Omics in Myopia
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
4.1. Experimental Models
4.1.1. Human Studies
4.1.2. Animal Models
4.2. Altered Pathways and Clinical Implications
4.2.1. Proteomics
4.2.2. Metabolomics
4.3. Analytical Aspects
4.3.1. Proteomics
4.3.2. Metabolomics
5. Conclusions
Funding
Conflicts of Interest
Abbreviations
| 2D-PAGE | two-dimensional gel electrophoresis |
| AH | aqueous humor |
| CE | capillary electrophoresis |
| DDA | data-dependent acquisition |
| DIA | data-independent acquisition |
| ERM | epiretinal membrane |
| FDM | form-deprived myopia |
| VH | vitreous humor |
| GC | gas chromatography |
| ICPL | isotope-coded protein label |
| iTRAQ | isobaric tags for relative and absolute quantitation |
| HM | high myopia |
| MH | macular hole |
| LC | liquid chromatography |
| LIM | lens-induced myopia |
| LF | label-free |
| MS | mass spectrometry |
| MALDI-TOF | matrix-assisted laser desorption/ionization |
| MRS | myopic retinoschisis |
| MRM | multiple reaction monitoring |
| NMR | nuclear magnetic resonance |
| PCV | polypoidal choroidal vasculopathy |
| PRM | parallel reaction monitoring |
| PM | pathological myopia |
| PPV | pars plana vitrectomy |
| PTM | post-translation modifications |
| RPE | retinal pigment epithelium |
| RRD | rhegmatogenous retinal detachment |
| SRM | selected reaction monitoring |
| SER | spherical equivalent refraction |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SWATH | sequential window acquisition of all theoretical fragment ion spectra |
| TMT | tandem mass tags |
References
- Ohno-Matsui, K.R.; Kawasaki, J.B.; Jonas, C.M.; Cheung, S.M.; Saw, V.J.; Verhoeven, C.C.; Klaver, M.; Moriyama, K.; Shinohara, Y.; Kawasaki, M.; et al. International photographic classification and grading system for myopic maculopathy. Am. J. Ophthalmol. 2015, 159, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Ohno-Matsui, K. Pathologic Myopia. Asia Pac. J. Ophthalmol. 2016, 5, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wu, A.; Zhang, L.; Wang, W.; Chen, X.; Yu, X.; Wang, K. The increasing prevalence of myopia and high myopia among high school students in Fenghua city, eastern China: A 15-year population-based survey. BMC Ophthalmol. 2018, 18, 159. [Google Scholar] [CrossRef] [Green Version]
- Rudnicka, A.R.; Kapetanakis, V.V.; Wathern, A.K.; Logan, N.S.; Gilmartin, B.; Whincup, P.H.; Cook, D.G.; Owen, C.G. Global variations and time trends in the prevalence of childhood myopia, a systematic review and quantitative meta-analysis: Implications for aetiology and early prevention. Br. J. Ophthalmol. 2016, 100, 882–890. [Google Scholar] [CrossRef] [Green Version]
- Holden, B.A.; Fricke, T.R.; Wilson, D.A.; Jong, M.; Naidoo, K.S.; Sankaridurg, P.; Wong, T.Y.; Naduvilath, T.J.; Resnikoff, S.G. Prevalence of Myopia and High Myopia and Temporal Trends from 2000 through 2050. Ophthalmology 2016, 123, 1036–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalczyk, T.; Ciborowski, M.; Kisluk, J.; Kretowski, A.; Barbas, C. Mass spectrometry based proteomics and metabolomics in personalized oncology. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165690. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.; Vorum, H.; Fagerholm, P.; Birkenkamp-Demtröder, K.; Honoré, B.; Ehlers, N.; Orntoft, T.F. Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study. Exp. Eye Res. 2006, 82, 201–219. [Google Scholar] [CrossRef]
- Cheung, J.K.; Li, K.K.; Zhou, L.; To, C.H.; Lam, T.C. Data on protein changes of chick vitreous during normal eye growth using data-independent acquisition (SWATH-MS). Data Brief 2020, 30, 105576. [Google Scholar] [CrossRef]
- Tse, J.S.; Lam, T.C.; Cheung, J.K.; Sze, Y.H.; Wong, T.K.; Chan, H.H. Data on assessment of safety and tear proteome change in response to orthokeratology lens—Insight from integrating clinical data and next generation proteomics. Data Brief 2020, 29, 105186. [Google Scholar] [CrossRef]
- Wei, Q.; Zhang, T.; Fan, J.; Jiang, R.; Chang, Q.; Hong, J.; Xu, G. Pathological myopia-induced antioxidative proteins in the vitreous humor. Ann. Transl. Med. 2020, 8, 193. [Google Scholar] [CrossRef]
