Association of MTHFR and DNMT-1 Gene Polymorphisms with Acute Coronary Syndrome in Patients Admitted to the Emergency Department
Abstract
1. Introduction
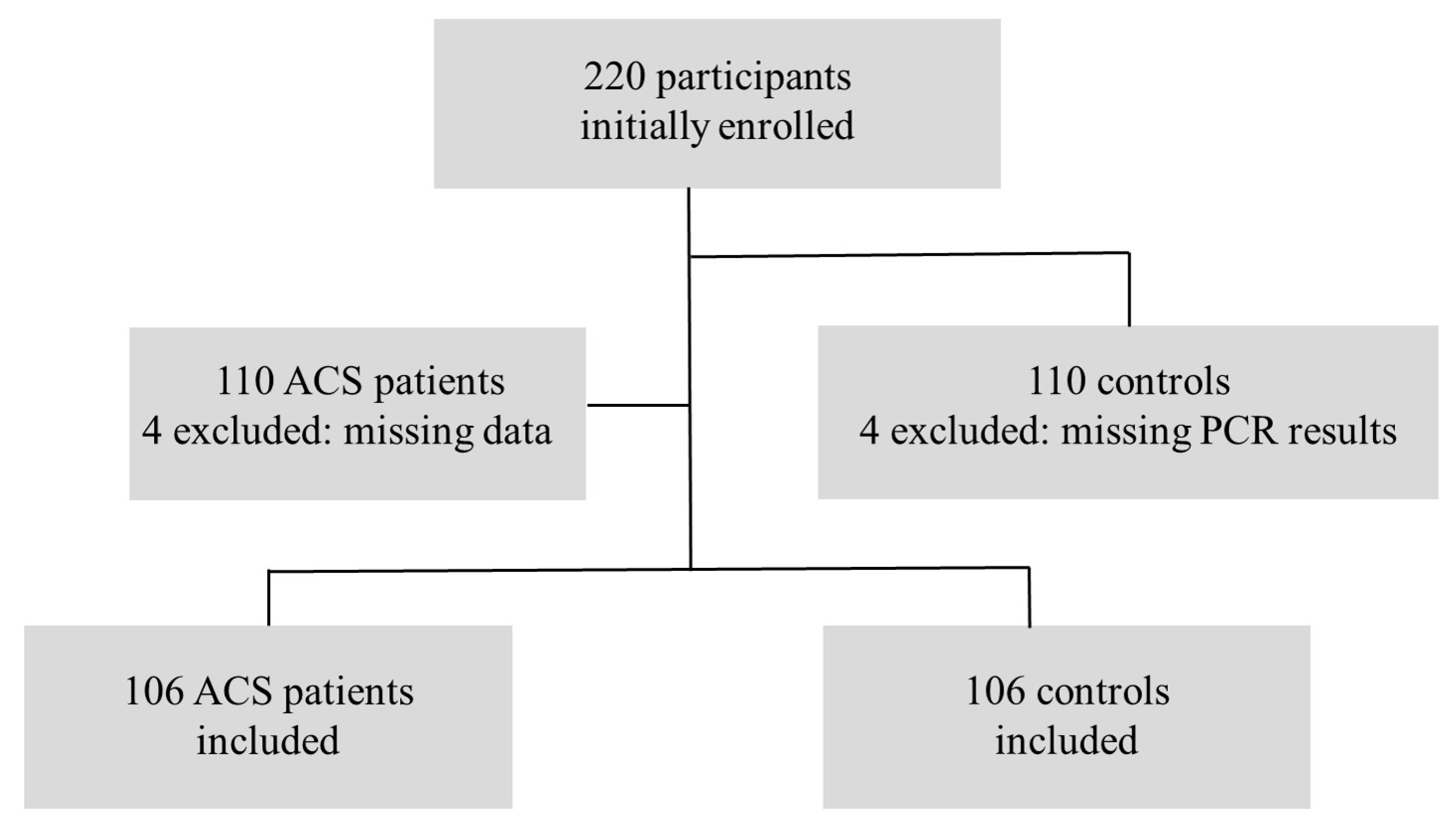
2. Materials and Methods
2.1. Study Design
2.2. Patient Selection
2.3. Data Collection
2.4. DNA Isolation and PCR-RFLP Methods
2.5. Sample Size
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACS | Acute coronary syndrome |
| MTHFR | Methylenetetrahydrofolate reductase |
| DNMT-1 | Deoxyribonucleic acid methyltransferase-1 |
| PCR | Polymerase chain reaction |
| RFLP | Restriction fragment length polymorphism |
| DNA | Deoxyribonucleic acid |
| OR | Odds ratio |
| CI | Confidence intervals |
| SCD | Sudden cardiac death |
| CAD | Coronary artery disease |
| ECG | Electrocardiogram |
| EDTA | Ethylenediaminetetraacetic acid |
| AST | Aspartate aminotransferase |
| ALT | Alanine aminotransferase |
| STE | ST-segment elevation |
| NSTEMI | Non-ST elevation myocardial infarction |
| STEMI | ST-elevation myocardial infarction |
| ESC | European Society of Cardiology |
| AHA | American Heart Association |
| ACC | American College of Cardiology |
| cTn | Cardiac troponin |
| SD | Standard deviation |
| ANOVA | One-way analysis of variance |
| χ2 | Chi-square |
References
- Wong, C.X.; Brown, A.; Lau, D.H.; Chugh, S.S.; Albert, C.M.; Kalman, J.M.; Sanders, P. Epidemiology of Sudden Cardiac Death: Global and Regional Perspectives. Heart Lung Circ. 2019, 28, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Avishay, D.M.; Jones, C.R.; Shaikh, J.D.; Kaur, R.; Aljadah, M.; Kichloo, A.; Shiwalkar, N.; Keshavamurthy, S. Sudden Cardiac Death: Epidemiology, Pathogenesis and Management. Rev. Cardiovasc. Med. 2021, 22, 147–158. [Google Scholar] [CrossRef]
- Święczkowski, M.; Dobrzycki, S.; Kuźma, Ł. Multi-City Analysis of the Acute Effect of Polish Smog on Cause-Specific Mortality (EP-PARTICLES Study). Int. J. Environ. Res. Public Health 2023, 20, 5566. [Google Scholar] [CrossRef] [PubMed]
