Biodiversity of Culturable Endophytic Actinobacteria Isolated from High Yield Camellia oleifera and Their Plant Growth Promotion Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Collection of Samples
2.3. Determination of Rhizosphere Soil Characteristics
2.4. Isolation of Endophytic Actinobacteria
2.5. Classification of Endophytic Actinobacteria
2.6. In Vitro Qualitative Assessment of Plant-Growth Promotion (PGP) Traits of Puried Endophytic Actinobacteria
2.7. Effects of the Potential Endophytic Actinobacteria on the Growth of C. oleifera Seedlings
2.8. Statistical Analysis
3. Results
3.1. Isolation of Culturable Endophytic Actinobacteria
3.2. Impact of Culture Media on Isolation
3.3. Composition of Endophytic Actinobacteria
3.4. Soil Characteristics
3.5. Effects of Soil Condition on Endophytic Actinobacterial Community Distribution
3.6. In Vitro Evaluation of Endophytic Actinobacteria with Plant Growth Promoting Traits
3.7. In Vivo Effects of Endophytic Actinobacteria on the Growth of C. oleifera Seedlings
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, J.; Li, Z.; Lv, X.; Yan, H.; Zhou, G.; Cao, L.; Yang, Q.; He, Y. Isolation and characterization of Bacillus subtilis strain 1-L-29, an endophytic bacteria from Camellia oleifera with antimicrobial activity and efficient plant-root colonization. PLoS ONE 2020, 15, e0232096. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Lu, J.; Zhang, Z.; Ma, L.; Liu, C.; Chen, Y. Global transcriptome and correlation analysis reveal cultivar-specific molecular signatures associated with fruit development and fatty acid determination in Camellia oleifera Abel. Int. J. Genom. 2020, 2020, 6162802. [Google Scholar] [CrossRef]
- Yu, J.; Wu, Y.; He, Z.; Li, M.; Zhu, K.; Gao, B. Diversity and antifungal activity of endophytic fungi associated with Camellia oleifera. Mycobiology 2018, 46, 85–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Chen, L.; Tang, W.; Peng, S.; Li, M.; Deng, N.; Chen, Y. Predicting potential distribution and evaluating suitable soil condition of oil tea Camellia in China. Forests 2018, 9, 487. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Ye, H.; Rui, Y.; Chen, G.; Zhang, N. Fatty acid composition of Camellia oleifera oil. J. Verbr. Lebensm. 2011, 6, 9–12. [Google Scholar] [CrossRef]
- Cheng, Y.; Wu, S.; Ho, C.; Huang, S.; Cheng, C.; Yen, G.C. Beneficial effects of Camellia Oil (Camellia oleifera Abel.) on ketoprofen-induced gastrointestinal mucosal damage through upregulation of HO-1 and VEGF. J. Agric. Food Chem. 2014, 62, 642–650. [Google Scholar] [CrossRef]