- Frost, R.M.; Norton, T.T. Differential protein expression in tree shrew sclera during development of lens-induced myopia and recovery. Mol. Vis. 2007, 13, 1580–1588. [Google Scholar] [PubMed]
- Kang, B.S.; Lam, T.C.; Cheung, J.K.; Li, K.K.; Kee, C.S. Data on corneal proteome and differentially expressed corneal proteins in highly myopic chicks using a data independent quantification approach. Data Brief 2019, 26, 104478. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wu, J.; Ding, H.; Liao, A.; He, H.; Stell, W.K.; Zhong, X. Flicker downregulates the content of crystallin proteins in form-deprived C57BL/6 mouse retina. Exp. Eye Res. 2012, 101, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jostrup, R.; Shen, W.; Burrows, J.T.; Sivak, J.G.; McConkey, B.J.; Singer, T.D. Identification of myopia-related marker proteins in tilapia retinal, RPE, and choroidal tissue following induced form deprivation. Curr. Eye Res. 2009, 34, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Cases, O.; Obry, A.; Ben-Yacoub, S.; Augustin, S.; Joseph, A.; Toutirais, G.; Simonutti, M.; Christ, A.; Cosette, P.; Kozyraki, R. Impaired vitreous composition and retinal pigment epithelium function in the FoxG1::LRP2 myopic mice. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1242–1254. [Google Scholar] [CrossRef]
- Lam, T.C.; Li, K.K.; Lo, S.C.; Guggenheim, J.A.; To, C.H. Application of fluorescence difference gel electrophoresis technology in searching for protein biomarkers in chick myopia. J. Proteome Res. 2007, 6, 4135–4149. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.J.; Lam, T.C.; Sze, A.Y.; Li, K.K.; Chun, R.K.; Shan, S.W.; To, C.H. Alteration of retinal metabolism and oxidative stress may implicate myopic eye growth: Evidence from discovery and targeted proteomics in an animal model. J. Proteom. 2020, 221, 103684. [Google Scholar] [CrossRef]
- Yu, F.J.; Lam, T.C.; Liu, L.Q.; Chun, R.K.; Cheung, J.K.; Li, K.K.; To, C.H. Isotope-coded protein label based quantitative proteomic analysis reveals significant up-regulation of apolipoprotein A1 and ovotransferrin in the myopic chick vitreous. Sci. Rep. 2017, 7, 12649. [Google Scholar] [CrossRef] [Green Version]
- Xiang, M.; Zhang, X.; Li, Q.; Wang, H.; Zhang, Z.; Han, Z.; Ke, M.; Chen, X. Identification of proteins in the aqueous humor associated with cataract development using iTRAQ methodology. Mol. Med. Rep. 2017, 15, 3111–3120. [Google Scholar] [CrossRef]
- Barathi, V.A.; Chaurasia, S.S.; Poidinger, M.; Koh, S.K.; Tian, D.; Ho, C.; Iuvone, P.M.; Beuerman, R.W.; Zhou, L. Involvement of GABA transporters in atropine-treated myopic retina as revealed by iTRAQ quantitative proteomics. J. Proteome Res. 2014, 13, 4647–4658. [Google Scholar] [CrossRef] [Green Version]
- Riddell, N.; Faou, P.; Crewther, S.G. Short term optical defocus perturbs normal developmental shifts in retina/RPE protein abundance. BMC Dev. Biol. 2018, 18, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ye, J.; Willcox, M.D.; Xie, R.; Jiang, L.; Lu, R.; Shi, J.; Bai, Y.; Qu, J. Changes in protein profiles of guinea pig sclera during development of form deprivation myopia and recovery. Mol. Vis. 2010, 16, 2163–2174. [Google Scholar] [PubMed]
- Frost, R.M.; Norton, T.T. Alterations in protein expression in tree shrew sclera during development of lens-induced myopia and recovery. Investig. Ophthalmol. Vis. Sci. 2012, 53, 322–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, S.W.; Tse, D.Y.; Zuo, B.; To, C.H.; Liu, Q.; McFadden, S.A.; Chun, R.K.; Bian, J.; Li, K.K.; Lam, T.C. Data on differentially expressed proteins in retinal emmetropization process in guinea pig using integrated SWATH-based and targeted-based proteomics. Data Brief 2018, 21, 1750–1755. [Google Scholar] [CrossRef]
- Bertrand, E.; Fritsch, C.; Diether, S.; Lambrou, G.; Müller, D.; Schaeffel, F.; Schindler, P.; Schmid, K.L.; van Oostrum, J.; Voshol, H. Identification of apolipoprotein A-I as a “STOP” signal for myopia. Mol. Cell. Proteom. 2006, 5, 2158–2166. [Google Scholar] [CrossRef] [Green Version]
- Ji, Y.; Rong, X.; Ye, H.; Zhang, K.; Lu, Y. Proteomic analysis of aqueous humor proteins associated with cataract development. Clin. Biochem. 2015, 48, 1304–1309. [Google Scholar] [CrossRef]