- Wołowiec, A.; Wołowiec, Ł.; Grześk, G.; Jaśniak, A.; Osiak, J.; Husejko, J.; Kozakiewicz, M. The Role of Selected Epigenetic Pathways in Cardiovascular Diseases as a Potential Therapeutic Target. Int. J. Mol. Sci. 2023, 24, 13723. [Google Scholar] [CrossRef]
- Rosenblatt, D.S. Methylenetetrahydrofolate Reductase. Clin. Investig. Med. 2001, 24, 56–59. [Google Scholar]
- Osian, G.; Procopciuc, L.; Vlad, L.; Iancu, C.; Mocan, T.; Mocan, L. C677T and A1298C Mutations in the MTHFR Gene and Survival in Colorectal Cancer. J. Gastrointestin. Liver Dis. 2009, 18, 455–460. [Google Scholar]
- Lee, H.A.; Park, E.A.; Cho, S.J.; Kim, H.S.; Kim, Y.J.; Lee, H.; Gwak, H.S.; Kim, K.N.; Chang, N.; Ha, E.H.; et al. Mendelian Randomization Analysis of the Effect of Maternal Homocysteine During Pregnancy, as Represented by Maternal MTHFR C677T Genotype, on Birth Weight. J. Epidemiol. 2013, 23, 371–375. [Google Scholar] [CrossRef]
- Sunder-Plassmann, G.; Födinger, M. Genetic Determinants of the Homocysteine Level. Kidney Int. Suppl. 2003, 84, 141–144. [Google Scholar] [CrossRef]
- Christen, W.G.; Ajani, U.A.; Glynn, R.J.; Hennekens, C.H. Blood Levels of Homocysteine and Increased Risks of Cardiovascular Disease: Causal or Casual? Arch. Intern. Med. 2000, 160, 422–434. [Google Scholar] [CrossRef]
- Cortese, C.; Motti, C. MTHFR gene polymorphism, homocysteine and cardiovascular disease. Public Health Nutr. 2001, 4, 493–497. [Google Scholar] [CrossRef]
- Costello, J.F.; Plass, C. Methylation matters. J. Med. Genet. 2001, 38, 285–303. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Qiu, Y.; Yang, J.; Bian, S.; Chen, G.; Deng, M.; Kang, H.; Huang, L. DNMT1-PPARγ pathway in macrophages regulates chronic inflammation and atherosclerosis development in mice. Sci. Rep. 2016, 6, 30053. [Google Scholar] [CrossRef] [PubMed]
- Ghaznavi, H.; Mahmoodi, K.; Soltanpour, M.S. A preliminary study of the association between the ABCA1 gene promoter DNA methylation and coronary artery disease risk. Mol. Biol. Res. Commun. 2018, 7, 59–65. [Google Scholar] [PubMed]
