Deep Learning-Based Classification of Inherited Retinal Diseases Using Fundus Autofluorescence
Abstract
:1. Introduction
2. Methods
2.1. Datasets
2.2. Development of a Deep Learning Classifier
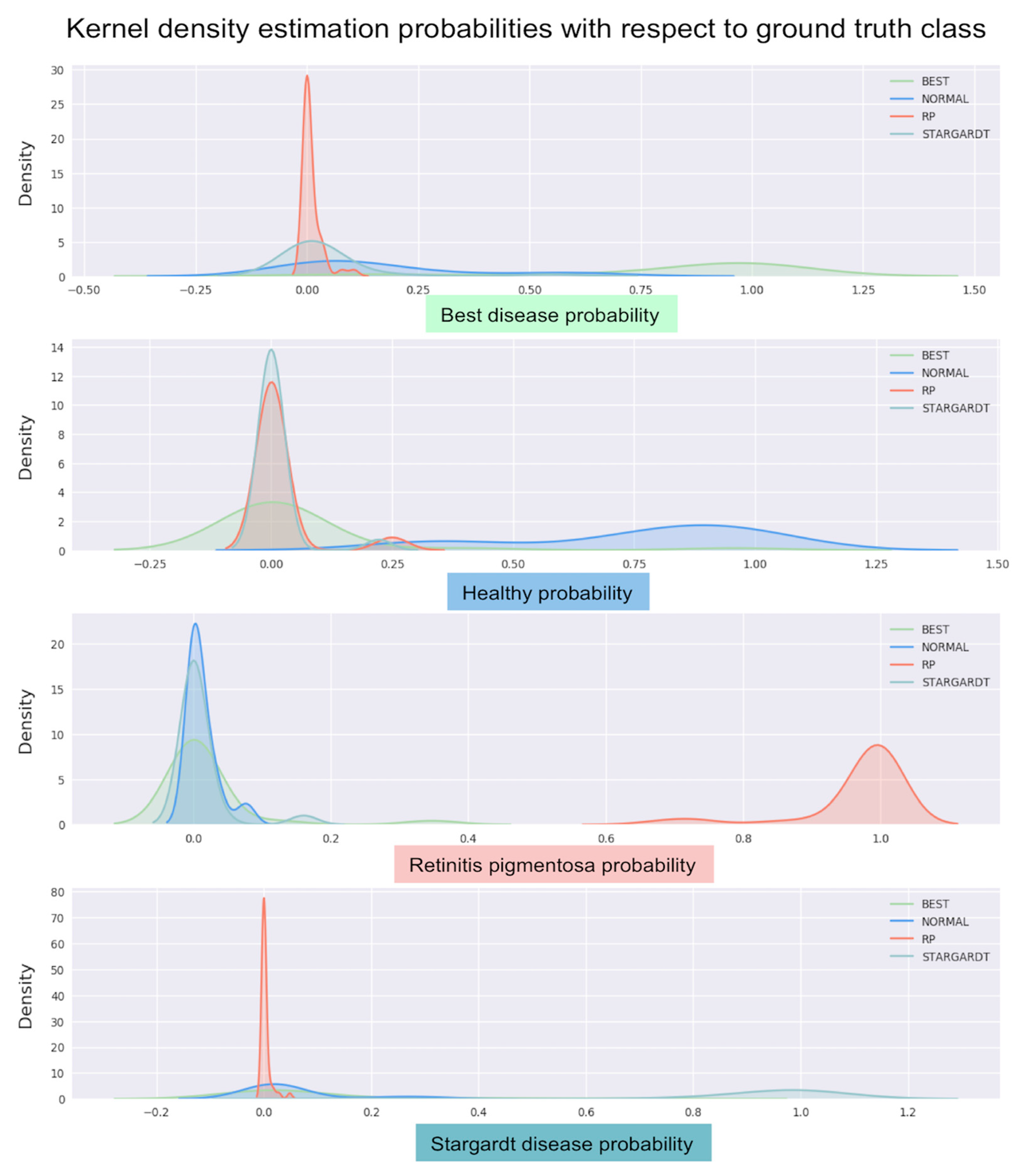
3. Results
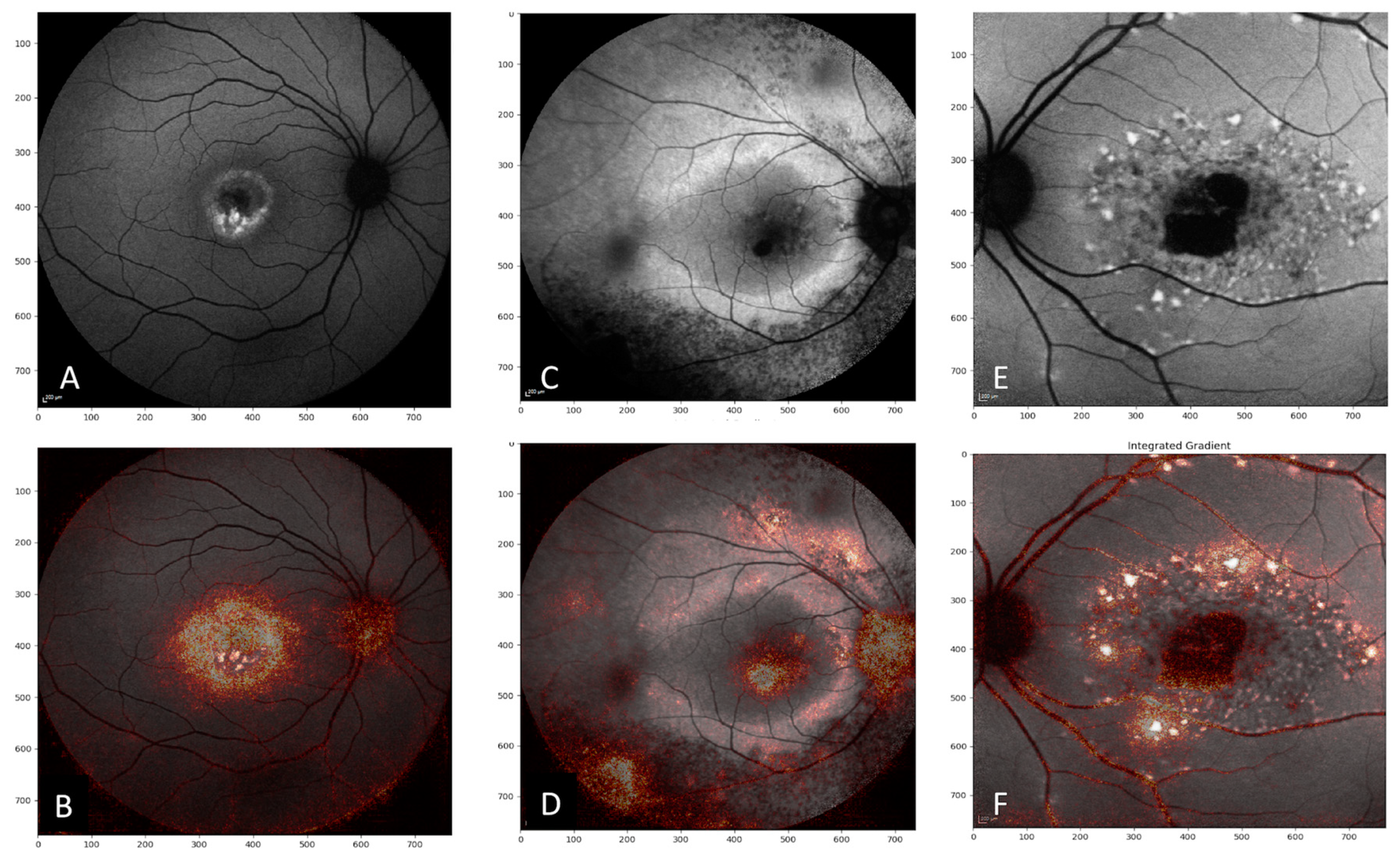
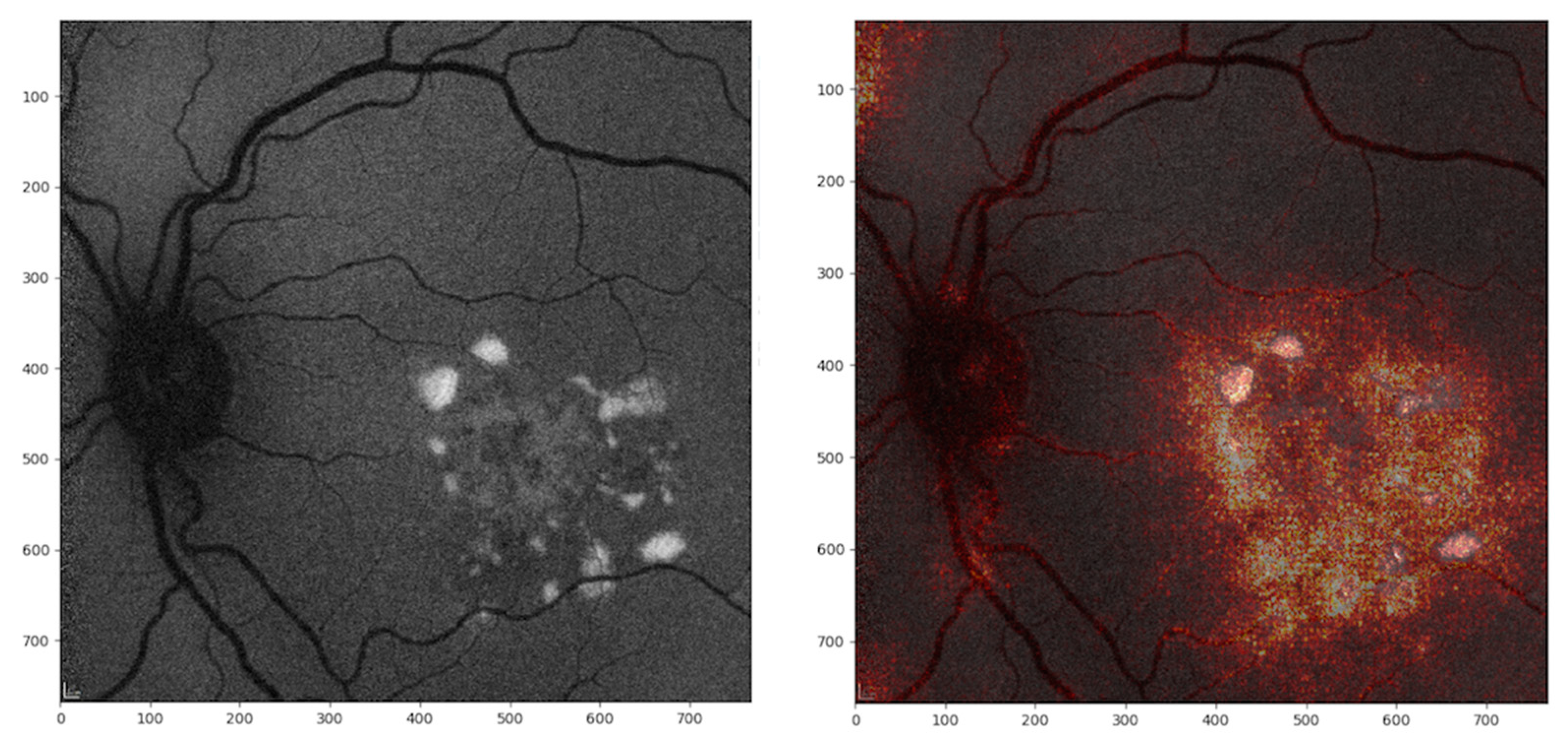
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Bessant, D.A.; Ali, R.R.; Bhattacharya, S.S. Molecular genetics and prospects for therapy of the inherited retinal dystrophies. Curr. Opin. Genet. Dev. 2001, 11, 307–316. [Google Scholar] [CrossRef]
- Sahel, J.A.; Marazova, K.; Audo, I. Clinical characteristics and current therapies for inherited retinal degenerations. Cold Spring Harb. Perspect. Med. 2014, 5, a017111. [Google Scholar] [CrossRef] [PubMed]
- Bundey, S.; Crews, S.J. A study of retinitis pigmentosa in the City of Birmingham. I Prevalence. J. Med. Genet. 1984, 21, 417–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holz, F.G.; Schmitz-Valckenberg, S.; Spaide, R.F.; Bird, A.C. Atlas of Fundus Autofluorescence Imaging; Springer: Berlin, Germany, 2007; pp. 41–45. [Google Scholar]
- Biarnés, M.; Arias, L.; Alonso, J.; Garcia, M.; Hijano, M.; Rodríguez, A.; Serrano, A.; Badal, J.; Muhtaseb, J.; Verdaguer, P.; et al. Increased Fundus Autofluorescence and Progression of Geographic Atrophy Secondary to Age-Related Macular Degeneration: The GAIN Study. Am. J. Ophthalmol. 2015, 160, 345–353. [Google Scholar] [CrossRef] [Green Version]
- Spaide, R.F.; Noble, K.; Morgan, A.; Freund, K.B. Vitelliform macular dystrophy. Ophthalmology 2006, 113, 1392–1400. [Google Scholar] [CrossRef]