- Riddell, N.; Faou, P.; Murphy, M.; Giummarra, L.; Downs, R.A.; Rajapaksha, H.; Crewther, S.G. The retina/RPE proteome in chick myopia and hyperopia models: Commonalities with inherited and age-related ocular pathologies. Mol. Vis. 2017, 23, 872–888. [Google Scholar]
- Duan, X.; Lu, Q.; Xue, P.; Zhang, H.; Dong, Z.; Yang, F.; Wang, N. Proteomic analysis of aqueous humor from patients with myopia. Mol. Vis. 2008, 14, 370–377. [Google Scholar]
- Wu, Y.; Lam, C.S.; Tse, D.Y.; Tom, C.H.; Liu, Q.; McFadden, S.A.; Chun, R.K.; Li, K.K.; Bian, J.; Lam, C. Early quantitative profiling of differential retinal protein expression in lens-induced myopia in guinea pig using fluorescence difference two-dimensional gel electrophoresis. Mol. Med. Rep. 2018, 17, 5571–5580. [Google Scholar] [CrossRef] [Green Version]
- Shan, S.W.; Tse, D.Y.; Zuo, B.; To, C.H.; Liu, Q.; McFadden, S.A.; Chun, R.K.; Bian, J.; Li, K.K.; Lam, T.C. Integrated SWATH-based and targeted-based proteomics provide insights into the retinal emmetropization process in guinea pig. J. Proteom. 2018, 181, 1–15. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Chun, R.K.M.; Wang, J.C.; Zuo, B.; Li, K.K.; Lam, T.C.; Liu, Q.; To, C.H. Proteomic analysis of chick retina during early recovery from lens-induced myopia. Mol. Med. Rep. 2018, 18, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Q.; Jiang, C.; Ye, X.; Huang, X.; Jin, H.; Xu, G. Vitreous Proteomics Provides New Insights into Antivascular Endothelial Growth Factor Therapy for Pathologic Myopia Choroid Neovascularization. J. Interferon Cytokine Res. 2019, 39, 786–796. [Google Scholar] [CrossRef]
- Barbas-Bernardos, C.; Armitage, E.G.; García, A.; Mérida, S.; Navea, A.; Bosch-Morell, F.; Barbas, C. Looking into aqueous humor through metabolomics spectacles-exploring its metabolic characteristics in relation to myopia. J. Pharm. Biomed. Anal 2016, 127, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Rao, J.; Rong, X.; Lou, S.; Zheng, Z.; Lu, Y. Metabolic characterization of human aqueous humor in relation to high myopia. Exp. Eye Res. 2017, 159, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Yang, W.; Qin, X.; Li, Y.; Cao, H.; Zhou, C.; Wang, Y. Serum metabolomics profiling and potential biomarkers of myopia using LC-QTOF/MS. Exp. Eye Res. 2019, 186, 107737. [Google Scholar] [CrossRef]
- Du, B.; Jin, N.; Zhu, X.; Lu, D.; Jin, C.; Li, Z.; Han, C.; Zhang, Y.; Lai, D.; Liu, K.; et al. A prospective study of serum metabolomic and lipidomic changes in myopic children and adolescents. Exp. Eye Res. 2020, 199, 108182. [Google Scholar] [CrossRef]
- Ke, C.; Xu, H.; Chen, Q.; Zhong, H.; Pan, C.W. Serum metabolic signatures of high myopia among older Chinese adults. Eye 2020, 2020, 1–8. [Google Scholar] [CrossRef]
- Liu, K.; Fang, J.; Jin, J.; Zhu, S.; Xu, X.; Xu, Y.; Ye, B.; Lin, S.H.; Xu, X. Serum Metabolomics Reveals Personalized Metabolic Patterns for Macular Neovascular Disease Patient Stratification. J. Proteome Res. 2020, 19, 699–707. [Google Scholar] [CrossRef]
- Kearney, S.; O’Donoghue, L.; Pourshahidi, L.K.; Cobice, D.; Saunders, K.J. Myopes have significantly higher serum melatonin concentrations than non-myopes. Ophthalmic. Physiol. Opt. 2017, 37, 557–567. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Reinach, P.S.; Zhang, S.; Pan, M.; Sun, W.; Liu, B.; Li, F.; Li, X.; Zhao, A.; Chen, T.; et al. Changes in retinal metabolic profiles associated with form deprivation myopia development in guinea pigs. Sci. Rep. 2017, 7, 2777. [Google Scholar] [CrossRef]
- Pietrowska, K.; Dmuchowska, D.A.; Krasnicki, P.; Mariak, Z.; Kretowski, A.; Ciborowski, M. Analysis of pharmaceuticals and small molecules in aqueous humor. J. Pharm. Biomed. Anal. 2018, 159, 23–36. [Google Scholar] [CrossRef]
- Pietrowska, K.; Dmuchowska, D.A.; Samczuk, P.; Kowalczyk, T.; Krasnicki, P.; Wojnar, M.; Skowronska, A.; Mariak, Z.; Kretowski, A.; Ciborowski, M. LC-MS-Based Metabolic Fingerprinting of Aqueous Humor. J. Anal. Methods Chem. 2017, 2017, 6745932. [Google Scholar] [CrossRef] [PubMed]
- Wallman, J.; Winawer, J. Homeostasis of eye growth and the question of myopia. Neuron 2004, 43, 447–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, L.K.; Wildsoet, C.F. Effects on the compensatory responses to positive and negative lenses of intermittent lens wear and ciliary nerve section in chicks. Vis. Res. 1996, 36, 1023–1036. [Google Scholar] [CrossRef]