- Zaina, S.; Heyn, H.; Carmona, F.J.; Varol, N.; Sayols, S.; Condom, E.; Ramírez-Ruz, J.; Gomez, A.; Gonçalves, I.; Moran, S.; et al. DNA methylation map of human atherosclerosis. Circ. Cardiovasc. Genet. 2014, 7, 692–700. [Google Scholar] [CrossRef]
- Damluji, A.A.; Forman, D.E.; Wang, T.Y.; Chikwe, J.; Kunadian, V.; Rich, M.W.; Young, B.A.; Page, R.L., 2nd; DeVon, H.A.; Alexander, K.P.; et al. Management of Acute Coronary Syndrome in the Older Adult Population: A Scientific Statement From the American Heart Association. Circulation 2023, 147, e32–e62. [Google Scholar] [CrossRef]
- Gulati, M.; Levy, P.D.; Mukherjee, D.; Amsterdam, E.; Bhatt, D.L.; Birtcher, K.K.; Blankstein, R.; Boyd, J.; Bullock-Palmer, R.P.; Conejo, T.; et al. 2021 AHA/ACC/ASE/CHEST/SAEM/SCCT/SCMR Guideline for the Evaluation and Diagnosis of Chest Pain: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2021, 144, e368–e454. [Google Scholar]
- Feng, L.G.; Song, Z.W.; Xin, F.; Hu, J. Association of Plasma Homocysteine and Methylenetetrahydrofolate Reductase C677T Gene Variant with Schizophrenia: A Chinese Han Population-Based Case-Control Study. Psychiatry Res. 2009, 168, 205–208. [Google Scholar] [CrossRef]
- Yang, S.M.; Huang, C.Y.; Shiue, H.S.; Pu, Y.S.; Hsieh, Y.H.; Chen, W.J.; Lin, Y.C.; Hsueh, Y.M. Combined Effects of DNA Methyltransferase 1 and 3A Polymorphisms and Urinary Total Arsenic Levels on the Risk for Clear Cell Renal Cell Carcinoma. Toxicol. Appl. Pharmacol. 2016, 305, 103–110. [Google Scholar] [CrossRef]
- Li, M.N.; Wang, H.J.; Zhang, N.R.; Xuan, L.; Shi, X.J.; Zhou, T.; Chen, B.; Zhang, J.; Li, H. MTHFR C677T gene polymorphism and the severity of coronary lesions in acute coronary syndrome. Medicine 2017, 96, e9044. [Google Scholar] [CrossRef]
- Lakkakula, B.V.K.S. Association between MTHFR 677C>T polymorphism and vascular complications in sickle cell disease: A meta-analysis. Transfus. Clin. Biol. J. Soc. Fr. Transfus. Sang. 2019, 26, 284–288. [Google Scholar] [CrossRef]