- Young, R.W. Pathophysiology of age-related macular degeneration. Surv. Ophthalmol. 1987, 31, 291–306. [Google Scholar] [CrossRef]
- Holz, F.G.; Schütt, F.; Kopitz, J.; Kruse, F.E.; Völcker, H.E.; Cantz, M. Inhibition of lysosomal degradative functions in RPE cells by a retinoid component of lipofuscin. Investig. Ophthalmol. Vis. Sci. 1999, 40, 737–743. [Google Scholar]
- Zhou, J.; Cai, B.; Jang, Y.P.; Pachydaki, S.; Schmidtm, A.M.; Sparrow, J.R. Mechanisms for the induction of HNE- MDA- and AGE-adducts, RAGE and VEGF in retinal pigment epithelial cells. Exp. Eye Res. 2005, 80, 567–580. [Google Scholar] [CrossRef]
- Holz, F.G.; Bindewald-Wittich, A.; Fleckenstein, M.; Dreyhaupt, J.; Scholl, H.P.N.; Schmitz-Valckenberg, S.; FAM-Study Group. Progression of geographic atrophy and impact of fundus autofluorescence patterns in age-related macular degeneration. Am. J. Ophthalmol. 2007, 143, 463–472. [Google Scholar] [CrossRef]
- Kuehlewein, L.; Hariri, A.H.; Ho, A.; Dustin, L.; Wolfson, Y.; Strauss, R.W.; Scholl, H.P.N.; Sadda, S.R. Comparison of manual and semiautomated fundus autofluorescence analysis of macular atrophy in Stargardt disease phenotype. Retina 2016, 36, 1216–1221. [Google Scholar] [CrossRef]
- Pichi, F.; Abboud, E.B.; Ghazi, N.G.; Khan, A.O. Fundus autofluorescence imaging in hereditary retinal diseases. Acta Ophthalmol. 2018, 96, e549–e561. [Google Scholar] [CrossRef] [PubMed]