- Zhang, P.; Cui, Z.; Guo, M.; Xi, R. Characteristics of the soil microbial community in the forestland of Camellia oleifera. PeerJ 2020, 8, e9117. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Kent, M.; Fang, X.F. Evergreen broad-leaved forest in Eastern China: Its ecology and conservation and the importance of resprouting in forest restoration. For. Ecol. Manag. 2007, 245, 76–87. [Google Scholar] [CrossRef]
- Liu, C.; Chen, L.; He, Z.; Zhang, Z.; Xu, Y.; Li, Z.; Peng, Y.; Deng, N.; Chen, Y. Integration and potential application ability of culturable functional microorganism in oil tea Camellia. Indian J. Microbiol. 2020, 61, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim, W.; Ebada, S.S.; Proksch, P. Bioprospecting of endophytes. In Endophyte Biotechnology: Potential for Agriculture and Pharmacology; Schouten, A., Ed.; CABI BioTechnology: Boston, MA, USA, 2019; pp. 145–163. [Google Scholar]
- Wallace, J.G.; May, G. Endophytes: The other maize genome. In The Maize Genome; Bennetzen, J., Flint-Garcia, S., Hirsch, C., Tuberosa, R., Eds.; Springer: Cham, Switzerland, 2018; pp. 213–246. [Google Scholar]
- Goudjal, Y.; Zamoum, M.; Sabaou, N.; Mathieu, F.; Zitouni, A. Potential of endophytic Streptomyces spp. for biocontrol of Fusarium root rot disease and growth promotion of tomato seedlings. Biocontrol Sci. Technol. 2016, 26, 1691–1705. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, M. Endophytic actinomycetes: Biocontrol agents and growth promoters. In Bacteria in Agrobiology: Plant Growth Responses; Maheshwari, D., Ed.; Springer: Heidelberg, Germany; Dordrecht, The Netherlands; New York, NY, USA; London, UK, 2011; pp. 201–220. [Google Scholar]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttila, A.M.; Compant, S.; Campisano, A.; Doring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strobel, G.A. Endophytes as sources of bioactive products. Microbes. Infect. 2003, 5, 535–544. [Google Scholar] [CrossRef]
- Barman, D.; Dkhar, M.S. Seasonal variation influence endophytic actinobacterial communities of medicinal plants from tropical deciduous forest of Meghalaya and characterization of their plant growth-promoting potentials. Curr. Microbiol. 2020, 77, 1689–1698. [Google Scholar] [CrossRef]
- Singh, R.; Dubey, A.K. Diversity and applications of endophytic actinobacteria of plants in special and other ecological niches. Front. Microbiol. 2018, 9, 1767. [Google Scholar] [CrossRef] [PubMed]
