Comparative Gene Expression Profiles in Parathyroid Adenoma and Normal Parathyroid Tissue
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Acquisition
2.2. RNA Sequencing
2.3. Identification of DEGs
2.4. Functional and KEGG Pathway Enrichment Analysis
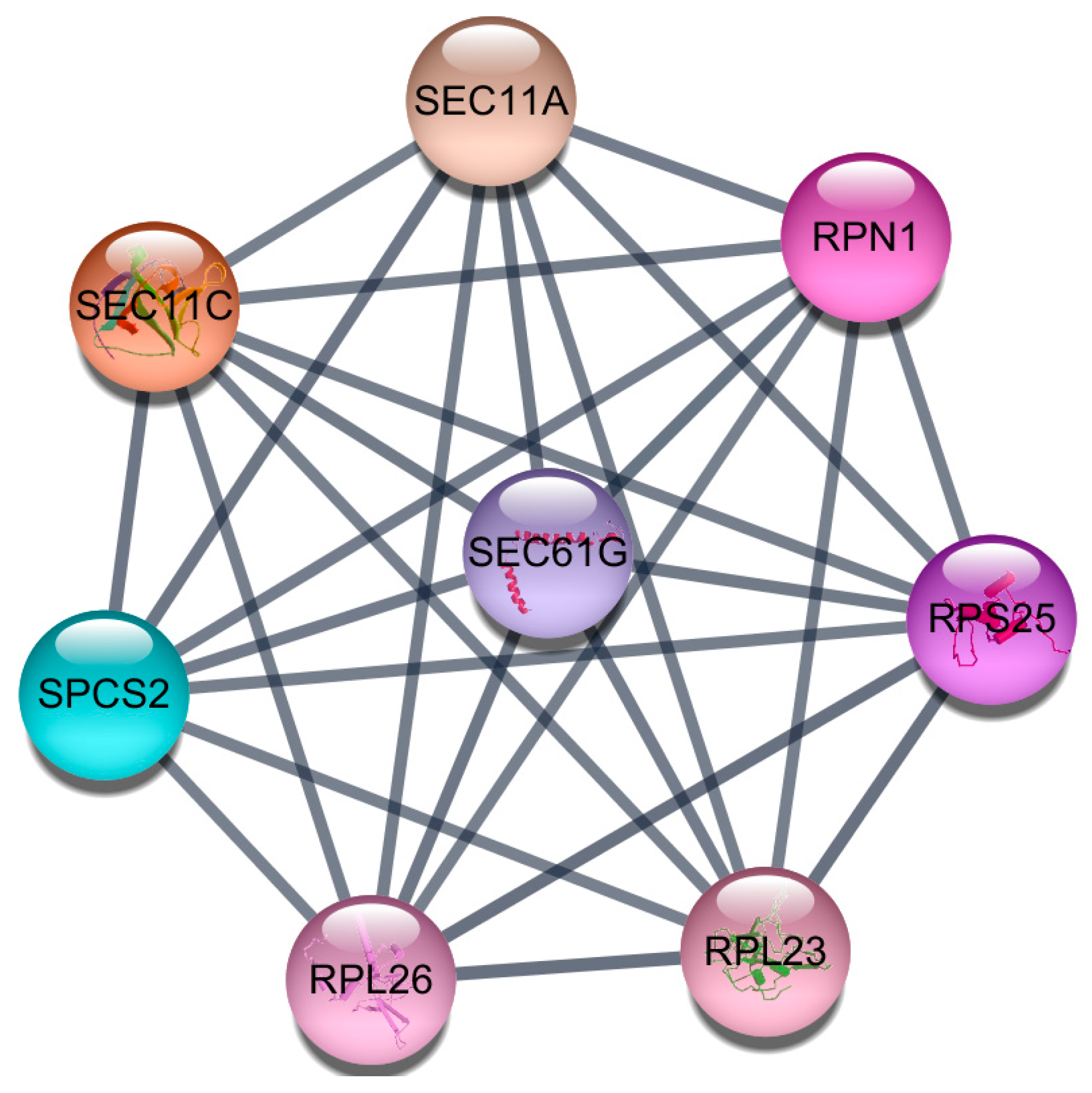
2.5. Protein-Protein Interaction (PPI) Network Construction and Module Analysis
3. Results
3.1. Characteristics of Study Subjects and Mutation Screening
3.2. Genes Differentially Expressed between PHPT and Normal Parathyroid Tissue
3.3. Gene Ontology (GO) Functional and KEGG Pathway Enrichment Analysis
3.4. Protein-Protein Interaction (PPI) Network Construction and Module Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Marcocci, C.; Cetani, F. Clinical practice. Primary hyperparathyroidism. N. Engl. J. Med. 2011, 365, 2389–2397. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Chai, Y.J.; Chung, J.K.; Hwang, K.T.; Heo, S.C.; Kim, S.J.; Choi, J.Y.; Yi, K.H.; Kim, S.W.; Cho, S.Y.; et al. The prevalence of primary hyperparathyroidism in Korea: A population-based analysis from patient medical records. Ann. Surg. Treat. Res. 2018, 94, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Wermers, R.A.; Khosla, S.; Atkinson, E.J.; Achenbach, S.J.; Oberg, A.L.; Grant, C.S.; Melton, L.J., III. Incidence of primary hyperparathyroidism in Rochester, Minnesota, 1993–2001: An update on the changing epidemiology of the disease. J. Bone Miner. Res. 2006, 21, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Wermers, R.A.; Khosla, S.; Atkinson, E.J.; Hodgson, S.F.; O’Fallon, W.M.; Melton, L.J., III. The rise and fall of primary hyperparathyroidism: A population-based study in Rochester, Minnesota, 1965–1992. Ann. Intern. Med. 1997, 126, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Yeh, M.W.; Ituarte, P.H.; Zhou, H.C.; Nishimoto, S.; Liu, I.L.; Harari, A.; Haigh, P.I.; Adams, A.L. Incidence and prevalence of primary hyperparathyroidism in a racially mixed population. J. Clin. Endocrinol. Metab. 2013, 98, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Donnan, P.T.; Murphy, M.J.; Leese, G.P. Epidemiology of primary hyperparathyroidism in Tayside, Scotland, UK. Clin. Endocrinol. (Oxf) 2009, 71, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, P.; Sadler, G.P.; Mihai, R. Persistent symptomatic improvement in the majority of patients undergoing parathyroidectomy for primary hyperparathyroidism. Langenbecks Arch. Surg. 2010, 395, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Romano, R.; Ellis, S.; Yu, N.; Bellizzi, J.; Brown, T.C.; Korah, R.; Carling, T.; Costa-Guda, J.; Arnold, A. Mutational Analysis of ZFY in Sporadic Parathyroid Adenomas. J. Endocr. Soc. 2017, 1, 313–316. [Google Scholar] [PubMed]