- Lois, N.; Halfyard, A.S.; Bird, A.C.; Holder, G.E.; Fitzke, F.W. Fundus autofluorescence in Stargardt macular dystrophy-fundus flavimaculatus. Am. J. Ophthalmol. 2004, 138, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Duncker, T.; Greenberg, J.P.; Ramachandran, R.; Hood, D.C.; Smith, R.T.; Hirose, T.; Woods, R.L.; Tsang, S.H.; Delori, F.C.; Sparrow, J.R. Quantitative fundus autofluorescence and optical coherence tomography in best vitelliform macular dystrophy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 1471–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robson, A.G.; Michaelides, M.; Saihan, Z.; Bird, A.C.; Webster, A.R.; Moore, A.T.; Fitzke, F.W.; Holder, G.E. Functional characteristics of patients with retinal dystrophy that manifest abnormal parafoveal annuli of high density fundus autofluorescence; a review and update. Doc. Ophthalmol. 2008, 116, 79–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalvin, L.A.; Pulido, J.S.; Marmorstein, A.D. Vitelliform dystrophies: Prevalence in Olmsted County, Minnesota, United States. Ophthalmic Genet. 2017, 38, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Tsang, S.H.; Sharma, T. Stargardt Disease. Adv. Exp. Med. Biol. 2018, 1085, 139–151. [Google Scholar]
- Hamel, C. Retinitis pigmentosa. Orphanet J. Rare Dis. 2006, 1, 40. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Lavin, P.T.; Birch, M.; Shah, N.; Folk, J.C. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit. Med. 2018, 1, 39. [Google Scholar] [CrossRef]
- Lee, C.S.; Baughman, D.M.; Lee, A.Y. Deep learning is effective for the classification of OCT images of normal versus Age-related Macular Degeneration. Ophthalmol. Retin. 2017, 1, 322–327. [Google Scholar] [CrossRef]