- Wildsoet, C.; Wallman, J. Choroidal and scleral mechanisms of compensation for spectacle lenses in chicks. Vis. Res. 1995, 35, 1175–1194. [Google Scholar] [CrossRef] [Green Version]
- Morgan, G.I.; Ashby, R.S.; Nickla, D.L. Form deprivation and lens-induced myopia: Are they different? Ophthalmic. Physiol. Opt. 2013, 33, 355–361. [Google Scholar] [CrossRef] [Green Version]
- Rose, K.A.; Morgan, I.G.; Ip, J.; Kifley, A.; Huynh, S.; Smith, W.; Mitchell, P. Outdoor activity reduces the prevalence of myopia in children. Ophthalmology 2008, 115, 1279–1285. [Google Scholar] [CrossRef]
- Li, S.M.; Li, H.; Li, S.Y.; Liu, L.R.; Kang, M.T.; Wang, Y.P.; Zhang, F.; Zhan, S.Y.; Gopinath, B.; Mitchell, P.; et al. Time Outdoors and Myopia Progression Over 2 Years in Chinese Children: The Anyang Childhood Eye Study. Investig. Ophthalmol. Vis. Sci. 2015, 56, 4734–4740. [Google Scholar] [CrossRef] [Green Version]
- Saw, S.M.; Hong, R.Z.; Zhang, M.Z.; Fu, Z.F.; Ye, M.; Tan, D.; Chew, S.J. Near-work activity and myopia in rural and urban schoolchildren in China. J. Pediatr. Ophthalmol. Strabismus 2001, 38, 149–155. [Google Scholar]
- Saw, S.M.; Chua, W.H.; Hong, C.Y.; Wu, H.M.; Chan, W.Y.; Chia, K.S.; Stone, R.A.; Tan, D. Nearwork in early-onset myopia. Investig. Ophthalmol. Vis. Sci. 2002, 43, 332–339. [Google Scholar]
- Chia, A.; Lu, Q.S.; Tan, D. Five-Year Clinical Trial on Atropine for the Treatment of Myopia 2: Myopia Control with Atropine 0.01% Eyedrops. Ophthalmology 2016, 123, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Yam, J.C.; Jiang, Y.; Tang, S.M.; Law, A.K.P.; Chan, J.J.; Wong, E.; Ko, S.T.; Young, A.L.; Tham, C.C.; Chen, L.J.; et al. Low-Concentration Atropine for Myopia Progression (LAMP) Study: A Randomized, Double-Blinded, Placebo-Controlled Trial of 0.05%, 0.025%, and 0.01% Atropine Eye Drops in Myopia Control. Ophthalmology 2019, 126, 113–124. [Google Scholar] [CrossRef]
- Barathi, S.R.; Weon, P.W.Y.; Rebekah, R.W. Beuerman Muscarinic Regulation of Epidermal Growth Factor Receptor in Mammalian Retinal Pigment Epithelial (RPE) Cells. Investig. Ophthalmol. Vis. Sci. 2008, 49, 3535. [Google Scholar]
- Atkinson, L.C.; Feng, J.; Zhang, D.Q. Functional integrity and modification of retinal dopaminergic neurons in the rd1 mutant mouse: Roles of melanopsin and GABA. J. Neurophysiol. 2013, 109, 1589–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.D.; Zhou, T.R.; McMahon, D.G. Functional heterogeneity of retinal dopaminergic neurons underlying their multiple roles in vision. J. Neurosci. 2007, 27, 692–699. [Google Scholar] [CrossRef] [Green Version]
- Mathis, U.; Feldkaemper, M.; Wang, M.; Schaeffel, F. Studies on retinal mechanisms possibly related to myopia inhibition by atropine in the chicken. Graefe’s Arch. Clin. Exp. Ophthalmol. 2020, 258, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Wiechmann, F.A.; Wirsig-Wiechmann, C.R. Multiple cell targets for melatonin action in Xenopus laevis retina: Distribution of melatonin receptor immunoreactivity. Vis. Neurosci. 2001, 18, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Wiechmann, F.A.; Rada, J.A. Melatonin receptor expression in the cornea and sclera. Exp. Eye Res. 2003, 77, 219–225. [Google Scholar] [CrossRef]
- Rada, J.A.; Wiechmann, A.F. Melatonin receptors in chick ocular tissues: Implications for a role of melatonin in ocular growth regulation. Investig. Ophthalmol. Vis. Sci. 2006, 47, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Zhou, J.; Lu, Y.; Chu, R. Effects of 530 nm green light on refractive status, melatonin, MT1 receptor, and melanopsin in the guinea pig. Curr. Eye Res. 2011, 36, 103–111. [Google Scholar] [CrossRef]
- Freedman, M.S.; Lucas, R.J.; Soni, B.; von Schantz, M.; Muñoz, M.; David-Gray, Z.; Foster, R. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science 1999, 284, 502–504. [Google Scholar] [CrossRef] [PubMed]
- Stormann, T.M.; Gdula, D.C.; Weiner, D.M.; Brann, M.R. Molecular cloning and expression of a dopamine D2 receptor from human retina. Mol. Pharmacol. 1990, 37, 1–6. [Google Scholar]