- Stalberg, P.; Carling, T. Familial parathyroid tumors: Diagnosis and management. World J. Surg. 2009, 33, 2234–2243. [Google Scholar] [CrossRef] [PubMed]
- Starker, L.F.; Delgado-Verdugo, A.; Udelsman, R.; Bjorklund, P.; Carling, T. Expression and somatic mutations of SDHAF2 (SDH5), a novel endocrine tumor suppressor gene in parathyroid tumors of primary hyperparathyroidism. Endocrine 2010, 38, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Cromer, M.K.; Starker, L.F.; Choi, M.; Udelsman, R.; Nelson-Williams, C.; Lifton, R.P.; Carling, T. Identification of somatic mutations in parathyroid tumors using whole-exome sequencing. J. Clin. Endocrinol. Metab. 2012, 97, E1774–E1781. [Google Scholar] [CrossRef] [PubMed]
- Farnebo, F.; The, B.T.; Kytola, S.; Svensson, A.; Phelan, C.; Sandelin, K.; Thompson, N.W.; Hoog, A.; Weber, G.; Farnebo, L.O.; et al. Alterations of the MEN1 gene in sporadic parathyroid tumors. J. Clin. Endocrinol. Metab. 1998, 83, 2627–2630. [Google Scholar] [CrossRef] [PubMed]
- Friedman, E.; De Marco, L.; Gejman, P.V.; Norton, J.A.; Bale, A.E.; Aurbach, G.D.; Spiegel, A.M.; Marx, S.J. Allelic loss from chromosome 11 in parathyroid tumors. Cancer Res. 1992, 52, 6804–6809. [Google Scholar] [PubMed]
- Yi, Y.; Nowak, N.J.; Pacchia, A.L.; Morrison, C. Chromosome 11 genomic changes in parathyroid adenoma and hyperplasia: Array CGH, FISH, and tissue microarrays. Genes Chromosomes Cancer 2008, 47, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Brewer, K.; Costa-Guda, J.; Arnold, A. Molecular genetic insights into sporadic primary hyperparathyroidism. Endocr. Relat. Cancer 2019, 26, R53–R72. [Google Scholar] [CrossRef] [PubMed]
- Newey, P.J.; Nesbit, M.A.; Rimmer, A.J.; Attar, M.; Head, R.T.; Christie, P.T.; Gorvin, C.M.; Stechman, M.; Gregory, L.; Mihai, R.; et al. Whole-exome sequencing studies of nonhereditary (sporadic) parathyroid adenomas. J. Clin. Endocrinol. Metab. 2012, 97, E1995–E2005. [Google Scholar] [CrossRef] [PubMed]
- Carling, T.; Rastad, J.; Szabo, E.; Westin, G.; Akerstrom, G. Reduced parathyroid vitamin D receptor messenger ribonucleic acid levels in primary and secondary hyperparathyroidism. J. Clin. Endocrinol. Metab. 2000, 85, 2000–2003. [Google Scholar] [CrossRef] [PubMed]
- FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 4 July 2017).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volders, P.J.; Helsens, K.; Wang, X.; Menten, B.; Martens, L.; Gevaert, K.; Vandesompele, J.; Mestdagh, P. LNCipedia: A database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013, 41, D246–D251. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.J.; Verheggen, K.; Menschaert, G.; Vandepoele, K.; Martens, L.; Vandesompele, J.; Mestdagh, P. An update on LNCipedia: A database for annotated human lncRNA sequences. Nucleic Acids Res. 2015, 43, 4363–4364. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006, 34, D322–D326. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinforma. 2003, 4, 2. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Knuesel, M.T.; Meyer, K.D.; Donner, A.J.; Espinosa, J.M.; Taatjes, D.J. The human CDK8 subcomplex is a histone kinase that requires MED12 for activity and can function independently of mediator. Mol. Cell Biol. 2009, 29, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Kampjarvi, K.; Makinen, N.; Kilpivaara, O.; Arola, J.; Heinonen, H.R.; Bohm, J.; Abdel-Wahab, O.; Lehtonen, H.J.; Pelttari, L.M.; Mehine, M.; et al. Somatic MED12 mutations in uterine leiomyosarcoma and colorectal cancer. Br. J. Cancer 2012, 107, 1761–1765. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.K.; Ong, C.K.; Tan, J.; Thike, A.A.; Ng, C.C.; Rajasegaran, V.; Myint, S.S.; Nagarajan, S.; Nasir, N.D.; McPherson, J.R.; et al. Exome sequencing identifies highly recurrent MED12 somatic mutations in breast fibroadenoma. Nat. Genet. 2014, 46, 877–880. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Rice, J.C. A new regulator of the cell cycle: The PR-Set7 histone methyltransferase. Cell Cycle 2011, 10, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Milite, C.; Feoli, A.; Viviano, M.; Rescigno, D.; Cianciulli, A.; Balzano, A.L.; Mai, A.; Castellano, S.; Sbardella, G. The emerging role of lysine methyltransferase SETD8 in human diseases. Clin. Epigenetics 2016, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, T.; Wang, Y.J.; Hu, J.Q.; Wang, Y.; Han, L.T.; Ma, B.; Shi, R.L.; Qu, N.; Wei, W.J.; Guan, Q.; et al. Histone methyltransferase KMT5A gene modulates oncogenesis and lipid metabolism of papillary thyroid cancer in vitro. Oncol. Rep. 2018, 39, 2185–2192. [Google Scholar] [CrossRef] [PubMed]
- Takawa, M.; Cho, H.S.; Hayami, S.; Toyokawa, G.; Kogure, M.; Yamane, Y.; Iwai, Y.; Maejima, K.; Ueda, K.; Masuda, A.; et al. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 2012, 72, 3217–3227. [Google Scholar] [CrossRef] [PubMed]
- Lisse, T.S.; Chun, R.F.; Rieger, S.; Adams, J.S.; Hewison, M. Vitamin D activation of functionally distinct regulatory miRNAs in primary human osteoblasts. J. Bone Miner. Res. 2013, 28, 1478–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deeb, K.K.; Trump, D.L.; Johnson, C.S. Vitamin D signalling pathways in cancer: Potential for anticancer therapeutics. Nat. Rev. Cancer 2007, 7, 684–700. [Google Scholar] [CrossRef] [PubMed]
- Ciro, M.; Prosperini, E.; Quarto, M.; Grazini, U.; Walfridsson, J.; McBlane, F.; Nucifero, P.; Pacchiana, G.; Capra, M.; Christensen, J.; et al. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009, 69, 8491–8498. [Google Scholar] [CrossRef] [PubMed]
