Vaccinomics and Adversomics in the Era of Precision Medicine: A Review Based on HBV, MMR, HPV, and COVID-19 Vaccines
Abstract
:1. Introduction
2. Vaccinomics
2.1. Vaccinomics of the Hepatitis B Virus Vaccine
2.2. Vaccinomics of the MMR Vaccine
2.3. Vaccinomics of the Human Papilloma Virus Vaccine
3. Adversomics
3.1. Adversomics of the Hepatitis B Vaccine
3.2. Adversomics of the MMR Vaccine
3.3. Adversomics of the Human Papilloma Virus Vaccine
3.4. Biological Sex Differences in Adversomics
3.5. Challenges and Future Perspectives in Adversomics
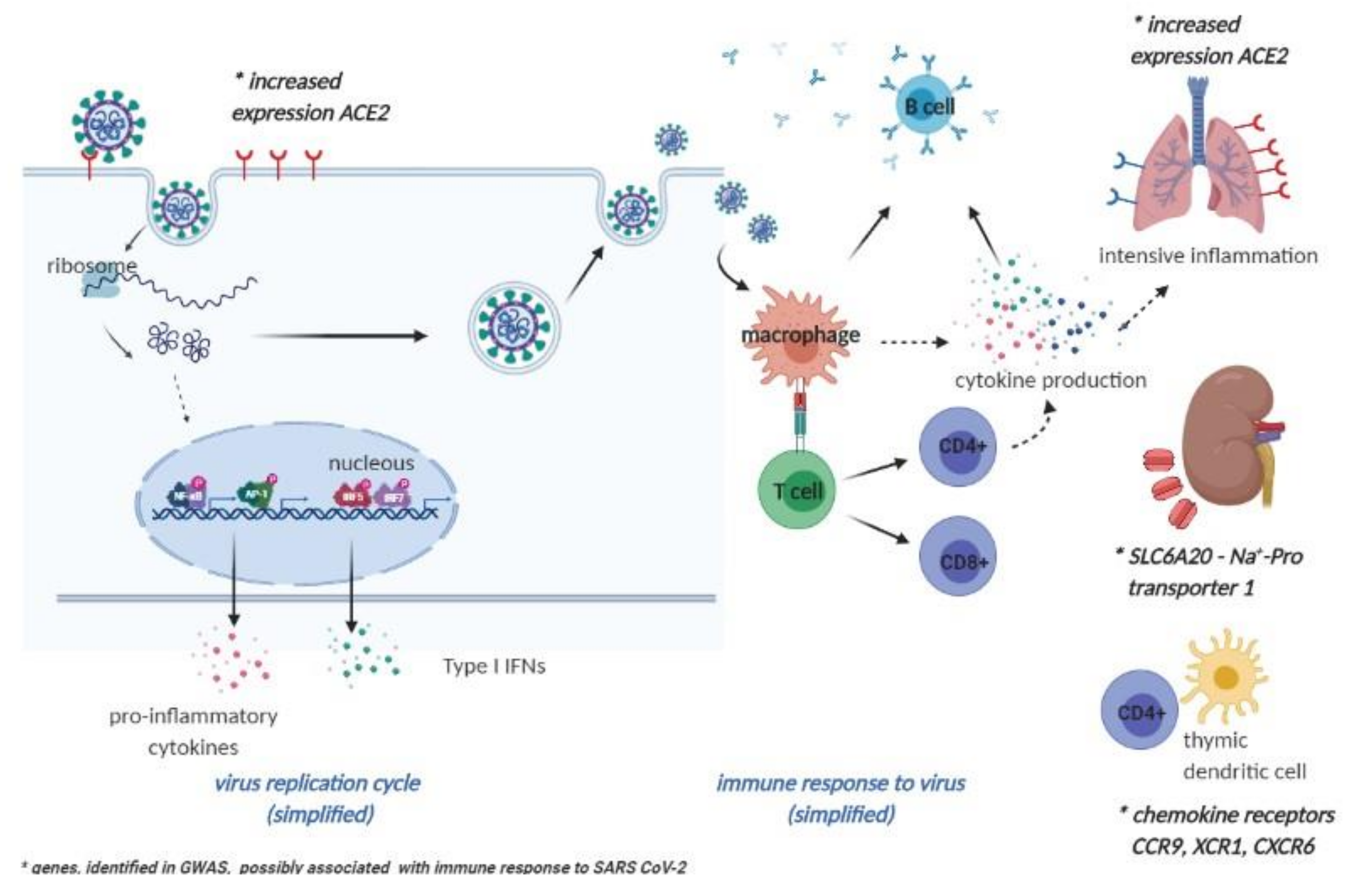
4. Future Perspectives on Vaccinomics and Adversomics of a COVID-19 Vaccine
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Precision Medicine Initiative. Available online: https://obamawhitehouse.archives.gov/precision-medicine (accessed on 25 September 2020).