- Hung, L.F.; Arumugam, B.; Ostrin, L.; Patel, N.; Trier, K.; Jong, M.; Smith, E.L., III. The Adenosine Receptor Antagonist, 7-Methylxanthine, Alters Emmetropizing Responses in Infant Macaques. Investig. Ophthalmol. Vis. Sci. 2018, 59, 472–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, H.H.; Huo, L.J.; Yang, X.; Gao, Z.Y.; Zeng, J.W.; Trier, K.; Cui, D.M. Effects of 7-methylxanthine on form-deprivation myopia in pigmented rabbits. Int. J. Ophthalmol. 2012, 5, 133–137. [Google Scholar] [PubMed]
- Trier, K.; Munk Ribel-Madsen, S.; Cui, D.; Brøgger Christensen, S. Systemic 7-methylxanthine in retarding axial eye growth and myopia progression: A 36-month pilot study. J. Ocul. Biol. Dis. Inform. 2008, 1, 85–93. [Google Scholar] [CrossRef] [Green Version]
- Erickson, B.K.; Rose, C.M.; Braun, C.R.; Erickson, A.R.; Knott, J.; McAlister, G.C.; Wühr, M.; Paulo, J.A.; Everley, R.A.; Gygi, S.P. A Strategy to Combine Sample Multiplexing with Targeted Proteomics Assays for High-Throughput Protein Signature Characterization. Mol. Cell 2017, 65, 361–370. [Google Scholar] [CrossRef] [Green Version]
- Neilson, K.A.; Ali, N.A.; Muralidharan, S.; Mirzaei, M.; Mariani, M.; Assadourian, G.; Lee, A.; van Sluyter, S.C.; Haynes, P.A. Less label, more free: Approaches in label-free quantitative mass spectrometry. Proteomics 2011, 11, 535–553. [Google Scholar] [CrossRef]
- Krasny, L.; Bland, P.; Kogata, N.; Wai, P.; Howard, B.A.; Natrajan, R.C.; Huang, P.H. SWATH mass spectrometry as a tool for quantitative profiling of the matrisome. J. Proteom. 2018, 189, 11–22. [Google Scholar] [CrossRef]
- Schmidlin, T.; Garrigues, L.; Lane, C.S.; Mulder, T.C.; van Doorn, S.; Post, H.; de Graaf, E.L.; Lemeer, S.; Heck, A.J.; Altelaar, A.F. Assessment of SRM, MRM(3), and DIA for the targeted analysis of phosphorylation dynamics in non-small cell lung cancer. Proteomics 2016, 16, 2193–2205. [Google Scholar] [CrossRef]
- Saraswathy, N.; Ramalingam, P. Introduction to Proteomics; Woodhead Publishing Series in Biomaterials; Woodhead Publishing: Cambridge, UK, 2011; pp. 147–158. [Google Scholar]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [Green Version]
- Filimoniuk, A.; Daniluk, U.; Samczuk, P.; Wasilewska, N.; Jakimiec, P.; Kucharska, M.; Lebensztejn, D.M.; Ciborowski, M. Metabolomic profiling in children with inflammatory bowel disease. Adv. Med Sci. 2020, 65, 65–70. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mihalik, S.J.; Michaliszyn, S.F.; de las Heras, J.; Bacha, F.; Lee, S.; Chace, D.H.; DeJesus, V.R.; Vockley, J.; Arslanian, S.A. Metabolomic profiling of fatty acid and amino acid metabolism in youth with obesity and type 2 diabetes: Evidence for enhanced mitochondrial oxidation. Diabetes Care 2012, 35, 605–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, Q.; Wang, W.; Wang, N.; Peng, Y.; Ma, W.; Dai, R. Quantification of Arachidonic Acid and Its Metabolites in Rat Tissues by UHPLC-MS/MS: Application for the Identification of Potential Biomarkers of Benign Prostatic Hyperplasia. PLoS ONE 2016, 11, e0166777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salek, R.M.; Steinbeck, C.; Viant, M.R.; Goodacre, R.; Dunn, W.B. The role of reporting standards for metabolite annotation and identification in metabolomic studies. GigaScience 2013, 2. [Google Scholar] [CrossRef]
| Type of Analytical Method | Type of Analysis | Type of Sample | Organism | Name of the Protein and/or Protein-Encoding Gene | Scientific Aspects | Ref. |
|---|---|---|---|---|---|---|
| SDS-PAGE MALDI TOF | untargeted | corneal epithelium | human | S100A4, KRT3, GSN, ENO1 | comparison of patients with keratoconus vs. myopic patients as the controls | [7] |
| SWATH-MS | untargeted | vitreous humor | white Leghorn chicks | - | creation of a proteome library in emmetropization | [8] |
| SWATH-MS | untargeted | tears | human | - | creation of a proteome library in patients wearing orthokeratology lenses as treatment of myopia | [9] |
| Label-free LC-MS | untargeted | vitreous humor | human | PTGDS, GPX3 | identification of expressed proteins in patients with pathological myopia and controls | [10] |
| 2D-PAGE MALDI TOF | untargeted | sclera | shrews (Tupaia belangeri) | pigment epithelium-derived factor, procollagen Iα1, procollagen Iα2, thrombospondin I, glucose-regulated protein | analysis of differences in the development of lens-induced myopia and recovery from this condition | [11] |