- Bjorklund, P.; Krajisnik, T.; Akerstrom, G.; Westin, G.; Larsson, T.E. Type I membrane klotho expression is decreased and inversely correlated to serum calcium in primary hyperparathyroidism. J. Clin. Endocrinol. Metab. 2008, 93, 4152–4157. [Google Scholar] [CrossRef] [PubMed]
- Hong, A.R.; Kim, Y.A.; Bae, J.H.; Min, H.S.; Kim, J.H.; Shin, C.S.; Kim, S.Y.; Kim, S.W. A Possible Link Between Parathyroid Hormone Secretion and Local Regulation of GABA in Human Parathyroid Adenomas. J. Clin. Endocrinol. Metab. 2016, 101, 2594–2601. [Google Scholar] [CrossRef] [PubMed]
- Olauson, H.; Mencke, R.; Hillebrands, J.L.; Larsson, T.E. Tissue expression and source of circulating alphaKlotho. Bone 2017, 100, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Liu, W.; Bi, R.; Densmore, M.J.; Sato, T.; Mannstadt, M.; Yuan, Q.; Zhou, X.; Olauson, H.; Larsson, T.E.; et al. Interrelated role of Klotho and calcium-sensing receptor in parathyroid hormone synthesis and parathyroid hyperplasia. Proc. Natl. Acad. Sci. USA 2018, 115, E3749–E3758. [Google Scholar] [CrossRef] [PubMed]
- Haglund, F.; Juhlin, C.C.; Kiss, N.B.; Larsson, C.; Nilsson, I.L.; Hoog, A. Diffuse parathyroid hormone expression in parathyroid tumors argues against important functional tumor subclones. Eur. J. Endocrinol. 2016, 174, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.J.; Russell, J.; Costanzo, M.K.; Karp, F.; Benjamin, M.; Hardy, M.A.; Feind, C.R. Relationships of parathyroid hormone, parathyroid secretory protein, parathyroid hormone messenger RNA, parathyroid secretory protein mRNA, and replication in human parathyroid adenoma and secondary hyperplasia tissues and cultures. Surgery 1992, 112, 1089–1094. [Google Scholar] [PubMed]
- Rao, C.V.S.; de Waelheyns, E.; Economou, A.; Anne, J. Antibiotic targeting of the bacterial secretory pathway. Biochim. Biophys. Acta. 2014, 1843, 1762–1783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, M.; Oiso, Y.; Murase, T.; Kondo, K.; Saito, H.; Chinzei, T.; Racchi, M.; Lively, M.O. Possible involvement of inefficient cleavage of preprovasopressin by signal peptidase as a cause for familial central diabetes insipidus. J. Clin. Investig. 1993, 91, 2565–2571. [Google Scholar] [CrossRef] [PubMed]
- Indridason, O.S.; Pieper, C.F.; Quarles, L.D. Predictors of short-term changes in serum intact parathyroid hormone levels in hemodialysis patients: Role of phosphorus, calcium, and gender. J. Clin. Endocrinol. Metab. 1998, 83, 3860–3866. [Google Scholar] [CrossRef] [PubMed]
| Group | Gender | Age, Years | Tumor Size, cm | Serum Intact PTH, mmol/L | Serum Calcium, mg/dL | Serum Phosphorous, mg/dL | Serum 25(OH)2D3, ng/mL | Serum 1,25(OH)2D3, pg/mL |
|---|---|---|---|---|---|---|---|---|
| PHPT group | Female | 52 | 2.5 | 350 | 12.6 | 1.9 | 11.5 | 57.2 |
| Female | 63 | 1.5 | 173 | 11.4 | 3.1 | 6.0 | 130.1 | |
| Female | 55 | 2.1 | 262 | 12.1 | 2.5 | 30.5 | 82.0 | |