- Poland, G.A.; Ovsyannikova, I.G.; Jacobson, R.M. Application of Pharmacogenomics to Vaccines. Pharmacogenomics 2009, 10, 837–852. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, S.L.; Rogers, W.O.; Carucci, D.J.; Venter, J.C. From Genomics to Vaccines: Malaria as a Model System. Nat. Med. 1998, 4, 1351–1353. [Google Scholar] [CrossRef]
- Poland, G.A.; Kennedy, R.B.; McKinney, B.A.; Ovsyannikova, I.G.; Lambert, N.D.; Jacobson, R.M.; Oberg, A.L. Vaccinomics, Adversomics, and the Immune Response Network Theory: Individualized Vaccinology in the 21st Century. Semin. Immunol. 2013, 25, 89–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poland, G.A.; Ovsyannikova, I.; Kennedy, R. Personalized Vaccinology: A Review. Vaccine 2018, 36, 5350–5357. [Google Scholar] [CrossRef]
- Whitaker, J.A.; Ovsyannikova, I.G.; Poland, G.A. Adversomics: A New Paradigm for Vaccine Safety and Design. Expert Rev. Vaccines 2015, 14, 935–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feenstra, B.; Pasternak, B.; Geller, F.; Carstensen, L.; Wang, T.; Huang, F.; Eitson, J.L.; Hollegaard, M.V.; Svanström, H.; Vestergaard, M.; et al. Common Variants Associated With General and MMR Vaccine–Related Febrile Seizures. Nat. Genet. 2014, 46, 1274–1282. [Google Scholar] [CrossRef] [Green Version]
- Kwok, R. Vaccines: The Real Issues in Vaccine Safety. Nature 2011, 473, 436–438. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, R.M.; Adegbenro, A.; Pankratz, V.; Poland, G.A. Adverse Events and Vaccination-the Lack of Power and Predictability of Infrequent Events in Pre-Licensure Study. Vaccine 2001, 19, 2428–2433. [Google Scholar] [CrossRef]
- Barker, C.I.S.; Snape, M.D. Pandemic Influenza a H1N1 Vaccines and Narcolepsy: Vaccine Safety Surveillance in Action. Lancet Infect. Dis. 2014, 14, 227–238. [Google Scholar] [CrossRef]
- Chen, D.-S. Hepatitis B Vaccination: The Key Towards Elimination and Eradication of Hepatitis B. J. Hepatol. 2009, 50, 805–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walayat, S.; Ahmed, Z.; Martin, D.; Puli, S.; Cashman, M.; Dhillon, S. Recent Advances in Vaccination of Non-Responders to Standard Dose Hepatitis B Virus Vaccine. World J. Hepatol. 2015, 7, 2503–2509. [Google Scholar] [CrossRef]
- Harrison, P. Hepatitis B Vaccine Ineffective in Most Patients with RA. In Proceedings of the European League Against Rheumatism (EULAR) Congress, Rome, Italy, 10–13 June 2015. [Google Scholar]
- Vitaliti, G.; Praticò, A.D.; Cimino, C.; Di Dio, G.; Lionetti, E.; La Rosa, M.; Leonardi, S. Hepatitis B Vaccine in Celiac Disease: Yesterday, Today and Tomorrow. World J. Gastroenterol. 2013, 19, 838–845. [Google Scholar] [CrossRef]
- Mormile, R.; Mormile, M.R. Hepatitis B Vaccine Non Response: A Predictor of Latent Autoimmunity? Med. Hypotheses 2017, 104, 45–47. [Google Scholar] [CrossRef]
- Newport, M.; The MRC Gambia Twin Study Group; Goetghebuer, T.; Weiss, H.A.; Whittle, H.; Siegrist, C.-A.; Marchant, A. Genetic Regulation of Immune Responses to Vaccines in Early Life. Genes Immun. 2004, 5, 122–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alper, C.A.; Kruskall, M.S.; Marcus-Bagley, D.; Craven, D.E.; Katz, A.J.; Brink, S.J.; Dienstag, J.L.; Awdeh, Z.; Yunis, E.J. Genetic Prediction of Nonresponse to Hepatitis B Vaccine. N. Engl. J. Med. 1989, 321, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Kruskall, M.S. The Major Histocompatibility Complex: The Value of Extended Haplotypes in the Analysis of Associated Immune Diseases and Disorders. Yale J. Boil. Med. 1990, 63, 477–486. [Google Scholar]
- Höhler, T.; Reuss, E.; Evers, N.; Dietrich, E.; Rittner, C.; Freitag, C.M.; Vollmar, J.; Schneider, P.M.; Fimmers, R. Differential Genetic Determination of Immune Responsiveness to Hepatitis B Surface Antigen and to Hepatitis a Virus: A Vaccination Study in Twins. Lancet 2002, 360, 991–995. [Google Scholar] [CrossRef]
- Desombere, I.; Willems, A.; Leroux-Roels, G. Response to Hepatitis B Vaccine: Multiple HLA Genes Are Involved. Tissue Antigens 1998, 51, 593–604. [Google Scholar] [CrossRef]
- Kruger, A.; Adams, P.; Hammer, J.; Böcher, W.O.; Schneider, P.M.; Rittner, C.; Hoehler, T. Hepatitis B Surface Antigen Presentation and HLA-DRB1*—Lessons from Twins and Peptide Binding Studies. Clin. Exp. Immunol. 2005, 140, 325–332. [Google Scholar] [CrossRef]
- Davila, S.; Froeling, F.E.M.; Tan, A.; Bonnard, C.; Boland, G.J.; Snippe, H.; Hibberd, M.L.; Seielstad, M. New Genetic Associations Detected in a Host Response Study to Hepatitis B Vaccine. Genes Immun. 2010, 11, 232–238. [Google Scholar] [CrossRef] [Green Version]
- Pan, L.; Zhang, L.; Zhang, W.; Wu, X.; Li, Y.; Yan, B.; Zhu, X.; Liu, X.; Yang, C.; Xu, J.; et al. A Genome-Wide Association Study Identifies Polymorphisms in the HLA-DR Region Associated with Non-Response to Hepatitis B Vaccination in Chinese Han Populations. Hum. Mol. Genet. 2013, 23, 2210–2219. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.-W.; Chen, C.-F.; Lai, S.-K.; Lin, H.H.; Chu, C.-C.; Wang, L.-Y. SNP rs7770370 inHLA-DPB1 Loci as a Major Genetic Determinant of Response to Booster Hepatitis B Vaccination: Results of a Genome-Wide Association Study. J. Gastroenterol. Hepatol. 2015, 30, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Nishida, N.; Sugiyama, M.; Sawai, H.; Nishina, S.; Sakai, A.; Ohashi, J.; Khor, S.-S.; Kakisaka, K.; Tsuchiura, T.; Hino, K.; et al. Key HLA-DRB1-DQB1 Haplotypes and Role of the BTNL2 Gene for Response to a Hepatitis B Vaccine. Hepatology 2018, 68, 848–858. [Google Scholar] [CrossRef]
- Höhler, T.; Stradmann-Bellinghausen, B.; Starke, R.; Sänger, R.; Victor, A.; Rittner, C.; Schneider, P.M. C4 A Deficiency and Nonresponse to Hepatitis B Vaccination. J. Hepatol. 2002, 37, 387–392. [Google Scholar] [CrossRef]
- Chen, J.; Liang, Z.; Lu, F.; Fang, X.; Liu, S.; Zeng, Y.; Zhu, F.; Chen, X.; Shen, T.; Li, J.; et al. Toll-Like Receptors and Cytokines/Cytokine Receptors Polymorphisms Associate With Non-Response to Hepatitis B Vaccine. Vaccine 2011, 29, 706–711. [Google Scholar] [CrossRef]
- Ensembl. Available online: https://www.ensembl.org/index.html (accessed on 1 March 2018).