- Mercedes Maldonado-Gonzalez, M.; Bakker, P.A.H.M.; Prieto, P.; Mercado-Blanco, J. Arabidopsis thaliana as a tool to identify traits involved in Verticillium dahliae biocontrol by the olive root endophyte Pseudomonas fluorescens PICF7. Front. Microbiol. 2015, 6, 266. [Google Scholar] [CrossRef]
- Mercedes Maldonado-Gonzalez, M.; Schiliro, E.; Prieto, P.; Mercado-Blanco, J. Endophytic colonization and biocontrol performance of Pseudomonas fluorescens PICF7 in olive (Olea europaea L.) are determined neither by pyoverdine production nor swimming motility. Environ. Microbiol. 2015, 17, 3139–3153. [Google Scholar] [CrossRef]
- Sunpapao, A.; Chairin, T.; Ito, S. The biocontrol by Streptomyces and Trichoderma of leaf spot disease caused by Curvularia oryzae in oil palm seedlings. Biol. Control 2018, 123, 36–42. [Google Scholar] [CrossRef]
- Zhou, S.; Yan, S.; Wu, Z.; Chen, S. Endophytic fungi associated with Macrosolen tricolor and its host Camellia oleifera. World J. Microbiol. Biotechnol. 2014, 30, 1775–1784. [Google Scholar] [CrossRef]
- Liu, C.; Deng, N.; He, Z.; Tang, W.; Wang, X.; Chen, L.; Wang, R.; Chen, Y. Effects of Camellia oleifera clone selection on soil nutrient and microbial community structure. Agron. J. 2021, 113, 200–209. [Google Scholar] [CrossRef]
- Vilanova, C.; Porcar, M. Are multi-omics enough? Nat. Microbiol. 2016, 1, 16101. [Google Scholar] [CrossRef]
- Chen, Y. Oil Tea Camellia Superior Germplasm Resources; China Forestry Publishing House: Beijing, China, 2008; pp. 25–104. [Google Scholar]
- Kader, M.; Lamb, D.T.; Correll, R.; Megharaj, M.; Naidu, R. Pore-water chemistry explains zinc phytotoxicity in soil. Ecotoxicol. Environ. Saf. 2015, 122, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Gaans, P.F.M.; Vriend, S.P.; Bleyerveld, S.; Schrage, G.; Vos, A. Assessing environmental soil quality in rural areas. Environ. Monit. Assess. 1995, 34, 73–102. [Google Scholar] [CrossRef] [PubMed]
- Tsiknia, M.; Tzanakakis, V.A.; Oikonomidis, D.; Paranychianakis, N.V.; Nikolaidis, N.P. Effects of olive mill wastewater on soil carbon and nitrogen cycling. Appl. Microbiol. Biot. 2014, 98, 2739–2749. [Google Scholar] [CrossRef] [PubMed]
- Kuo, S. Phosphorus. In Methods of Soil Analysis, Part 3. Chemical Methods; Sparks, D.L., Ed.; SSSA and ASA: Madison, WI, USA, 1996; pp. 869–919. [Google Scholar]
- Yanu, P.; Jakmunee, J. Flow injection with in-line reduction column and conductometric detection for determination of total inorganic nitrogen in soil. Talanta 2015, 144, 263–267. [Google Scholar] [CrossRef]