| Female | 47 | 2.2 | 286 | 12.0 | 2.4 | 12.9 | 53.0 | |
| Female | 51 | 3.5 | 1596 | 14.2 | 2.5 | 8.7 | 30.3 | |
| Female | 56 | 3.8 | 204 | 12.6 | 2.3 | 12.4 | 69.2 | |
| Female | 47 | 0.9 | 120 | 11.3 | 2.6 | 21.1 | 59.5 | |
| Female | 56 | 0.6 | 213 | 12.8 | 2.5 | 22.9 | 69.0 | |
| Female | 50 | 2.1 | 267 | 13.9 | 2.9 | 17.9 | 69.4 | |
| Female | 53 | 3.8 | 1133 | 13.8 | 1.8 | 10.0 | 34.3 | |
| Normal group | Female | 61 | 40 | 9.3 | 3.9 | - | 15.6 | |
| Female | 50 | 29 | 9.0 | 3.3 | 20.6 | 69.9 | ||
| Female | 46 | 28 | 8.5 | 3.3 | - | 30.0 | ||
| Female | 41 | 61 | 8.0 | 4.1 | - | 51.5 | ||
| Female | 50 | 15.7 | 8.9 | 3.6 | 19.1 | 25.2 |
| Expression | Gene | Fold Change | p Value × 10 | FDR |
|---|---|---|---|---|
| Up-regulated | BMP2K (BMP2 Inducible Kinase) | 5.67 | 5.90 × 10−8 | 6.39 × 10−5 |
| MED12 (Mediator Complex Subunit 12) | 4.79 | 5.38 × 10−8 | 6.39 × 10−5 | |
| NUFIP1 (Nuclear FMR1 Interacting Protein 1) | 2.47 | 1.33 × 10−7 | 8.55 × 10−5 | |
| KRBOX4 (KRAB Box Domain Containing 4) | 3.60 | 2.96 × 10−7 | 0.00011 | |
| ATAD2 (ATPase Family, AAA Domain Containing 2) | 3.70 | 5.41 × 10−7 | 0.000156 | |
| GPBP1 (GC-Rich Promoter Binding Protein 1) | 2.85 | 6.49 × 10−7 | 0.000158 | |
| LUC7L (LUC7 Like) | 2.07 | 1.16 × 10−6 | 0.000176 | |
| TCHP (Trichoplein Keratin Filament Binding) | 2.76 | 1.20 × 10−6 | 0.000176 | |
| GOLGA8Q (Golgin A8 Family Member Q) | 2.80 | 2.69 × 10−6 | 0.00025 | |
| CCDC174 (Coiled-Coil Domain Containing 174) | 2.02 | 2.97 × 10−6 | 0.000262 | |
| ZNF674 (Zinc Finger Protein 674) | 3.63 | 3.09 × 10−6 | 0.000264 | |
| CTTNBP2 (Cortactin Binding Protein 2) | 5.49 | 4.82 × 10−6 | 0.00035 | |
| ARIH2 (Ariadne RBR E3 Ubiquitin Protein Ligase 2) | 5.14 | 5.16 × 10−6 | 0.000363 | |
| GOLGA8O (Golgin A8 Family Member O) | 2.71 | 6.00 × 10−6 | 0.00039 | |
| PPM1B (Protein Phosphatase, Mg2+/Mn2+ Dependent 1B) | 4.14 | 6.18 × 10−6 | 0.000392 | |
| ZNF605 (Zinc Finger Protein 605) | 2.94 | 6.92 × 10−6 | 0.000401 | |
| COPS7B (COP9 Signalosome Subunit 7B) | 3.61 | 8.75 × 10−6 | 0.000451 | |
| KMT5A (Lysine Methyltransferase 5A) | 3.00 | 9.42 × 10−6 | 0.000459 | |
| NPIPA7 (Nuclear Pore Complex Interacting Protein Family Member A7) | 2.22 | 9.86 × 10−6 | 0.000459 | |
| SLTM (SAFB Like Transcription Modulator) | 2.52 | 9.34 × 10−6 | 0.000459 | |
| Down-regulated | DEGS1 (Delta 4-Desaturase, Sphingolipid 1) | −6.93 | 1.95 × 10−8 | 6.39 × 10−5 |
| TMBIM6 (Transmembrane BAX Inhibitor Motif Containing 6) | −7.28 | 4.65 × 10−8 | 6.39 × 10−5 | |
| SSBP3 (Single Stranded DNA Binding Protein 3) | −7.37 | 6.67 × 10−8 | 6.39 × 10−5 | |
| SNORA74A (Small Nucleolar RNA, H/ACA Box 74A) | −46.50 | 7.37 × 10−8 | 6.39 × 10−5 | |
| DPYD (Dihydropyrimidine Dehydrogenase) | −5.02 | 9.45 × 10−8 | 7.02 × 10−5 | |
| ALG5 (ALG5, Dolichyl-Phosphate Beta-Glucosyltransferase) | −15.16 | 1.48 × 10−7 | 8.55 × 10−5 | |