- Ovsyannikova, I.G.; Pankratz, V.S.; Vierkant, R.A.; Jacobson, R.M.; Poland, G.A. Consistency of HLA Associations Between Two Independent Measles Vaccine Cohorts: A Replication Study. Vaccine 2012, 30, 2146–2152. [Google Scholar] [CrossRef] [Green Version]
- Ovsyannikova, I.G.; Poland, G.A. Vaccinomics: Current Findings, Challenges and Novel Approaches for Vaccine Development. AAPS J. 2011, 13, 438–444. [Google Scholar] [CrossRef] [Green Version]
- Belloni, C.; Avanzini, M.A.; De Silvestri, A.; Martinetti, M.; Pasi, A.; Coslovich, E.; Autelli, M.; Masanti, M.L.; Cuccia, M.; Tinelli, C.; et al. No Evidence of Autoimmunity in 6-Year-Old Children Immunized at Birth with Recombinant Hepatitis B Vaccine. Pediatrics 2002, 110, e4. [Google Scholar] [CrossRef] [Green Version]
- Miller, J.D.; Whitehair, L.H. HypothesisConcurrent HLA- Related Response Factors Mediate Recombinant Hepatitis B Vaccine Major Adverse Events. Autoimmunity 2005, 38, 181–194. [Google Scholar] [CrossRef]
- Scepanovic, P.; Alanio, C.; Hammer, C.; Hodel, F.; Bergstedt, J.; Patin, E.; Thorball, C.W.; Chaturvedi, N.; Charbit, B.; Abel, L.; et al. Human Genetic Variants and Age Are the Strongest Predictors of Humoral Immune Responses to Common Pathogens and Vaccines. Genome Med. 2018, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Dhiman, N.; Ovsyannikova, I.G.; Vierkant, R.A.; Pankratz, V.S.; Jacobson, R.M.; Poland, G.A. Associations Between Cytokine/Cytokine Receptor Single Nucleotide Polymorphisms and Humoral Immunity to Measles, Mumps and Rubella in a Somali Population. Tissue Antigens 2008, 72, 211–220. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, N.; Bucciol, G.; Moens, L.; Le Pen, J.; Shahrooei, M.; Goudouris, E.; Shirkani, A.; Changi-Ashtiani, M.; Rokni-Zadeh, H.; Sayar, E.H.; et al. Inherited IFNAR1 Deficiency in Otherwise Healthy Patients With Adverse Reaction to Measles and Yellow Fever Live Vaccines. J. Exp. Med. 2019, 216, 2057–2070. [Google Scholar] [CrossRef] [PubMed]
- Duncan, C.J.A.; Mohamad, S.M.B.; Young, D.F.; Skelton, A.J.; Leahy, T.R.; Munday, D.C.; Butler, K.M.; Morfopoulou, S.; Brown, J.R.; Hubank, M.; et al. Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci. Transl. Med. 2015, 7, 307ra154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ovsyannikova, I.G.; Haralambieva, I.H.; Vierkant, R.A.; O’Byrne, M.M.; Jacobson, R.M.; Poland, G.A. Effects of Vitamin A and D Receptor Gene Polymorphisms/Haplotypes on Immune Responses to Measles Vaccine. Pharm. Genom. 2012, 22, 20–31. [Google Scholar] [CrossRef] [Green Version]
- Ovsyannikova, I.G.; Haralambieva, I.H.; Vierkant, R.A.; O’Byrne, M.M.; Poland, G.A. Associations Between Polymorphisms in the Antiviral TRIM Genes and Measles Vaccine Immunity. Hum. Immunol. 2013, 74, 768–774. [Google Scholar] [CrossRef] [Green Version]
- Voigt, E.; Haralambieva, I.H.; Larrabee, B.R.; Kennedy, R.B.; Ovsyannikova, I.G.; Schaid, D.J.; Poland, G.A. Polymorphisms in the Wilms Tumor Gene Are Associated With Interindividual Variations in Rubella Virus-Specific Cellular Immunity After Measles-Mumps-Rubella II Vaccination. J. Infect. Dis. 2018, 217, 560–566. [Google Scholar] [CrossRef]
- Burns, C.; Cheung, A.; Stark, Z.; Choo, S.; Downie, L.; White, S.; Conyers, R.; Cole, T. A Novel Presentation of Homozygous Loss-of-Function STAT-1 Mutation in an Infant with Hyperinflammation—A Case Report and Review of the Literature. J. Allergy Clin. Immunol. Pract. 2016, 4, 777–779. [Google Scholar] [CrossRef] [PubMed]
- Hambleton, S.; Goodbourn, S.; Young, D.F.; Dickinson, P.; Mohamad, S.M.B.; Valappil, M.; McGovern, N.; Cant, A.J.; Hackett, S.J.; Ghazal, P.; et al. STAT2 Deficiency and Susceptibility to Viral Illness in Humans. Proc. Natl. Acad. Sci. USA 2013, 110, 3053–3058. [Google Scholar] [CrossRef] [Green Version]
- Moens, L.; Van Eyck, L.; Jochmans, D.; Mitera, T.; Frans, G.; Bossuyt, X.; Matthys, P.; Neyts, J.; Ciancanelli, M.; Zhang, S.-Y.; et al. A Novel Kindred with Inherited STAT2 Deficiency and Severe Viral Illness. J. Allergy Clin. Immunol. 2017, 139, 1995–1997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahni, R.; Cale, C.M.; Anderson, G.; Osellame, L.D.; Hambleton, S.; Jacques, T.S.; Wedatilake, Y.; Taanman, J.-W.; Chan, E.; Qasim, W.; et al. Signal Transducer and Activator of Transcription 2 Deficiency Is a Novel Disorder of Mitochondrial Fission. Brain 2015, 138, 2834–2846. [Google Scholar] [CrossRef] [Green Version]
- Ciancanelli, M.J.; Huang, S.X.L.; Luthra, P.; Garner, H.; Itan, Y.; Volpi, S.; Lafaille, F.G.; Trouillet, C.; Schmolke, M.; Albrecht, R.A.; et al. Life-Threatening Influenza and Impaired Interferon Amplification in Human IRF7 Deficiency. Science 2015, 348, 448–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, N.; Melki, I.; Jing, H.; Habib, T.; Huang, S.S.; Danielson, J.; Kula, T.; Drutman, S.; Belkaya, S.; Rattina, V.; et al. Life-Threatening Influenza Pneumonitis in a Child With Inherited IRF9 Deficiency. J. Exp. Med. 2018, 215, 2567–2585. [Google Scholar] [CrossRef]