- Frosst, P.; Blom, H.J.; Milos, R.; Goyette, P.; Sheppard, C.A.; Matthews, R.G.; Boers, G.J.; den Heijer, M.; Kluijtmans, L.A.; van den Heuvel, L.P. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995, 10, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Homocysteine Studies Collaboration. Homocysteine and risk of ischemic heart disease and stroke: A meta-analysis. JAMA 2002, 288, 2015–2022. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Deng, Q.; Li, Z.; Xiong, C.; Li, C.; Zheng, F. Risk-Association of DNMT1 Gene Polymorphisms with Coronary Artery Disease in Chinese Han Population. Int. J. Mol. Sci. 2014, 15, 22694–22705. [Google Scholar] [CrossRef]
- Xu, H.; Liu, C.; Wang, Q. Plaque Image Characteristics, Hyperhomocysteinemia, and Gene Polymorphism of Homocysteine Metabolism-Related Enzyme (MTHFR C677T) in Acute Coronary Syndrome. Cell Biochem. Biophys. 2013, 66, 403–407. [Google Scholar] [CrossRef]
- Chen, H.L.; Li, Z.M.; Liu, J.F.; Han, B.; Wu, Z.X.; Mao, Y.Q.; Sun, K.Y.; Wang, L.S. Polymorphism of the DNA methyltransferase 1 gene is associated with the susceptibility to essential hypertension in male. Clin. Exp. Hypertens. 2018, 40, 695–701. [Google Scholar] [CrossRef]
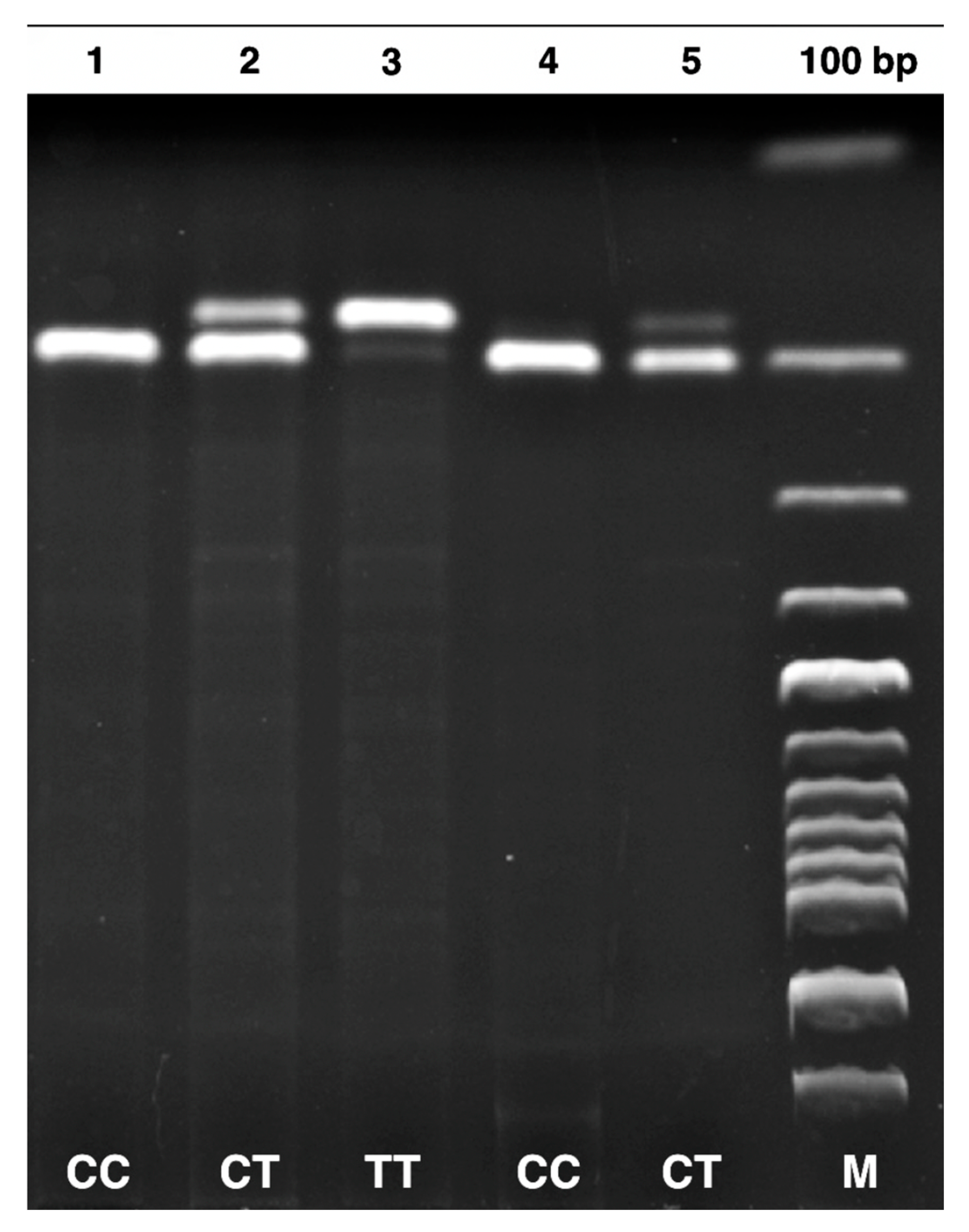

| SNP | Prime Sequence (5′-3′) | Tm (°C) | PCR Product Size | Restriction Enzyme | Genotyping (bp) |
|---|---|---|---|---|---|
| MTHFR 677 C/T (rs1801133) | F-TGAAGGAGAAGGTGTCTGCGGGA R-AGGACGGTGCGGTGAGAGTG | 61 °C | 198 bp | HinfI | CC: 198 CT: 175–198 TT: 175 |
| DNMT-1 +32204 A/G (rs2228611) | F-CTGATCTGAAGTCTGCACGAAG R-TCCAGGCTCGTCTCAAACTC | 58 °C | 329 bp | Alw26I (BsmAI) | GG: 143–186 GA: 143–186–329 AA: 329 |
| Characteristic | Groups | p-Value | |
|---|---|---|---|
| Control (n = 106) | ACS (n = 106) | ||
| Age | 51.0 ± 19.1 | 61.5 ± 14.1 | <0.001 * |
| Sex | |||
| Female [n (%)] | 42 (51.9) | 39 (48.1) | 0.778 |
| Male [n (%)] | 64 (48.9) | 67 (51.1) | |