| ZNF552 (Zinc Finger Protein 552) | −5.20 | 1.96 × 10−7 | 0.000102 | |
| NBEAL1 (Neurobeachin Like 1) | −3.90 | 2.46 × 10−7 | 0.00011 | |
| OGN (Osteoglycin) | −18.30 | 2.67 × 10−7 | 0.00011 | |
| CALR (Calreticulin) | −6.21 | 2.90. × 10−7 | 0.00011 | |
| ZNF33A (Zinc Finger Protein 33A) | −4.17 | 3.83. × 10−7 | 0.000133 | |
| SPDYE16 (Speedy/RINGO Cell Cycle Regulator Family Member E16) | −12.79 | 4.24 × 10−7 | 0.000138 | |
| FZD6 (Frizzled Class Receptor 6) | −3.90 | 4.53 × 10−7 | 0.000139 | |
| ATP6AP2 (ATPase H+ Transporting Accessory Protein 2) | −21.89 | 5.72 × 10−7 | 0.000157 | |
| PIGG (Phosphatidylinositol Glycan Anchor Biosynthesis Class G) | −3.80 | 6.47 × 10−7 | 0.000158 | |
| UBAP1 (Ubiquitin Associated Protein 1) | −3.57 | 6.68 × 10−7 | 0.000158 | |
| ST13 (ST13, Hsp70 Interacting Protein) | −3.20 | 7.75 × 10−7 | 0.000175 | |
| YTHDC2 (YTH Domain Containing 2) | −3.57 | 8.17 × 10−7 | 0.000176 | |
| CPE (Carboxypeptidase E) | −10.34 | 9.18 × 10−7 | 0.000176 | |
| PTH (Parathyroid Hormone) | −6.62 | 9.36 × 10−7 | 0.000176 |
| Gene | Parathyroid Adenomas | Normal Parathyroid | Fold Change | p Value | Adjusted p Value |
|---|---|---|---|---|---|
| CASR (Calcium Sensing Receptor) | 174.02 (59.59, 345.05) | 394.47 (265.2, 601.57) | 0.44 | 0.004 | 0.004 |
| FGFR1 (Fibroblast Growth Factor receptor1) | 4.75 (0.10, 30.60) | 7.99 (2.66, 9.54) | 0.59 | 0.9 | 0.25 |
| FGFR2 (Fibroblast Growth Factor Receptor2) | 5.37(1.94, 10.06) | 16.02 (10.00, 23.48) | 0.34 | 0.01 | 0.007 |
| KL (Klotho) | 3.33 (0.23, 7.76) | 55.02 (19.47, 89.49) | 0.06 | <0.001 | <0.001 |
| PTH (Parathyroid Hormone) | 2216.46 (1083.31, 5942.02) | 16,100 (12,266.3, 24,066.2) | 0.14 | <0.001 | <0.001 |
| VDR (Vitamin D Receptor) | 12.86 (4.23, 44.80) | 13.05 (10.38, 36.49) | 0.99 | 0.54 | 0.168 |
| Category | GO Term ID | GO Term | Count | p Value |
|---|---|---|---|---|
| Biological process ontology | GO:0006351 | transcription, DNA-templated | 14 | <0.001 |
| GO:0050684 | regulation of mRNA processing | 3 | <0.001 | |
| GO:0006325 | chromatin organization | 3 | 0.004 | |
| GO:0045893 | positive regulation of transcription, DNA-templated | 6 | 0.005 | |
| GO:0018026 | peptidyl-lysine monomethylation | 2 | 0.017 | |
| GO:0006355 | regulation of transcription, DNA-templated | 8 | 0.038 | |
| Cellular component ontology | GO:0005654 | nucleoplasm | 19 | <0.001 |
| GO:0005634 | nucleus | 26 | <0.001 | |
| Molecular function ontology | GO:0003676 | nucleic acid binding | 10 | <0.001 |
| GO:0003682 | chromatin binding | 6 | 0.002 | |
| GO:0003713 | transcription coactivator activity | 5 | 0.002 | |
| GO:0000166 | nucleotide binding | 5 | 0.008 | |
| GO:0003677 | DNA binding | 10 | 0.012 | |
| GO:0003690 | double-stranded DNA binding | 3 | 0.015 | |