- Haralambieva, I.H.; Ovsyannikova, I.G.; Kennedy, R.B.; Larrabee, B.R.; Zimmermann, M.T.; Grill, D.E.; Schaid, D.J.; Poland, G.A. Genome-Wide Associations of CD46 and IFI44L Genetic Variants with Neutralizing Antibody Response to Measles Vaccine. Hum. Genet. 2017, 136, 421–435. [Google Scholar] [CrossRef] [Green Version]
- Ovsyannikova, I.G.; Haralambieva, I.H.; Vierkant, R.A.; O’Byrne, M.M.; Jacobson, R.M.; Poland, G.A. The Association of CD46, SLAM and CD209 Cellular Receptor Gene SNPs with Variations in Measles Vaccine-Induced Immune Responses: A Replication Study and Examination of Novel Polymorphisms. Hum. Hered. 2011, 72, 206–223. [Google Scholar] [CrossRef] [Green Version]
- Ovsyannikova, I.G.; Haralambieva, I.H.; Vierkant, R.A.; Pankratz, V.S.; Jacobson, R.M.; Poland, G.A. The Role of Polymorphisms in Toll-Like Receptors and Their Associated Intracellular Signaling Genes in Measles Vaccine Immunity. Hum. Genet. 2011, 130, 547–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, P.-L.; Jacobson, R.M.; Poland, G.A.; Jacobsen, S.J.; Pankratz, V. Twin Studies of Immunogenicity—Determining the Genetic Contribution to Vaccine Failure. Vaccine 2001, 19, 2434–2439. [Google Scholar] [CrossRef]
- Schaid, D.J.; Haralambieva, I.H.; Larrabee, B.R.; Ovsyannikova, I.G.; Kennedy, R.B.; Poland, G.A. Heritability of Vaccine-Induced Measles Neutralizing Antibody Titers. Vaccine 2017, 35, 1390–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoggins, J.W.; Wilson, S.J.; Panis, M.; Murphy, M.Y.; Jones, C.T.; Bieniasz, P.D.; Rice, C.M. A Diverse Range of Gene Products Are Effectors of the Type I Interferon Antiviral Response. Nature 2011, 472, 481–485. [Google Scholar] [CrossRef]
- Mora, J.R.; Iwata, M.; Von Andrian, U.H. Vitamin Effects on the Immune System: Vitamins A and D Take Centre Stage. Nat. Rev. Immunol. 2008, 8, 685–698. [Google Scholar] [CrossRef] [Green Version]
- Nisole, S.; Stoye, J.P.; Saïb, A. TRIM Family Proteins: Retroviral Restriction and Antiviral Defence. Nat. Rev. Microbiol. 2005, 3, 799–808. [Google Scholar] [CrossRef]
- Haralambieva, I.H.; Zimmermann, M.T.; Ovsyannikova, I.G.; Grill, D.E.; Oberg, A.L.; Kennedy, R.B.; Poland, G.A. Whole Transcriptome Profiling Identifies CD93 and Other Plasma Cell Survival Factor Genes Associated with Measles-Specific Antibody Response after Vaccination. PLoS ONE 2016, 11, e0160970. [Google Scholar] [CrossRef] [Green Version]
- Haralambieva, I.H.; Kennedy, R.B.; Simon, W.L.; Goergen, K.M.; Grill, D.E.; Ovsyannikova, I.G.; Poland, G.A. Differential miRNA expression in B cells is associated with inter-individual differences in humoral immune response to measles vaccination. PLoS ONE 2018, 13, e0191812. [Google Scholar] [CrossRef] [Green Version]
- Haralambieva, I.H.; Kennedy, R.B.; Ovsyannikova, I.G.; Whitaker, J.A.; Poland, G.A. Variability in Humoral Immunity to Measles Vaccine: New Developments. Trends Mol. Med. 2015, 21, 789–801. [Google Scholar] [CrossRef] [Green Version]
- Leo, P.J.; Madeleine, M.M.; Wang, S.; Schwartz, S.M.; Newell, F.; Pettersson-Kymmer, U.; Hemminki, K.; Hallmans, G.; Tiews, S.; Steinberg, W.; et al. Defining the Genetic Susceptibility to Cervical Neoplasia—A Genome-Wide Association Study. PLoS Genet. 2017, 13, e1006866. [Google Scholar] [CrossRef] [Green Version]
- Mainali, B.; Schabath, M.B.; Sudenga, S.L.; Ye, Y.; Wiener, H.W.; Villa, L.L.; Giuliano, A.R.; Shrestha, S. Variants in Immune-Related Genes and Genital HPV 16 Persistence in Men. Papillomavirus Res. 2019, 7, 11–14. [Google Scholar] [CrossRef]
- Guan, X.; Sturgis, E.M.; Lei, D.; Liu, Z.; Dahlstrom, K.R.; Wei, Q.; Li, G. Association of TGF- Beta1 Genetic Variants with HPV16-Positive Oropharyngeal Cancer. Clin. Cancer Res. 2010, 16, 1416–1422. [Google Scholar] [CrossRef] [Green Version]
- Levovitz, C.; Chen, D.; Ivansson, E.; Gyllensten, U.; Finnigan, J.P.; Alshawish, S.; Zhang, W.; Schadt, E.E.; Posner, M.R.; Genden, E.M.; et al. TGFbeta Receptor 1: An Immune Susceptibility Gene in HPV-Associated Cancer. Cancer Res. 2014, 74, 6833–6844. [Google Scholar]
- Chambuso, R.S.; Rebello, G.; Kaambo, E. Personalized Human Papillomavirus Vaccination for Persistence of Immunity for Cervical Cancer Prevention: A Critical Review with Experts’ Opinions. Front. Oncol. 2020, 10, 548. [Google Scholar] [CrossRef]
- Council for International Organisations of Medical Sciences, World Health Organisation. Available online: https://cioms.ch/ (accessed on 25 September 2020).