- Treder, M.; Lauermann, J.L.; Eter, N. Deep learning-based detection and classification of geographic atrophy using a deep convolutional neural network classifier. Graefes. Arch. Clin. Exp. Ophthalmol. 2018, 256, 2053–2060. [Google Scholar] [CrossRef]
- Li, Z.; He, Y.; Keelm, S.; Mengm, W.; Changm, R.T.; He, M. Efficacy of a Deep Learning System for Detecting Glaucomatous Optic Neuropathy Based on Color Fundus Photographs. Ophthalmology 2018, 125, 1199–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt-Erfurth, U.; Sadeghipour, A.; Gerendas, B.S.; Waldstein, S.M.; Bogunović, H. Artificial intelligence in retina. Prog. Retin. Eye Res. 2018, 67, 1–29. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27 June–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Akil, M.; Elloumi, Y.; Kachouri, R. Detection of Retinal Abnormalities in Fundus Image Using CNN Deep Learning Networks. 2020. Available online: https://hal-upec-upem.archives-ouvertes.fr/hal-02428351/document (accessed on 13 October 2020).
- Ometto, G.; Montesano, G.; Sadeghi-Afgeh, S.; Lazaridis, G.; Liu, X.; Keane, P.A.; Crabb, D.P.; Denniston, A.K. Merging Information From Infrared and Autofluorescence Fundus Images for Monitoring of Chorioretinal Atrophic Lesions. Transl. Vis. Sci. Technol. 2020, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sadda, S.V.; Hu, Z. Deep learning for automated screening and semantic segmentation of age-related and juvenile atrophic macular degeneration. In Proceedings of the SPIE 10950, Medical Imaging 2019: Computer-Aided Diagnosis, 109501Q, San Diego, CA, USA, 16–21 February 2019. [Google Scholar]