| Glucose (mg/dL) | 122.1 ± 62.6 | 158.1 ± 76.8 | <0.001 * |
| BUN (mg/dL) | 36.2 ± 22.0 | 40.9 ± 24.3 | 0.138 |
| Creatine (mg/dL) | 1.04 ± 0.90 | 1.03 ± 0.51 | 0.938 |
| Sodium (mmol/L) | 138.7 ± 2.81 | 138.1 ± 3.68 | 0.218 |
| Potassium (mmol/L) | 4.22 ± 0.55 | 4.45 ± 0.58 | 0.004 * |
| ALT (U/L) | 27.7 ± 48.5 | 34.2 ± 58.1 | 0.375 |
| AST (U/L) | 27.1 ± 28.6 | 53.9 ± 88.7 | 0.003 * |
| Troponin I (ng/L) | |||
| <100 [n (%)] | 88 (79.3) | 23 (20.7) | 0.000 * |
| >100 [n (%)] | 18 (17.8) | 83 (82.2) | |
| Polymorphic Site | Genotype/Allele | Control (n = 106) | ACS (n = 106) | OR (95% CI) | p–Value |
|---|---|---|---|---|---|
| MTHFR 677 C/T (rs1801133) | CC | 33 (36.7%) | 57 (63.3%) | 7.34 (2.28–23.6) | 0.000 * |
| CT | 56 (55.4%) | 45 (44.6%) | 3.42 (1.07–10.8) | 0.03 * | |
| TT | 17 (81.0%) | 4 (19.0%) | 1 (reference) | ||
| χ2 = 15.6 df = 2 p = 0.000 * | |||||
| C | 122 (43.4%) | 159 (56.6%) | 2.21 (1.46–3.35) | 0.000 * | |
| T | 90 (62.9%) | 53 (37.1%) | 1 (reference) | ||
| χ2 = 14.4 df = 1 p = 0.000 * | |||||
| DNMT-1 +32204 A/G (rs2228611) | GG | 33 (57.9%) | 24 (42.1%) | 1 (reference) | |
| GA | 48 (47.5%) | 53 (52.5%) | 1.52 (0.79–2.92) | 0.210 | |
| AA | 25 (46.3%) | 29 (53.7%) | 1.60 (0.75–3.38) | 0.221 | |
| χ2 = 1.96 df = 2 p = 0.374 | |||||
| G | 114 (53.0%) | 101 (47.0%) | 1 (reference) | ||
| A | 98 (46.9%) | 111 (53.1%) | 1.28 (0.87–1.87) | 0.207 | |
| χ2 = 1.59 df = 1 p = 0.207 | |||||
| Characteristic | MTHFR 677 C/T Genotypes | p-Value | DNMT-1 +32204 A/G Genotypes | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | GG | GA | AA | |||
| Age | 51.6 ± 20.2 | 50.4 ± 18.2 | 51.8 ± 20.7 | 0.944 | 47.3 ± 17.2 | 51.7 ± 17.2 | 54.8 ± 24.2 | 0.322 |
| Sex | ||||||||
| Female [n (%)] | 10 (30.3) | 25 (44.6) | 7 (41.2) | 0.406 | 15 (45.5) | 16 (33.3) | 11 (44.0) | 0.481 |
| Male [n (%)] | 23 (69.7) | 31 (55.4) | 10 (58.8) | 18 (54.5) | 32 (66.7) | 14 (56.0) | ||
| Glucose (mg/dL) | 121.7 ± 71.7 | 121.9 ± 51.2 | 123.4 ± 80.1 | 0.996 | 112.2 ± 47.1 | 130.8 ± 70.9 | 118.4 ± 63.4 | 0.401 |