- Maglione, M.A.; Das, L.; Raaen, L.; Smith, A.; Chari, R.; Newberry, S.; Shanman, R.; Perry, T.; Goetz, M.B.; Gidengil, C. Safety of Vaccines Used for Routine Immunization of U.S. Children: A Systematic Review. Pediatrics 2014, 134, 325–337. [Google Scholar] [PubMed] [Green Version]
- Stratton, K. Immunization Safety Review: Hepatitis B Vaccine and Demyelinating Neurological Disordres. In Immunization Safety Review: Hepatitis B Vaccine and Demyelinating Neurological Disordres; National Academic Press: Washington, DC, USA, 2002. [Google Scholar]
- Mailand, M.T.; Frederiksen, J.L. Vaccines and Multiple Sclerosis: A Systematic Review. J. Neurol. 2017, 264, 1035–1050. [Google Scholar] [CrossRef]
- Salemi, S.; D’Amelio, R. Could Autoimmunity Be Induced by Vaccination? Int. Rev. Immunol. 2010, 29, 247–269. [Google Scholar] [CrossRef]
- Porobič, J.M.; Avcin, T.; Božič, B.; Kuhar, M.; Cucnik, S.; Zupancic, M.; Prosenc, K.; Kveder, T.; Rozman, B. Anti-Phospholipid Antibodies Following Vaccination with Recombinant Hepatitis B Vaccine. Clin. Exp. Immunol. 2005, 142, 377–380. [Google Scholar] [CrossRef]
- Ravel, G.; Christ, M.; Horand, F.; Descotes, J. Autoimmunity, Environmental Exposure and Vaccination: Is There a Link? Toxicology 2004, 196, 211–216. [Google Scholar] [CrossRef]
- De Wolf, A.C.M.T.; Van Aalst, S.; Ludwig, I.S.; Bodinham, C.L.; Lewis, D.J.; Van Der Zee, R.; Van Eden, W.; Broere, F. Regulatory T Cell Frequencies and Phenotypes Following Anti-Viral Vaccination. PLoS ONE 2017, 12, e0179942. [Google Scholar] [CrossRef]
- Fourati, S.; Cristescu, R.; Loboda, A.; Talla, A.; Filali, A.; Railkar, R.; Schaeffer, A.K.; Favre, D.; Gagnon, D.; Peretz, Y.; et al. Pre-Vaccination Inflammation and B-Cell Signalling Predict Age-Related Hyporesponse to Hepatitis B Vaccination. Nat. Commun. 2016, 7, 10369. [Google Scholar] [CrossRef]
- Pöyhönen, L.; Bustamante, J.; Casanova, J.-L.; Jouanguy, E.; Zhang, Q. Life-Threatening Infections Due to Live-Attenuated Vaccines: Early Manifestations of Inborn Errors of Immunity. J. Clin. Immunol. 2019, 39, 376–390. [Google Scholar] [CrossRef]
- Hur, J.; Özgür, A.; Xiang, Z.; Ehe, Y.O. Identification of Fever and Vaccine-Associated Gene Interaction Networks Using Ontology-Based Literature Mining. J. Biomed. Semant. 2012, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Center for Disease Control and Prevention. Human Papilloma Virus—HPV Vaccination Is Safe and Effective. 2019. Available online: https://www.cdc.gov/hpv/parents/vaccinesafety.html (accessed on 18 July 2020).
- Klumb, E.; Pinto, A.; Jesus, G.; Araujo, M.; Jascone, L.; Gayer, C.; Ribeiro, F.; Albuquerque, E.; Macedo, J. Are Women with Lupus at Higher Risk of HPV Infection? Lupus 2010, 19, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- Soldevilla, H.; Briones, S.; Navarra, S. Systemic Lupus Erythematosus Following HPV Immunization or Infection? Lupus 2012, 21, 158–161. [Google Scholar] [CrossRef] [PubMed]
- García-Carrasco, M.; Mendoza-Pinto, C.; Rojas-Villarraga, A.; Molano-González, N.; Vallejo-Ruiz, V.; Munguía-Realpozo, P.; Colombo, A.L.; Cervera, R. Corrigendum to ‘Prevalence of Cervical HPV Infection in Women with Systemic Lupus Erythematosus: A Systematic Review and Meta-Analysis. Autoimmun. Rev. 2019, 18, 437. [Google Scholar] [CrossRef]
- Blitshteyn, S.; Brinth, L.; Hendrickson, J.E.; Martínez-Lavín, M. Autonomic Dysfunction and HPV Immunization: An Overview. Immunol. Res. 2018, 66, 744–754. [Google Scholar] [CrossRef]
- Martínez-Lavín, M. HPV Vaccination Syndrome: A Clinical Mirage, or a New Tragic Fibromyalgia Model. Reumatol. Clín. 2018, 14, 211–214. [Google Scholar] [CrossRef]