| SWATH-MS | untargeted | cornea | white Leghorn chicks | - | creation of a corneal proteome library in high myopia | [12] |
| 2D-PAGE MALDI TOF | untargeted | retina | mouse | Cryga, Cryba2, Cryba1 | analysis of differences after exposure to various light conditions | [13] |
| 2D-PAGE LC-MS | untargeted | retina, RPE, choroid | tilapia (Oreochromis niloticus) | annexin A5, gelsolin, TCP-1 | analysis of differences in the protein profiles found in induced myopia | [14] |
| Label-free LC-MS | untargeted | vitreous humor | mouse | over 30 differentiating proteins | analysis of high myopia profiles with the low-density lipoprotein receptor-related protein 2 | [15] |
| 2D-PAGE MALDI-TOF | untargeted | retina | white Leghorn chicks | VIL1, DPYSL2, SARS, SEPTIN2, PGAM1 tubulin α, tubulin β2, tubulin α-chain, β-tubulin | analysis of differences in retinal proteins from lens-induced myopic chicks and controls | [16] |
| ICPL LC-MS, MRM LC-MS | untargeted, targeted | retina | white Leghorn chicks | VIM, APOA1, GSTM2 | identification of proteins in myopic chicks and their association with excessive eye elongation | [17] |
| ICPL LC-MS | untargeted | vitreous humor | white Leghorn chicks | APOA1, TF, purpurin | identification of proteins differentiating myopia from hyperopia | [18] |
| iTRAQ LC-MS | untargeted | aqueous humor | human | ATP8A1, KRT2, KRT10, CRYAA, CRYBA4, CRYAA, CRYBB1, CRYBB2, CRYBA1, KRT6B, KRT6A, KRT14, KRT16 | comparison of protein profiles in patients undergoing cataract surgery with concomitant myopia, glaucoma, or diabetes and controls | [19] |
| iTRAQ LC-MS | untargeted | retina | mouse | over 25 differentiating proteins | analysis of atropine effect on retina proteome in myopic mice | [20] |
| Label-free LC-MS | untargeted | retina | chicks | analysis of a biochemical pathway | identification of pathways involved in myopia and hyperopia | [21] |
| 2D-PAGE MALDI-TOF | untargeted | sclera | guinea pig | Cryab, CryaA | analysis of changes in protein profiles during the development of form-deprivation myopia and recovery from this condition | [22] |
| 2D-PAGE LC-MS | untargeted | sclera | tree shrew | over 50 differentiating proteins | analysis of changes in the protein profiles of lens-induced myopia and recovery from this condition | [23] |
| SWATH, MRM-MS | untargeted, targeted | retina | guinea pigs | - | creation of a proteome library in emmetropization | [24] |
| 2D-PAGE MALDI-TOF | untargeted | retina and fibrous sclera | chicks | APOA1, CRMP-62, CKB, ENO2, tubulin α-1 chain, VIM | study of emmetropization | [25] |
| iTRAQ LC-MS | untargeted | aqueous humor | human | over 200 differentiating proteins | identification of proteins contributing to the development of cataract in myopic patients | [26] |
| Label-free LC-MS | untargeted | retina, retinal pigment epithelium | chicks (White Leghorn/New Hampshire) | over 65 differentiating proteins | analysis of proteomic responses to early optical defocus in relation to transcriptome-level changes | [27] |
| 2D-PAGE MALDI-TOF | untargeted | aqueous humor | human | ALB, TTR, GC | comparison of the proteome in high myopia patients and controls | [28] |
| 2D-PAGE MALDI-TOF | untargeted | retina | guinea pig | ACTB, MDH1, Rab-11B, PKM2, ACP1 | analysis of differential protein expression in response to lens-induced myopia | [29] |
| SWATH-MS, MRM-MS | untargeted targeted | retina | guinea pig | - | creation of a spectral library of protein profile changes during emmetropization | [30] |
| 2D-PAGE LC-MS | untargeted | retina | chick | ARR3, Rab-11B, PSMD14, β-tubulin, PRDX6, UCH-L1 | proteome study during early recovery from lens-induced myopia | [31] |
| Label-free LC-MS | untargeted | vitreous humor | human | over 50 differentiating proteins | analysis of protein expression profiles in vitreous humor from patients with pathologic myopic retinoschisis with/without intravitreal antivascular endothelial growth factor therapy | [32] |
| Type of Analytical Method | Type of Analysis | Type of Sample | Organism | Type of Analytical Method | Potential Biomarkers or Altered Pathways | Ref. |
|---|---|---|---|---|---|---|