| BUN (mg/dL) | 37.0 ± 16.7 | 36.6 ± 26.7 | 33.0 ± 13.2 | 0.815 | 29.9 ± 13.0 | 38.8 ± 26.2 | 39.4 ± 21.8 | 0.145 |
| Creatine (mg/dL) | 1.00 ± 0.36 | 1.10 ± 1.20 | 0.93 ± 0.34 | 0.749 | 0.87 ± 0.30 | 1.18 ± 1.28 | 1.01 ± 0.40 | 0.328 |
| Sodium (mmol/L) | 139.1± 2.16 | 138.5 ± 3.25 | 138.7 ± 2.44 | 0.625 | 138.6 ± 2.17 | 138.4 ± 2.53 | 139.4 ± 3.89 | 0.354 |
| Potassium (mmol/L) | 4.29 ± 0.46 | 4.22 ± 0.62 | 4.07 ± 0.44 | 0.423 | 4.22 ± 0.44 | 4.19 ± 0.49 | 4.29 ± 0.76 | 0.766 |
| ALT (U/L) | 22.9 ± 12.6 | 32.2 ± 65.1 | 22.1 ± 20.6 | 0.602 | 33.7 ± 62.9 | 29.1 ± 49.3 | 16.9 ± 9.49 | 0.416 |
| AST (U/L) | 24.3 ± 9.72 | 30.5 ± 38.3 | 21.1 ± 8.57 | 0.401 | 31.7 ± 34.5 | 27.2 ± 31.1 | 20.7 ± 6.45 | 0.357 |
| Troponin I (ng/L) | ||||||||
| <100 [n (%)] | 27 (81.8) | 46 (82.1) | 15 (88.2) | 0.822 | 28 (84.8) | 41 (85.4) | 19 (76.0) | 0.563 |
| >100 [n (%)] | 6 (18.2) | 10 (17.9) | 2 (11.8) | 5 (15.2) | 7 (14.6) | 6 (24.0) | ||
| Characteristic | MTHFR 677 C/T Genotypes | p-Value | DNMT-1 +32204 A/G Genotypes | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | GG | GA | AA | |||
| Age | 62.1 ± 12.8 | 61.4 ± 15.6 | 54.2 ± 16.8 | 0.560 | 59.0 ± 13.6 | 63.5 ± 12.8 | 60.1 ± 16.7 | 0.348 |
| Sex | ||||||||
| Female [n (%)] | 20 (35.1) | 19 (42.2) | 0 (0) | 0.226 | 7 (29.2) | 24 (45.3) | 8 (27.6) | 0.192 |
| Male [n (%)] | 37 (64.9) | 26 (57.8) | 4 (100) | 17 (70.8) | 29 (54.7) | 21 (72.4) | ||
| Glucose (mg/dL) | 146.4 ± 56.2 | 168.7 ± 92.3 | 207.0 ± 122.5 | 0.150 | 144.7 ± 68.9 | 164.1 ± 87.0 | 158.3 ± 62.9 | 0.597 |
| BUN (mg/dL) | 38.1 ± 23.6 | 45.0 ± 25.8 | 35.1 ± 4.59 | 0.322 | 39.8 ± 26.0 | 41.7 ± 19.4 | 40.5 ± 30.9 | 0.945 |
| Creatine (mg/dL) | 0.99 ± 0.37 | 1.10 ± 0.66 | 1.04 ± 0.23 | 0.564 | 1.06 ± 0.65 | 1.05 ± 0.50 | 0.99 ± 0.40 | 0.835 |
| Sodium (mmol/L) | 138.3 ± 3.01 | 137.6 ± 4.26 | 141.0 ± 4.83 | 0.193 | 137.8 ± 2.98 | 138.4 ± 4.09 | 137.9 ± 3.47 | 0.744 |