- Ryabkova, V.A.; Churilov, L.P.; Shoenfeld, Y. Neuroimmunology: What Role for Autoimmunity, Neuroinflammation, and Small Fiber Neuropathy in Fibromyalgia, Chronic Fatigue Syndrome, and Adverse Events after Human Papillomavirus Vaccination? Int. J. Mol. Sci. 2019, 20, 5164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, K.D.; Gelbart, T.; Whisenant, T.C.; Waalen, J.; Mondala, T.S.; Iklé, D.N.; Salomon, D.R.; Bennett, R.M.; Kurian, S.M. Genome-Wide Expression Profiling in the Peripheral Blood of Patients with Fibromyalgia. Clin. Exp. Rheumatol. 2016, 34, S89–S98. [Google Scholar] [PubMed]
- Park, E.; Kim, J.-Y.; Choi, S.; Kim, D.S.; Oh, Y.L. Carcinogenic Risk of Human Papillomavirus (HPV) Genotypes and Potential Effects of HPV Vaccines in Korea. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Flanagan, K.L.; Fink, A.L.; Plebanski, M.; Klein, S.L. Sex and Gender Differences in the Outcomes of Vaccination over the Life Course. Annu. Rev. Cell Dev. Biol. 2017, 33, 577–599. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L.; Flanagan, K.L. Sex Differences in Immune Responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef]
- Garly, M.-L.; Jensen, H.; Martins, C.L.; Balé, C.; Baldé, M.A.; Lisse, I.M.; Aaby, P. Hepatitis B Vaccination Associated with Higher Female Than Male Mortality in Guinea-Bissau: An Observational Study. Pediatric Infect. Dis. J. 2004, 23, 1086–1092. [Google Scholar]
- Khalil, M.K.; Al-Mazrou, Y.Y.; Al-Ghamdi, Y.S.; Tumsah, S.; Al-Jeffri, M.; Meshkhas, A. Effect of Gender on Reporting of MMR Adverse Events in Saudi Arabia. East. Mediterr. Health J. 2003, 9, 152–158. [Google Scholar]
- Martins, S.; Livramento, A.D.; Andrigueti, M.; Kretzer, I.F.; Machado, M.J.; Spada, C.; Treitinger, A. The Prevalence of Hepatitis B Virus Infection Markers and Socio-Demographic Risk Factors in HIV-Infected Patients in Southern Brazil. Rev. Soc. Bras. Med. Trop. 2014, 47, 552–558. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, I.; Lleo, A.; Gershwin, M.E.; Invernizzi, P. The X Cromosome and Immune Associated Genes. J. Autoimmun. 2012, 38, 187–192. [Google Scholar] [CrossRef]
- Schurz, H.; Salie, M.; Tromp, G.; Hoal, E.G.; Kinnear, C.J.; Möller, M. The X Chromosome and Sex-Specific Effects in Infectious Disease Susceptibility. Hum. Genom. 2019, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Klein, S.L.; Jedlicka, A.; Pekosz, A. The Xs and Y of Immune Responses to Viral Vaccines. Lancet Infect. Dis. 2010, 10, 338–349. [Google Scholar] [CrossRef]
- Klein, S.L.; Pekosz, A. Sex-Based Biology and the Rational Design of Influenza Vaccination Strategies. J. Infect. Dis. 2014, 209, S114–S119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; Mcmahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA Vaccine Protection Against SARS-Cov-2 in Rhesus Macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584, 437–442. [Google Scholar] [CrossRef]
- The COVID-19 Host Genetics Initiative. A Global Initiative to Elucidate the Role of Host Genetic Factors in Susceptibility and Severity of the SARS-Cov-2 Virus Pandemic. Eur. J. Hum. Genet. 2020, 28, 715–718. [Google Scholar] [CrossRef]
- Cao, Y.; Li, L.; Feng, Z.; Wan, S.; Huang, P.; Sun, X.; Wen, F.; Huang, X.; Ning, G.; Wang, W. Comparative Genetic Analysis of the Novel Coronavirus (2019-nCov/SARS-Cov-2) Receptor ACE2 in Different Populations. Cell Discov. 2020, 6, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Latz, C.A.; Decarlo, C.; Boitano, L.; Png, C.Y.M.; Patell, R.; Conrad, M.F.; Eagleton, M.; Dua, A. Blood type and outcomes in patients with COVID-19. Ann. Hematol. 2020, 99, 2113–2118. [Google Scholar] [CrossRef] [PubMed]
- Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Agustin, A.; Invernizzi, P.; Fernandez, J.; Prati, D.; Baselli, G.; Asselta, R.; et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- The COVID-19 Sex-Disaggregated Data Tracker. Available online: https://globalhealth5050.org/the-sex-gender-and-covid-19-project/the-data-tracker/ (accessed on 31 October 2020).