| GO:0003729 | mRNA binding | 3 | 0.034 | |
| GO:0016279 | protein-lysine N-methyltransferase activity | 2 | 0.039 |
| Category | GO Term ID | GO Term | Count | p Value |
|---|---|---|---|---|
| Biological process ontology | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 5 | <0.001 |
| GO:0061077 | chaperone-mediated protein folding | 5 | <0.001 | |
| GO:0006465 | signal peptide processing | 4 | 0.002 | |
| GO:0006506 | GPI anchor biosynthetic process | 4 | 0.003 | |
| GO:0044829 | positive regulation by host of viral genome replication | 3 | 0.003 | |
| Cellular component ontology | GO:0070062 | extracellular exosome | 79 | <0.001 |
| GO:0016020 | membrane | 64 | <0.001 | |
| GO:0005789 | endoplasmic reticulum membrane | 36 | <0.001 | |
| GO:0005783 | endoplasmic reticulum | 31 | <0.001 | |
| GO:0042470 | melanosome | 12 | <0.001 | |
| Molecular function ontology | GO:0044822 | poly(A) RNA binding | 23 | 0.003 |
| GO:0005515 | protein binding | 110 | 0.004 | |
| GO:0001540 | beta-amyloid binding | 4 | 0.005 | |
| GO:0019904 | protein domain specific binding | 8 | 0.006 | |
| GO:0051087 | chaperone binding | 5 | 0.01 |
| Term ID | Term | Count | p Value | Genes |
|---|---|---|---|---|
| hsa04141 | Protein processing in endoplasmic reticulum | 15 | <0.001 | SEC63, DNAJA1, EIF2AK1, XBP1, UGGT2, CANX, SEC61G, STT3A, PDIA6, HYOU1, SEC13, RPN1, PDIA3, CALR, BCAP31 |
| hsa03060 | Protein export | 5 | <0.001 | SEC63, SPCS2, SEC61G, SEC11A, SEC11C |
| hsa03013 | RNA transport | 8 | 0.008 | STRAP, XPO1, EIF3E, SUMO1, EIF2S3, NUP205, SEC13, RGPD2 |
| hsa00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 3 | 0.043 | PIGU, PIGG, PIGP |
| hsa00240 | Pyrimidine metabolism | 5 | 0.049 | CTPS2, NT5C3A, NME7, POLR2H, DPYD |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, Y.J.; Chae, H.; Kim, K.; Lee, H.; Choi, S.; Lee, K.E.; Kim, S.W. Comparative Gene Expression Profiles in Parathyroid Adenoma and Normal Parathyroid Tissue. J. Clin. Med. 2019, 8, 297. https://doi.org/10.3390/jcm8030297
Chai YJ, Chae H, Kim K, Lee H, Choi S, Lee KE, Kim SW. Comparative Gene Expression Profiles in Parathyroid Adenoma and Normal Parathyroid Tissue. Journal of Clinical Medicine. 2019; 8(3):297. https://doi.org/10.3390/jcm8030297
Chicago/Turabian StyleChai, Young Jun, Heejoon Chae, Kwangsoo Kim, Heonyi Lee, Seongmin Choi, Kyu Eun Lee, and Sang Wan Kim. 2019. "Comparative Gene Expression Profiles in Parathyroid Adenoma and Normal Parathyroid Tissue" Journal of Clinical Medicine 8, no. 3: 297. https://doi.org/10.3390/jcm8030297
APA StyleChai, Y. J., Chae, H., Kim, K., Lee, H., Choi, S., Lee, K. E., & Kim, S. W. (2019). Comparative Gene Expression Profiles in Parathyroid Adenoma and Normal Parathyroid Tissue. Journal of Clinical Medicine, 8(3), 297. https://doi.org/10.3390/jcm8030297