- Shi, R. Agricultural Chemistry Analyses of Soils; China Agricultural Press: Beijing, China, 1996; pp. 37–39. [Google Scholar]
- Olsen, S.R.; Sommers, L.E. Phosphorus. In Methods of Soil Analysis; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy: Madison, WI, USA, 1982; pp. 403–430. [Google Scholar]
- Knudsen, D.; Peterson, G.; Pratt, P. Lithium, sodium, and potassium. In Methods of Soil Analysis, Part 2, Chemical and Microbiological Properties; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy, Soil Science Society of American: Madison, WI, USA, 1982; pp. 225–246. [Google Scholar]
- Wang, H.; Yan, S.; Ren, T.; Yuan, Y.; Kuang, G.; Wang, B.; Yun, F.; Feng, H.; Ji, X.; Yuan, X.; et al. Novel environmental factors affecting microbial responses and physicochemical properties by sequentially applied biochar in black soil. Environ. Sci. Pollut. R. 2020, 27, 37432–37443. [Google Scholar] [CrossRef]
- Xiong, X.; Liao, H.; Ma, J.; Liu, X.; Zhang, L.; Shi, X.; Yang, X.; Lu, X.; Zhu, Y. Isolation of a rice endophytic bacterium, Pantoea sp. Sd-1, with ligninolytic activity and characterization of its rice straw degradation ability. Lett. Appl. Microbiol. 2014, 58, 123–129. [Google Scholar] [CrossRef]
- Xu, W.; Wang, F.; Zhang, M.; Ou, T.; Wang, R.; Strobel, G.; Xiang, Z.; Zhou, Z.; Xie, J. Diversity of cultivable endophytic bacteria in mulberry and their potential for antimicrobial and plant growth-promoting activities. Microbiol. Res. 2019, 229, 126328. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Pande, S.; Sharma, M.; Humayun, P.; Kiran, B.K.; Sandeep, D.; Vidya, M.S.; Deepthi, K.; Rupela, O. Evaluation of actinomycete isolates obtained from herbal vermicompost for the biological control of Fusarium wilt of chickpea. Crop Protect. 2011, 30, 1070–1078. [Google Scholar] [CrossRef] [Green Version]
- Ben-Abdallah, D.; Frikha-Gargouri, O.; Tounsi, S. Rizhospheric competence, plant growth promotion and biocontrol efficacy of Bacillus amyloliquefaciens subsp. plantarum strain 32a. Biol. Control 2018, 124, 61–67. [Google Scholar] [CrossRef]
- Coombs, J.T.; Franco, C.M.M. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 2003, 69, 5603–5608. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Bai, J.; Yang, H.; Zhang, W.; Xiong, Y.; Ding, P.; Qin, S. Phylogenetic diversity and investigation of plant growth-promoting traits of actinobacteria in coastal salt marsh plant rhizospheres from Jiangsu, China. Syst. Appl. Microbiol. 2018, 41, 516–527. [Google Scholar] [CrossRef]