- Scully, E.P.; Haverfield, J.; Ursin, R.L.; Tannenbaum, C.; Klein, S.L. Considering How Biological Sex Impacts Immune Responses and COVID-19 Outcomes. Nat. Rev. Immunol. 2020, 20, 442–447. [Google Scholar] [CrossRef]
- Takahashi, T.; Yale IMPACT Research Team; Ellingson, M.K.; Wong, P.; Israelow, B.; Lucas, C.; Klein, J.; Silva, J.; Mao, T.; Oh, J.E.; et al. Sex Differences in Immune Responses That Underlie COVID-19 Disease Outcomes. Nature 2020, 1–9. [Google Scholar] [CrossRef] [PubMed]
| Gene | Gene Description | Genomic Location | Function of Gene Product | Associated Disease Phenotypes–Not Vaccine Related [28] | Studies on Associated Vaccinomic Phenotypes | Studies on Associated Adversomics Phenotypes | |
|---|---|---|---|---|---|---|---|
| HBV | MMR | HBV | |||||
| HLA-DRB1 | Major histocompatibility complex, class II, DR β 1 | 6p21.32 | HLA class II β chain paralogue, presenting peptides derived from extracellular proteins | Sarcoidosis, multiple sclerosis, rheumatoid arthritis, autism/schizophrenia | [17,18,19,20,21,23,25] | [29,30] | [31,32] |
| HLA-DQB1 | Major histocompatibility complex, class II, DQ β 1 | 6p21.32 | HLA class II β chain paralogue, presenting peptides derived from extracellular proteins | Celiac disease, multiple sclerosis, Creutzfeldt-Jakob disease, systemic lupus erythematosus, autism/schizophrenia | [17,18,19,20,22,25] | [30] | [31] |
| HLA-DPB1 | Major histocompatibility complex, class II, DP β 1 | 6p21.32 | HLA class II β chain paralogue, presenting peptides derived from extracellular proteins | Chronic beryllium disease, Wegener granulomatosis, chronic hepatitis B infection | [24,25] | [30,33] | None |
| HLA-B | Major histocompatibility complex, class I, B | 6p21.33 | HLA class I heavy chain paralogue, presenting peptides derived from the endoplasmic reticulum lumen | Behcet disease, pulmonary arterial hypertension, toxic epidermal necrolysis, spondyloarthropathy, Stevens-Johnson syndrome, Takayasu arteritis | None | [29,30] | None |
| HLA-DQA1 | Major histocompatibility complex, class II, DQ α 1 | 6p21.32 | HLA class II α chain paralogue, presenting peptides derived from extracellular proteins | Myasthenia gravis, celiac disease, idiopathic achalasia | None | [29,30] | None |
| HLA-A | Major histocompatibility complex, class I, A | 6p22.1 | HLA class I heavy chain paralogue, presenting peptides derived from the endoplasmic reticulum lumen | Cancers, birdshot chorioretinopathy, myelodysplastic syndrome, toxic epidermal necrolysis | None | [30] | None |
| HLA-DPA1 | Major histocompatibility complex, class II, DP α 1 | 6p21.32 | HLA class II α chain paralogue, presenting peptides derived from extracellular proteins | Granulomatosis with polyangiitis | None | [30] | None |
| Gene | Gene Description | Genomic Location | Gene Product Function | Associated Disease Phenotypes–Not Vaccine Related [28] | Studies on Associated Vaccinomic Phenotypes | Studies on Associated Adversomics Phenotypes | |
|---|---|---|---|---|---|---|---|
| HBV | MMR | MMR | |||||
| TNF | Tumour necrosis factor | 6p21.33 | Cytokine, regulation of cell proliferation, differentiation, apoptosis, lipid metabolism, coagulation | Asthma, malaria susceptibility, migraine | None | [34] | None |
| TNFRSF1A | TNF receptor superfamily member 1A | 12p13.31 | TNF receptor, cell survival, apoptosis, and inflammation | Intermittent hydrarthrosis, multiple sclerosis, familial periodic fever, TNF receptor 1 associated periodic syndrome | None | [34] | None |
| IL-6 | Interleukin 6 | 7p15.3 | Cytokine, inflammation, and maturation of B cells | Arteriovenous malformations of the brain, diabetes mellitus type I, Crohn’s disease, Kaposi sarcoma, juvenile rheumatoid arthritis, juvenile idiopathic arthritis | None | [34] | None |
| IL-1β | Interleukin 1β | 2q14.1 | Cytokine, mediator of inflammatory responses | Gastric cancer risk (Helicobacter pylori induced), periodontal disease | [27] | [34] | None |
| IL-13 | Interleukin 13 | 5q31.1 | Cytokine, B cell maturation, promotion of IgE isotype switching of B cells | Asthma, allergic rhinitis | [27] | None | None |
| IL-4 | Interleukin 4 | 5q31.1 | Cytokine, B cell activation, IgE secretion | Allergic bronchopulmonary aspergillosis, schistosomiasis | [27] | None | None |
| IL4RA | Interleukin 4 receptor | 16p12.1 | Receptor for IL-4 and IL-13, promotes Th2 differentiation | Atopy, human immunodeficiency virus -1 resistance | [27] | None | None |
| IL2RA | Interleukin 2 receptor subunit α | 10p15.1 | Part of IL-2 receptor, T cell-mediated immune responses | Diabetes mellitus type I, immunodeficiency, juvenile idiopathic arthritis | None | [34] | None |
| IL2RB | Interleukin 2 receptor subunit β | 22q12.3 | Part of IL2 receptor, T cell-mediated immune response | Juvenile idiopathic arthritis, immunodeficiency | None | [34] | None |
| IFNB1 | Interferon β1 | 9p21.3 | Cytokine, defense against viral infections, cell differentiation, anti-tumor defense | Multiple sclerosis | None | [34] | None |
| INFAR1 | Interferon α/β receptor subunit 1 | 21q22.11 | One of two chains of a receptor for INFα and INFβ, activation of receptor stimulates Janus protein kinases, which phosphorylate STAT1 and STAT2 | Hepatitis C susceptibility, measles susceptibility | None | None | [35] |
| INFAR2 | Interferon α/β receptor subunit 2 | 21q22.11 | One of two chains of a receptor for INFα and INFβ, activation of receptor stimulates Janus protein kinases, which phosphorylate STAT1 and STAT2 | Measles susceptibility, immunodeficiency 45 | None | None | [36] |
| Gene | Gene Description | Genomic Location | Gene Product Function | Associated Disease Phenotypes–Not Vaccine Related [28] | Studies on Associated Vaccinomic Phenotype: MMR | Studies on Associated Adversomics Phenotypes: MMR |
|---|---|---|---|---|---|---|
| RARB | Retinoic acid receptor β | 3p24.2 | Nuclear transcriptional regulator, binds retinoic acid | Matthew-Wood syndrome, microphthalmia, diaphragmatic hernia | [37] | None |
| RXRA | Retinoid X receptor α | 9q34.2 | Nuclear receptor, involvement in retinoic-acid-mediated gene activation | Colon adenoma, recessive dystrophic Epidermolysis bullosa | [37] | None |