- Canziani, A.; Paszke, A.; Culurciello, E. An Analysis of Deep Neural Network Models for Practical Applications. 2016. Available online: https://arxiv.org/abs/1605.07678 (accessed on 5 May 2020).
- Kingma, P.; Ba, J. A Method for Stochastic Optimization. 2019. Available online: https://arxiv.org/abs/1412.6980 (accessed on 5 May 2020).
- Mukund, S.; Taly, A.; Yan, Q. Axiomatic Attribution for Deep Networks. 2017. Available online: https://arxiv.org/abs/1703.01365 (accessed on 5 May 2020).
- Gass, J.D. Inherited macular disease. In Stereoscopic Atlas of Macular Diseases: Diagnosis and treatment, 3rd ed.; Gass, J.D., Ed.; Mosby: St. Louis, MO, USA, 1997; Volume 1, pp. 98–99. [Google Scholar]
- Smith, R.T. Fundus autofluorescence patterns in stargardt disease over time. Arch. Ophthalmol. 2012, 130, 1354–1355. [Google Scholar] [CrossRef]
- Murakami, T.; Akimoto, M.; Ooto, S.; Ikeda, H.; Kawagoe, N.; Takahashi, M.; Yoshimura, N. Association between abnormal autofluorescence and photoreceptor disorganization in retinitis pigmentosa. Am. J. Ophthalmol. 2008, 145, 687–694. [Google Scholar] [CrossRef]
- Fujinami-Yokokawa, Y.; Pontikos, N.; Yang, L.; Tsunoda, K.; Yoshitake, K.; Iwata, T.; Miyata, H.; Fujinami, K.; Japan Eye Genetics Consortiu. Prediction of Causative Genes in Inherited Retinal Disorders from Spectral-Domain Optical Coherence Tomography Utilizing Deep Learning Techniques. J. Ophthalmol. 2019, 2019, 1691064. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In Proceedings of the 31st AAAI Conference on Artificial Intelligence, 2017, San Francisco, CA, USA, 4–9 February 2017; Available online: https://arxiv.org/abs/1602.07261 (accessed on 6 May 2020).