- Nguyen, M.P.; Lehosmaa, K.; Martz, F.; Koskimäki, J.J.; Pirttilä, A.M.; Häggman, H. Host species shape the community structure of culturable endophytes in fruits of wild berry species (Vaccinium myrtillus L., Empetrum nigrum L., and Vaccinium vitis-idaea L.). FEMS Microbiol. Ecol. 2021, 97, fiab097. [Google Scholar] [CrossRef] [PubMed]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The soil microbiome influences grapevine-associated microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinesh, R.; Srinivasan, V.; Sheeja, T.E.; Anandaraj, M.; Srambikkal, H. Endophytic actinobacteria: Diversity, secondary metabolism and mechanisms to unsilence biosynthetic gene clusters. Crit. Rev. Microbiol. 2017, 43, 546–566. [Google Scholar] [CrossRef] [PubMed]
- Govindasamy, V.; Raina, S.K.; George, P.; Kumar, M.; Rane, J.; Minhas, P.S.; Vittal, K.P.R. Functional and phylogenetic diversity of cultivable rhizobacterial endophytes of sorghum [Sorghum bicolor (L.) Moench]. Anton. Leeuw. 2017, 110, 925–943. [Google Scholar] [CrossRef]
- Dimpka, C.; Weinand, T.; Asch, F. Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 2009, 32, 1682–1694. [Google Scholar] [CrossRef]
- Gururani, M.A.; Upadhyaya, C.P.; Baskar, V.; Venkatesh, J.; Nookaraju, A.; Park, S.W. Plant growth-promoting rhizobacteria enhance abiotic stress tolerance in Solanum tuberosum through inducing changes in the expression of ROS-scavenging enzymes and improved photosynthetic performance. J. Plant Growth Regul. 2013, 32, 245–258. [Google Scholar] [CrossRef]
- Borah, A.; Thakur, D. Phylogenetic and functional characterization of culturable endophytic actinobacteria associated with Camellia spp. for growth promotion in commercial tea cultivars. Front. Microbiol. 2020, 11, 318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.; Zhang, C.; Ju, X.; Xiong, Y.; Xing, K.; Qin, S. Community composition and metabolic potential of endophytic actinobacteria from coastal salt marsh plants in Jiangsu, China. Front. Microbiol. 2019, 10, 1063. [Google Scholar] [CrossRef]
- Mitra, A.; Santra, S.C.; Mukherjee, J. Distribution of actinomycetes, their antagonistic behaviour and the physico-chemical characteristics of the world’s largest tidal mangrove forest. Appl. Microbiol. Biotechnol. 2008, 80, 685–695. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Giovanardi, D.; Stefani, E. Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, G.Y.A.; Goodfellow, M.; Genus, V. Amycolatopsis Lechevalier, Prauser, Labeda and Ruan 1986 emend Lee 2009, 1403. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K., Ludwig, W., Eds.; Springer: New York, NY, USA, 2012; Volume 5, pp. 1334–1358. [Google Scholar]