| Potassium (mmol/L) | 4.36 ± 0.55 | 4.57 ± 0.61 | 4.42 ± 0.51 | 0207 | 4.42 ± 0.58 | 4.52 ± 0.55 | 4.35 ± 0.65 | 0.462 |
| ALT (U/L) | 29.6 ± 48.9 | 38.8 ± 69.9 | 47.2 ± 30.4 | 0.661 | 23.7 ± 9.91 | 43.7 ± 80.1 | 25.6 ± 18.4 | 0.246 |
| AST (U/L) | 42.6 ± 53.2 | 67.0 ± 121.2 | 67.7 ± 43.2 | 0.373 | 43.5 ± 35.0 | 119.2 ± 16.3 | 37.1 ± 6.90 | 0.256 |
| Troponin I (ng/L) | ||||||||
| <100 [n (%)] | 18 (31.6) | 5 (11.1) | 0 (0) | 0.025 * | 5 (20.8) | 11 (20.8) | 7 (24.1) | 0.932 |
| >100 [n (%)] | 39 (68.4) | 40 (88.9) | 83 (100) | 19 (79.2) | 42 (79.2) | 22 (75.9) | ||
| Diagnosis | ||||||||
| STEMI [n (%)] | 11 (19.3) | 7 (15.6) | 1 (25) | 0.321 | 4 (13.3) | 9 (16.9) | 6 (20.7) | 0.434 |
| NSTEMI [n (%)] | 35 (61.4) | 35 (77.8) | 2 (50) | 13 (63.3) | 39 (73.6) | 20 (69.0) | ||
| Unstable angina [n (%)] | 11 (19.3) | 3 (6.6) | 1 (25) | 7 (23.6) | 5 (9.5) | 3 (10.3) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yukcu, F.; Kaya, M.; Akcilar, R.; Can, F.; Yildirim, H. Association of MTHFR and DNMT-1 Gene Polymorphisms with Acute Coronary Syndrome in Patients Admitted to the Emergency Department. J. Clin. Med. 2025, 14, 2767. https://doi.org/10.3390/jcm14082767
Yukcu F, Kaya M, Akcilar R, Can F, Yildirim H. Association of MTHFR and DNMT-1 Gene Polymorphisms with Acute Coronary Syndrome in Patients Admitted to the Emergency Department. Journal of Clinical Medicine. 2025; 14(8):2767. https://doi.org/10.3390/jcm14082767
Chicago/Turabian StyleYukcu, Fulya, Murtaza Kaya, Raziye Akcilar, Fatmagul Can, and Harun Yildirim. 2025. "Association of MTHFR and DNMT-1 Gene Polymorphisms with Acute Coronary Syndrome in Patients Admitted to the Emergency Department" Journal of Clinical Medicine 14, no. 8: 2767. https://doi.org/10.3390/jcm14082767
APA StyleYukcu, F., Kaya, M., Akcilar, R., Can, F., & Yildirim, H. (2025). Association of MTHFR and DNMT-1 Gene Polymorphisms with Acute Coronary Syndrome in Patients Admitted to the Emergency Department. Journal of Clinical Medicine, 14(8), 2767. https://doi.org/10.3390/jcm14082767