| CE-TOF-MS | untargeted | aqueous humor | human | comparison of patients with high myopia (n = 12) and low myopia (n = 24) | aminooctanoic acid, L-arginine, citrulline, aminoundecanoic acid, L-cysteinylglycine disulfide | [33] |
| LC-QTOF-MS | untargeted | aqueous humor | human | comparison of patients with high myopia (n = 12) and low myopia (n = 24) | trihydroxyphenyl-gamma-valerolactone, dihydropteroic acid, dodecanedioic acid, aminocyclohexanecarboxylic acid, butyryl-L-carnitine, pantothenic acid, didehydro-retinoic acid, sphinganine, histidinyl-phenylalanine, dimethylnonanoyl carnitine, PC(O-32:2)//PC(P-32:1), PC (42:6), C24 sulfatide, PC(P-42:2)//PC(O-42:3), LacCer(d40:0), trihexosylceramide (d36:2), NeuAcaGalCer(d42:2) | [33] |
| GC-TOF-MS | untargeted | aqueous humor | human | metabolic profiling in patients with high myopia (n = 20) and controls (n = 20) | glutamine, N-alpha-acetyl-L-ornithine, nicotinoylglycine, oxalacetic acid, o-hydroxyhippuric acid, oxalic acid, ribose, cis-gondoic acid, linoleic acid methyl ester, thymidine, phosphate, indole-3-acetamide, 2-aminophenol, 2-ketoadipate, 3-phenyllactic acid, cis-phytol, conduritol b epoxide, salicin | [34] |
| LC-QTOF-MS | untargeted | serum | human | metabolic profiling in high myopia cases (n = 30) and controls (n = 30) | γ-glutamyltyrosine and 12-oxo-20-trihydroxy-leukotriene B4 | [35] |
| UHPLC-MS | untargeted | serum | human | metabolomics profiling in myopia cases (n = 108) and controls (n = 103) | steroid biosynthesis, lysine degradation, arginine and proline metabolism, glycerolipid metabolism, glycerophospholipid metabolism, arachidonic acid metabolism, linoleic acid metabolism, sphingolipid metabolism | [36] |
| UHPLC-MS | untargeted | serum | human | lipid profiling in myopia cases (n = 108) and controls (n = 103) | steroid biosynthesis, lysine degradation, glycerolipid metabolism, glycerophospholipid metabolism, arachidonic acid metabolism, linoleic acid metabolism, alpha-linolenic acid metabolism, sphingolipid metabolism | [36] |
| GC-TOF-MS | untargeted | serum | human | metabolomic analysis of patients with high myopia (n = 40) and low myopia (n = 40) | alanine, mannose, itaconic acid, aconitic acid, O-acetylserine, phthalic acid, abietic acid, salicin, citric acid, aminomalonic acid, palmitoleic acid, conduritol b epoxide, shikimic acid, 4-hydroxyphenylacetic acid, hesperitin, anandamide, oxalacetic acid, pimelic acid, 2-ketoadipate, N-ethylmaleamic acid | [37] |
| GC-TOF-MS | untargeted | serum | human | metabolic profiling in patients with pathological myopia (n = 57) and controls (n = 81) | hypoxanthine, L-2-amino-3-(1-pyrazolyl)propanoic acid, linoleic acid, maleic acid, ribonolactone | [38] |
| LC-On-Line SPE-MS/MS | targeted | serum | human | myopia patients (n = 25) and controls (n = 29) at the baseline and after 18-month follow-up (22 patients and 23 controls) | melatonin, dopamine | [39] |
| GC-TOF-MS | untargeted | retina | guinea pig | time-dependent form-deprivation myopia, T = 3 days, 12 cases and 5 controls | mannose, urea, glucose, arabinose, tyrosine, glutamic acid, threonine, valine, isoleucine, malic acid, alanine | [40] |
| GC-TOF-MS | untargeted | retina | guinea pig | time-dependent form-deprivation myopia, T = 2 weeks, 12 cases and 6 controls | threonine, valine, isoleucine, malic acid, alanine, arachidic acid (20:0), octadecenoic acid (18:1), octadecanoic acid (18:0), arachidonic acid (20:4), cholesterol, ethanolamine, hexadecanoic acid (16:0), tetradecanoic acid (14:0), octadecadienoic acid (18:2), 2-ketoglutaric acid, GABA | [40] |
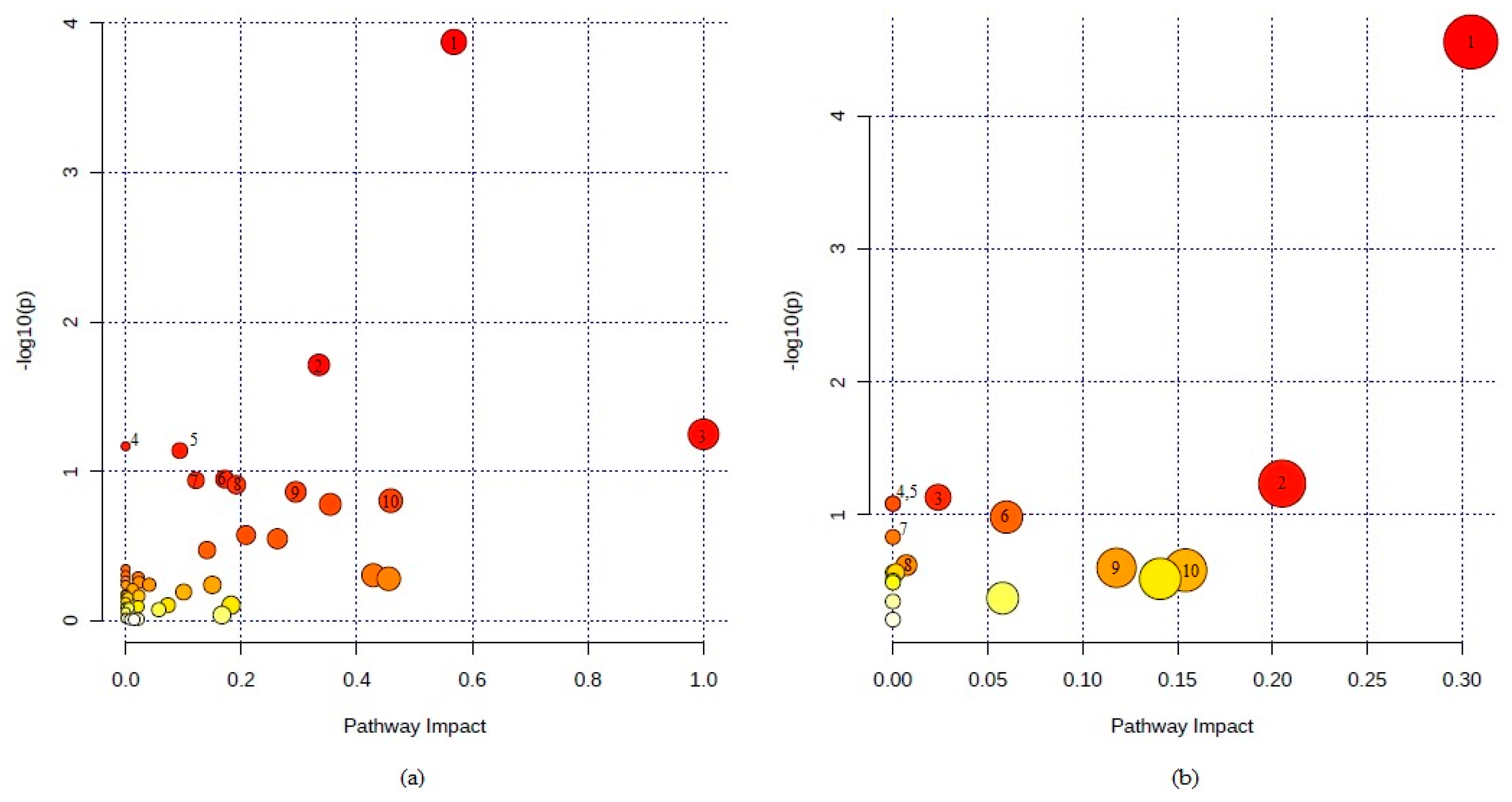
| Pathway | No. of Metabolites in the Pathway | No. of Metabolites Detected in Serum | p-Value | Pathway Impact |
|---|---|---|---|---|
| Sphingolipid metabolism | 21 | 8 | 0.00013 | 0.57 |
| Citrate cycle (TCA cycle) | 20 | 5 | 0.019 | 0.33 |
| Linoleic acid metabolism | 5 | 2 | 0.056 | 1.0 |
| Biosynthesis of unsaturated fatty acids | 36 | 6 | 0.068 | 0.0 |
| Alanine, aspartate and glutamate metabolism | 28 | 5 | 0.072 | 0.094 |
| Tryptophan metabolism | 41 | 6 | 0.11 | 0.17 |
| Glyoxylate and dicarboxylate metabolism | 32 | 5 | 0.11 | 0.12 |
| Tyrosine metabolism | 42 | 6 | 0.12 | 0.19 |
