at the Academic Cancer Center Network to Unveil the Structure and Function of the Gut Microbiome. Journal of Clinical Medicine, 14(6), 2040. https://doi.org/10.3390/jcm14062040