- Shah, M.; Roomans Ledo, A.; Rittscher, J. Automated classification of normal and Stargardt disease optical coherence tomography images using deep learning. Acta Ophthalmol. 2020. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, e52. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |

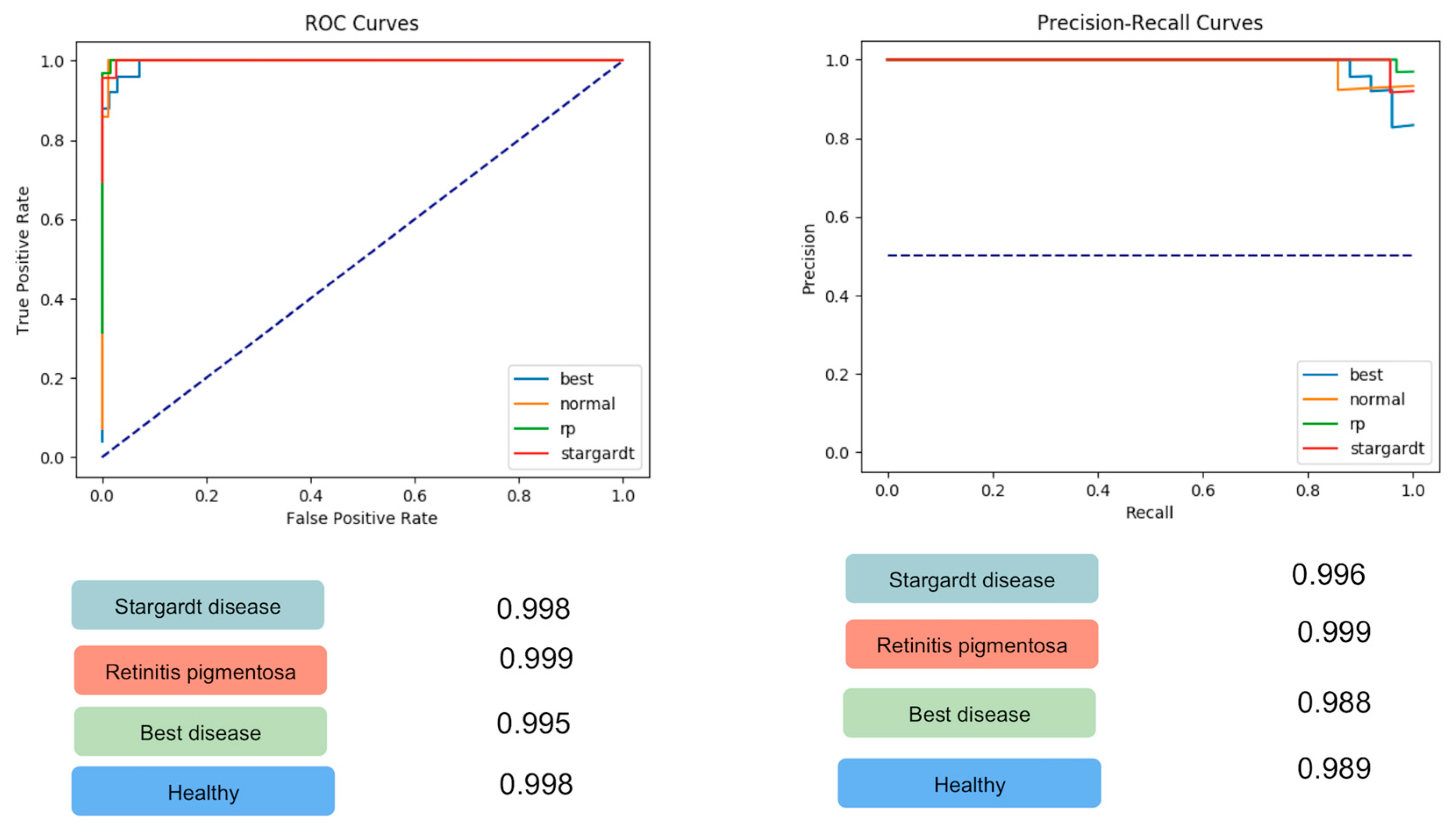

| Class | ROC-AUC | PRC-AUC | Sensitivity | Specificity |
|---|---|---|---|---|
| Stargardt disease FAF | 0.998 | 0.986 | 0.96 | 1 |
| Retinitis pigmentosa FAF | 0.999 | 0.999 | 1 | 0.97 |
| Best disease FAF | 0.995 | 0.988 | 0.92 | 0.97 |
| Healthy controls FAF | 0.998 | 0.989 | 0.86 | 0.99 |
| Ground Truth Class | Predicted Class | |||
|---|---|---|---|---|
| Stargardt Disease | Retinitis Pigmentosa | Best Disease | Healthy Control | |
| Stargardt Disease | 22 | 0 | 1 | 0 |
| Retinitis Pigmentosa | 0 | 32 | 0 | 0 |
| Best Disease | 0 | 1 | 23 | 1 |
| Healthy Control | 0 | 1 | 1 | 12 |
| 30 × 30 Degree-Field-of-View | ||||
| Ground Truth Class | Predicted Class | |||
| Stargardt Disease | Retinitis Pigmentosa | Best Disease | Healthy Control | |
| Stargardt Disease | 10 | 0 | 1 | 0 |
| Retinitis pigmentosa | 0 | 24 | 0 | 0 |
| Best disease | 0 | 0 | 17 | 0 |
| Healthy control | 0 | 0 | 2 | 11 |
| 55 × 55 Degree-Field-of-View | ||||
| Ground Truth Class | Predicted Class | |||
| Stargardt Disease | Retinitis Pigmentosa | Best Disease | Healthy Control | |
| Stargardt disease | 9 | 0 | 0 | 0 |
| Retinitis pigmentosa | 0 | 17 | 0 | 0 |