| Glycerolipid metabolism | 16 | 3 | 0.14 | 0.29 |
| Retinol metabolism | 17 | 3 | 0.16 | 0.46 |
| Glycerophospholipid metabolism | 36 | 5 | 0.17 | 0.35 |
| Pyruvate metabolism | 22 | 3 | 0.27 | 0.21 |
| Glycine, serine and threonine metabolism | 33 | 4 | 0.28 | 0.26 |
| Lysine degradation | 25 | 3 | 0.34 | 0.14 |
| Thiamine metabolism | 7 | 1 | 0.45 | 0.0 |
| Ascorbate and aldarate metabolism | 8 | 1 | 0.50 | 0.0 |
| Taurine and hypotaurine metabolism | 8 | 1 | 0.50 | 0.43 |
| Cysteine and methionine metabolism | 33 | 3 | 0.52 | 0.021 |
| Beta-alanine metabolism | 21 | 2 | 0.52 | 0.45 |
| One carbon pool by folate | 9 | 1 | 0.54 | 0.0 |
| Steroid hormone biosynthesis | 85 | 7 | 0.56 | 0.023 |
| Propanoate metabolism | 23 | 2 | 0.57 | 0.041 |
| Phenylalanine metabolism | 10 | 1 | 0.57 | 0.0 |
| Biotin metabolism | 10 | 1 | 0.57 | 0.15 |
| Arginine and proline metabolism | 38 | 3 | 0.61 | 0.012 |
| Glycolysis or Gluconeogenesis | 26 | 2 | 0.64 | 0.10 |
| Galactose metabolism | 27 | 2 | 0.66 | 0.0 |
| Alpha-linolenic acid metabolism | 13 | 1 | 0.67 | 0.0 |
| Glutathione metabolism | 28 | 2 | 0.68 | 0.023 |
| Arginine biosynthesis | 14 | 1 | 0.70 | 0.0 |
| Glycosylphosphatidylinositol(GPI)-anchor biosynthesis | 14 | 1 | 0.70 | 0.0040 |
| Histidine metabolism | 16 | 1 | 0.75 | 0.0 |
| Starch and sucrose metabolism | 18 | 1 | 0.79 | 0.073 |
| Terpenoid backbone biosynthesis | 18 | 1 | 0.79 | 0.18 |
| Pantothenate and CoA biosynthesis | 19 | 1 | 0.80 | 0.021 |
| Arachidonic acid metabolism | 36 | 2 | 0.81 | 0.0 |
| Fructose and mannose metabolism | 20 | 1 | 0.82 | 0.0 |
| Selenocompound metabolism | 20 | 1 | 0.82 | 0.0 |
| Ether lipid metabolism | 20 | 1 | 0.82 | 0.0 |
| Amino sugar and nucleotide sugar metabolism | 37 | 2 | 0.82 | 0.0058 |
| Pyrimidine metabolism | 39 | 2 | 0.84 | 0.057 |
| Steroid biosynthesis | 42 | 2 | 0.87 | 0.0 |
| Aminoacyl-tRNA biosynthesis | 48 | 2 | 0.92 | 0.17 |
| Drug metabolism - cytochrome P450 | 55 | 2 | 0.95 | 0.0 |
| Fatty acid elongation | 39 | 1 | 0.96 | 0.0 |
| Fatty acid degradation | 39 | 1 | 0.96 | 0.0 |
| Purine metabolism | 65 | 2 | 0.97 | 0.022 |
| Primary bile acid biosynthesis | 46 | 1 | 0.98 | 0.0076 |
| Fatty acid biosynthesis | 47 | 1 | 0.98 | 0.015 |
| Pathway | No. of Metabolites in the Pathway | No. of Metabolites Detected in AH | p-Value | Pathway Impact |
|---|---|---|---|---|
| Arginine biosynthesis | 14 | 4 | 0.000028 | 0.30 |
| Alanine, aspartate and glutamate metabolism | 28 | 2 | 0.058 | 0.21 |
| Glyoxylate and dicarboxylate metabolism | 32 | 2 | 0.074 | 0.024 |
| D-Glutamine and D-glutamate metabolism | 6 | 1 | 0.082 | 0.0 |
| Nitrogen metabolism | 6 | 1 | 0.082 | 0.0 |
| Pyrimidine metabolism | 39 | 2 | 0.10 | 0.06 |
| Aminoacyl-tRNA biosynthesis | 48 | 2 | 0.15 | 0.0 |
| Pantothenate and CoA biosynthesis | 19 | 1 | 0.24 | 0.0071 |
| Citrate cycle (TCA cycle) | 20 | 1 | 0.25 | 0.12 |
| Sphingolipid metabolism | 21 | 1 | 0.26 | 0.15 |
| Pentose phosphate pathway | 22 | 1 | 0.27 | 0.0 |
| Pyruvate metabolism | 22 | 1 | 0.27 | 0.0016 |
| Lysine degradation | 25 | 1 | 0.30 | 0.14 |
| Glycolysis or Gluconeogenesis | 26 | 1 | 0.31 | 0.0 |
| Folate biosynthesis | 27 | 1 | 0.32 | 0.0 |
| Arginine and proline metabolism | 38 | 1 | 0.42 | 0.056 |
| Tryptophan metabolism | 41 | 1 | 0.45 | 0.0 |
| Purine metabolism | 65 | 1 | 0.61 | 0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grochowski, E.T.; Pietrowska, K.; Kowalczyk, T.; Mariak, Z.; Kretowski, A.; Ciborowski, M.; Dmuchowska, D.A. Omics in Myopia. J. Clin. Med. 2020, 9, 3464. https://doi.org/10.3390/jcm9113464
Grochowski ET, Pietrowska K, Kowalczyk T, Mariak Z, Kretowski A, Ciborowski M, Dmuchowska DA. Omics in Myopia. Journal of Clinical Medicine. 2020; 9(11):3464. https://doi.org/10.3390/jcm9113464
Chicago/Turabian StyleGrochowski, Emil Tomasz, Karolina Pietrowska, Tomasz Kowalczyk, Zofia Mariak, Adam Kretowski, Michal Ciborowski, and Diana Anna Dmuchowska. 2020. "Omics in Myopia" Journal of Clinical Medicine 9, no. 11: 3464. https://doi.org/10.3390/jcm9113464
APA StyleGrochowski, E. T., Pietrowska, K., Kowalczyk, T., Mariak, Z., Kretowski, A., Ciborowski, M., & Dmuchowska, D. A. (2020). Omics in Myopia. Journal of Clinical Medicine, 9(11), 3464. https://doi.org/10.3390/jcm9113464