| VDR | Vitamin D receptor | 12q13.11 | Ligand-inducible transcription factor, also a receptor for the secondary bile acid, lithocholic acid | Hypocalcaemic vitamin D-resistant rickets, osteoporosis, vitamin D-dependent rickets (type 2a) | [37] | None |
| TRIM25 | Tripartite motif-containing 25 | 17q23.1 | Transcription factor, mediates estrogen actions in breast cancer | Influenza, swine influenza | [38] | None |
| WT1 | WT1 transcription factor | 11p13 | Transcription factor, development of urogenital system, tumor suppressor gene | Gonadal dysgenesis, cancers, aniridia, Denys-Drash syndrome, Frasier syndrome, genetic steroid-resistant nephrotic syndrome, Meacham syndrome, ulcerative colitis, Wilms tumor, aniridia, genitourinary anomalies, and retardation syndrome | [39] | None |
| STAT1 | Signal transducer and activator of transcription 1 | 2q32.2 | Transcription activator, mediates expression of a variety of genes, which is thought to be important for cell viability in response to pathogens, can be activated by IFNα and IFNγ | Autoimmune enteropathy and endocrinopathy, immunodeficiency 31A, 31B, and 31C, susceptibility to viral and mycobacterial infections | None | [40] |
| STAT2 | Signal transducer and activator of transcription 2 | 12q13.3 | Transcription activator, in response to IFN, forms a complex with STAT1 and ISGF3G | Immunodeficiency 44, primary immunodeficiency with post-measles-mumps-rubella vaccine viral infection | None | [41,42,43] |
| IRF7 | Interferon regulatory factor 7 | 11p15.5 | Transcriptional activation of virus-inducible cellular genes, including IFNβ chain genes | Immunodeficiency 39 | None | [44] |
| IRF9 | Interferon regulatory factor 9 | 14q12 | Transcription factor, mediates signaling of IFNα and IFNβ, IRF9/ISGF3G associates with the phosphorylated STAT1:STAT2 dimer to form ISGF3 transcription factor | Immunodeficiency 65, susceptibility to viral infections | None | [45] |
| Gene | Gene Description | Genomic Location | Gene Product Function | Associated Disease Phenotypes–Not Vaccine Related [28] | Studies on Associated Vaccinomic Phenotypes | Studies on Associated Adversomics Phenotypes | |
|---|---|---|---|---|---|---|---|
| HBV | MMR | MMR | |||||
| CD46 | CD46 molecule | 1q32.2 | Cofactor activity for inactivation of complement components C3b and C4b by serum factor I | Atypical hemolytic uremic syndrome with complement gene abnormality, hemolysis, elevated liver enzymes, and a low platelet count syndrome | None | [46,47] | [7] |
| BTNL2 | Butyrophilin-like 2 | 6p21.32 | MHC-II-associated, transmembrane protein, negative T cell regulator | Sarcoidosis, multiple sclerosis, autism/schizophrenia | [22,23,25] | None | None |
| SLAM | Signaling lymphocytic activation molecule family member 1 | 1q23.3 | Self-ligand receptor of signaling lymphocytic activation molecule, modulating the activation and differentiation of a wide variety of immune cells | Measles susceptibility, subacute sclerosing panencephalitis | None | [47] | None |
| IFI44L | Interferon-induced protein 44 like | 1p31.1 | Unknown, shown a low antiviral activity against hepatitis C virus | Lymph node tuberculosis, Aicardi-Goutieres syndrome | None | [46] | [7] |
| TLR4 | Toll-like receptor 4 | 9q33.1 | Pathogen recognition and activation of innate immunity | Behcet’s disease | None | [48] | None |
| TLR2 | Toll-like receptor 2 | 4q31.3 | Pathogen recognition and activation of innate immunity | Leprosy susceptibility, tuberculosis susceptibility | [27] | [48] | None |
| KIAA1542 | PHD and ring finger domains 1 | 11p15.5 | Protein domain-specific binding and RNA polymerase binding | Systemic lupus erythematosus | None | [48] | None |
| TRIM5 | Tripartite motif-containing 5 | 11p15.4 | E3 ubiquitin-ligase, may have role in retroviral restriction | Rubella susceptibility, immune deficiency disease | None | [38] | None |
| TRIM22 | Tripartite motif-containing 22 | 11p15.4 | Mediates interferon antiviral effects | Rubella susceptibility, hepatitis B susceptibility | None | [38] | None |
| SCN1A | Sodium voltage-gated channel α subunit 1 | 2q24.3 | Sodium channel α subunit, regulates sodium exchange between intracellular and extracellular spaces, generation of action potentials in muscle cells and neurons | Dravet syndrome, early infantile epileptic encephalopathy 6, familial or sporadic hemiplegic migraine, generalized epilepsy with febrile seizures plus (type 2), Lennox-Gastaut syndrome | None | None | [7] |
| SCN2A | Sodium voltage-gated channel α subunit 2 | 2q24.3 | Sodium channel α subunit, regulates sodium exchange between intracellular and extracellular spaces, generation of action potentials in muscle cells and neurons | Benign familial infantile epilepsy, benign familial neonatal-infantile seizures, Dravet syndrome, early infantile epileptic encephalopathy, generalized epilepsy with febrile seizures-plus, West syndrome | None | None | [7] |
| ANO3 | Anoctamin 3 | 11p14.2 | Membrane protein, Ca2+-activated chloride channel | Cranio-cervical dystonia with laryngeal and upper-limb involvement | None | None | [7] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omersel, J.; Karas Kuželički, N. Vaccinomics and Adversomics in the Era of Precision Medicine: A Review Based on HBV, MMR, HPV, and COVID-19 Vaccines. J. Clin. Med. 2020, 9, 3561. https://doi.org/10.3390/jcm9113561
Omersel J, Karas Kuželički N. Vaccinomics and Adversomics in the Era of Precision Medicine: A Review Based on HBV, MMR, HPV, and COVID-19 Vaccines. Journal of Clinical Medicine. 2020; 9(11):3561. https://doi.org/10.3390/jcm9113561
Chicago/Turabian StyleOmersel, Jasna, and Nataša Karas Kuželički. 2020. "Vaccinomics and Adversomics in the Era of Precision Medicine: A Review Based on HBV, MMR, HPV, and COVID-19 Vaccines" Journal of Clinical Medicine 9, no. 11: 3561. https://doi.org/10.3390/jcm9113561
APA StyleOmersel, J., & Karas Kuželički, N. (2020). Vaccinomics and Adversomics in the Era of Precision Medicine: A Review Based on HBV, MMR, HPV, and COVID-19 Vaccines. Journal of Clinical Medicine, 9(11), 3561. https://doi.org/10.3390/jcm9113561