| Best disease | 2 | 0 | 4 | 0 |
| Healthy control | 0 | 0 | 0 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miere, A.; Le Meur, T.; Bitton, K.; Pallone, C.; Semoun, O.; Capuano, V.; Colantuono, D.; Taibouni, K.; Chenoune, Y.; Astroz, P.; et al. Deep Learning-Based Classification of Inherited Retinal Diseases Using Fundus Autofluorescence. J. Clin. Med. 2020, 9, 3303. https://doi.org/10.3390/jcm9103303
Miere A, Le Meur T, Bitton K, Pallone C, Semoun O, Capuano V, Colantuono D, Taibouni K, Chenoune Y, Astroz P, et al. Deep Learning-Based Classification of Inherited Retinal Diseases Using Fundus Autofluorescence. Journal of Clinical Medicine. 2020; 9(10):3303. https://doi.org/10.3390/jcm9103303
Chicago/Turabian StyleMiere, Alexandra, Thomas Le Meur, Karen Bitton, Carlotta Pallone, Oudy Semoun, Vittorio Capuano, Donato Colantuono, Kawther Taibouni, Yasmina Chenoune, Polina Astroz, and et al. 2020. "Deep Learning-Based Classification of Inherited Retinal Diseases Using Fundus Autofluorescence" Journal of Clinical Medicine 9, no. 10: 3303. https://doi.org/10.3390/jcm9103303
APA StyleMiere, A., Le Meur, T., Bitton, K., Pallone, C., Semoun, O., Capuano, V., Colantuono, D., Taibouni, K., Chenoune, Y., Astroz, P., Berlemont, S., Petit, E., & Souied, E. (2020). Deep Learning-Based Classification of Inherited Retinal Diseases Using Fundus Autofluorescence. Journal of Clinical Medicine, 9(10), 3303. https://doi.org/10.3390/jcm9103303