- Bull, A.T.; Asenjo, J.A.; Goodfellow, M.; Go’mez-Silva, B. The Atacama Desert: Technical resources and the growing importance of novel microbial diversity. Annu. Rev. Microbial. 2016, 70, 215–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busarakam, K.; Brown, R.; Bull, A.T.; Tan, G.Y.; Zucchi, T.D.; da Silva, L.J.; de Souza, W.R.; Goodfellow, M. Classification of thermophilic actinobacteria isolated from arid desert soils, including the description of Amycolatopsis deserti sp. nov. Anton. Leeuw. 2016, 109, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Mingma, R.; Inahashi, Y.; Matsumoto, A.; Takahashi, Y.; Duangmal, K. Amycolatopsis pithecelloba sp. nov., a novel actinomycete isolated from roots of Pithecellobium dulce in Thailand. J. Antibiot. 2020, 73, 230–235. [Google Scholar] [CrossRef]
- Kaewkla, O.; Franco, C.M.M. Amycolatopsis pittospori sp. nov., an endophytic actinobacterium isolated from native apricot tree and genome mining revealed the biosynthesis potential as antibiotic producer and plant growth promoter. Anton. Leeuw. 2021, 114, 365–377. [Google Scholar] [CrossRef]
- Idris, H.; Nouioui, I.; Pathomaree, W.; Castro, J.F.; Bull, A.T.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M. Amycolatopsis vastitatis sp. nov., an isolate from a high altitude subsurface soil on Cerro Chajnantor, northern Chile. Anton. Leeuwe. 2018, 111, 1523–1533. [Google Scholar] [CrossRef] [PubMed]
- Vo, Q.A.T.; Ballard, R.A.; Barnett, S.J.; Franco, C.M.M. Isolation and characterisation of endophytic actinobacteria and their effect on the growth and nodulation of chickpea (Cicer arietinum). Plant Soil 2021, 466, 357–371. [Google Scholar] [CrossRef]
- Martínez-Hidalgo, P.; Galindo-Villardón, P.; Trujillo, M.; Igual, J.M.; Martínez-Molina, E. Micromonospora from nitrogen fixing nodules of alfalfa (Medicago sativa L.). A new promising plant probiotic bacteria. Sci. Rep. 2014, 4, 6389. [Google Scholar] [CrossRef] [Green Version]
- Ghodhbane-Gtari, F.; Nouioui, I.; Hezbri, K.; Lundstedt, E.; D’Angelo, T.; McNutt, Z.; Laplaze, L.; Gherbi, H.; Vaissayre, V.; Svistoonoff, S.; et al. The plant-growth-promoting actinobacteria of the genus Nocardia induces root nodule formation in Casuarina glauca. Anton. Leeuw. 2019, 112, 75–90. [Google Scholar] [CrossRef]

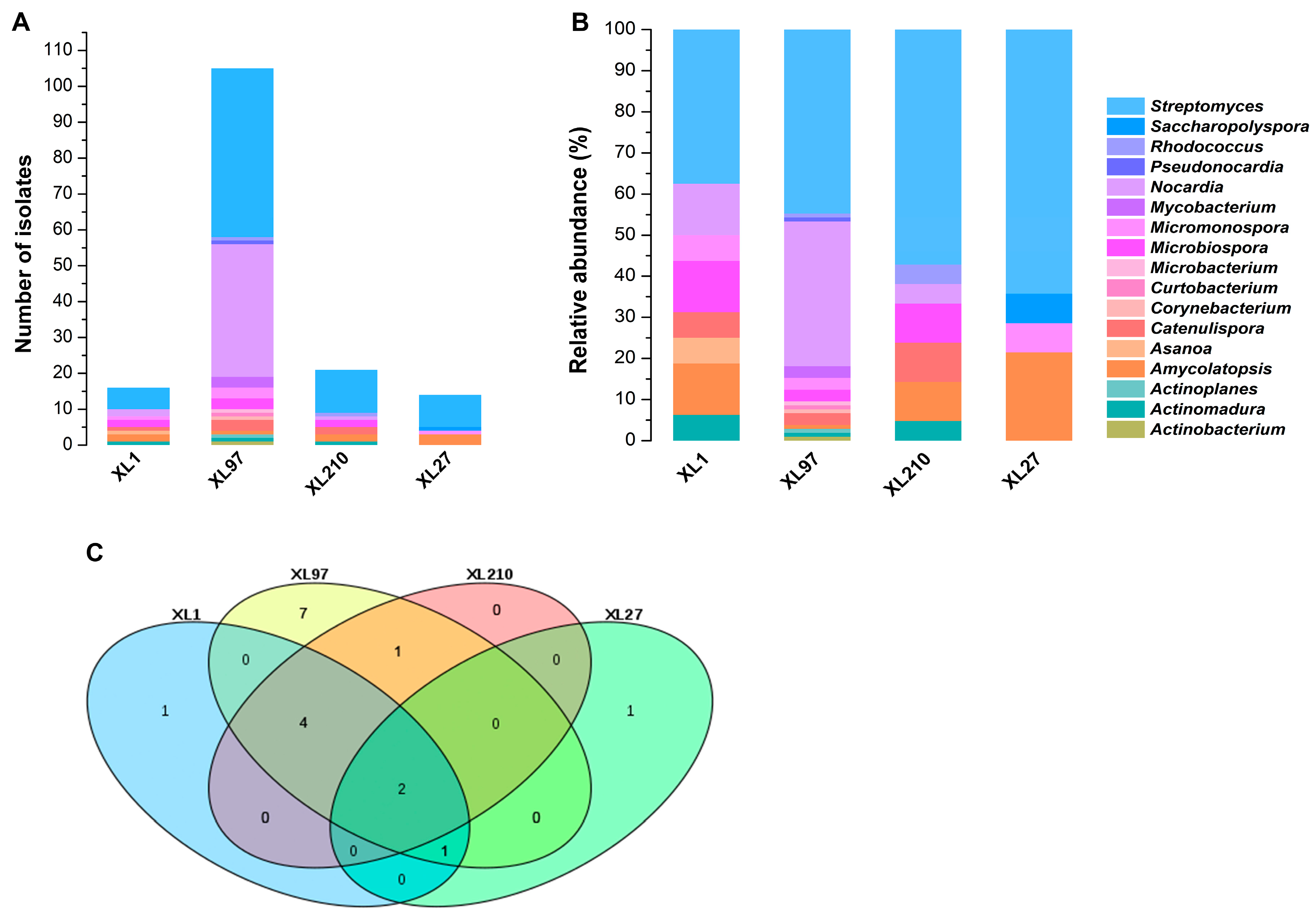
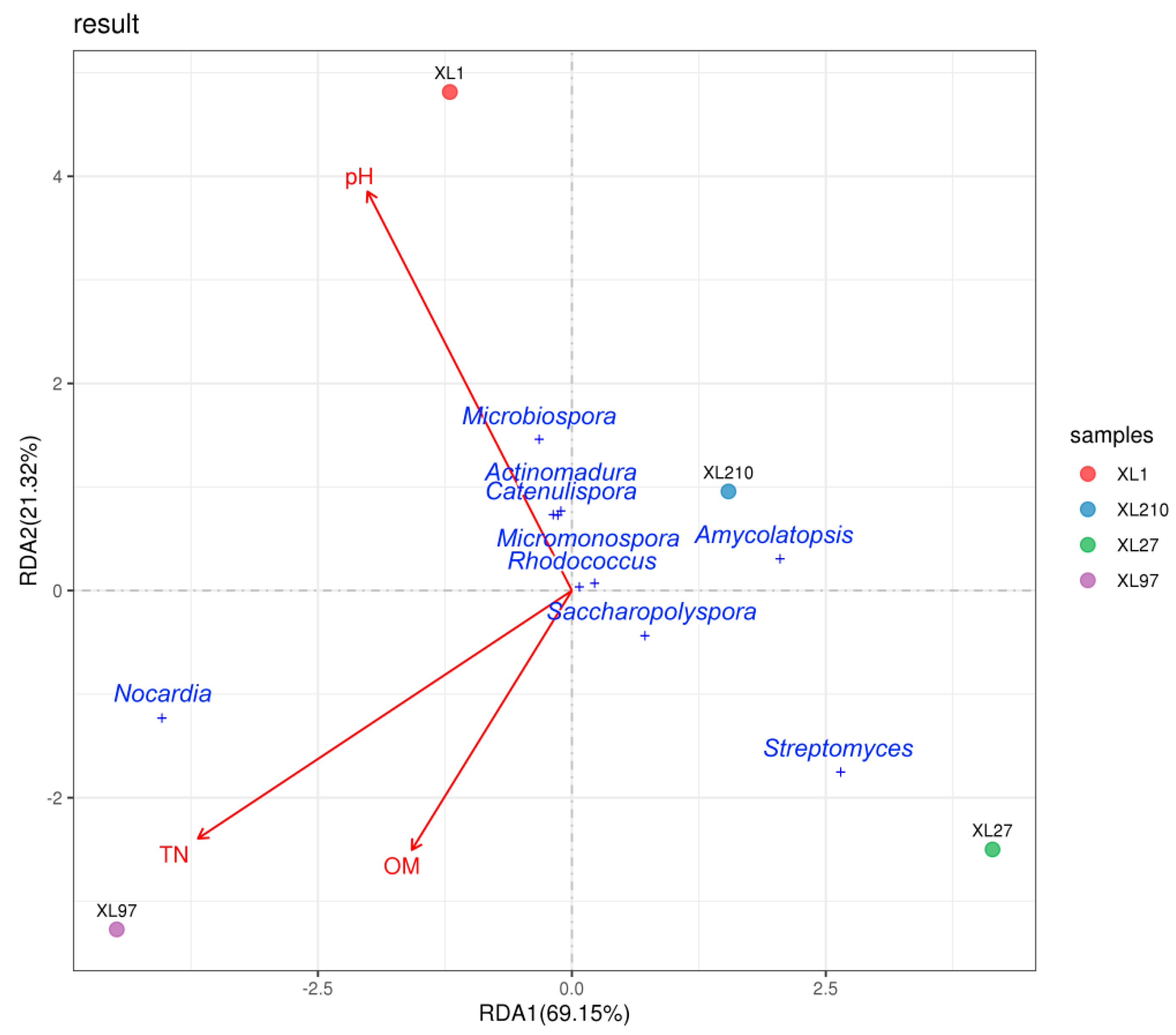
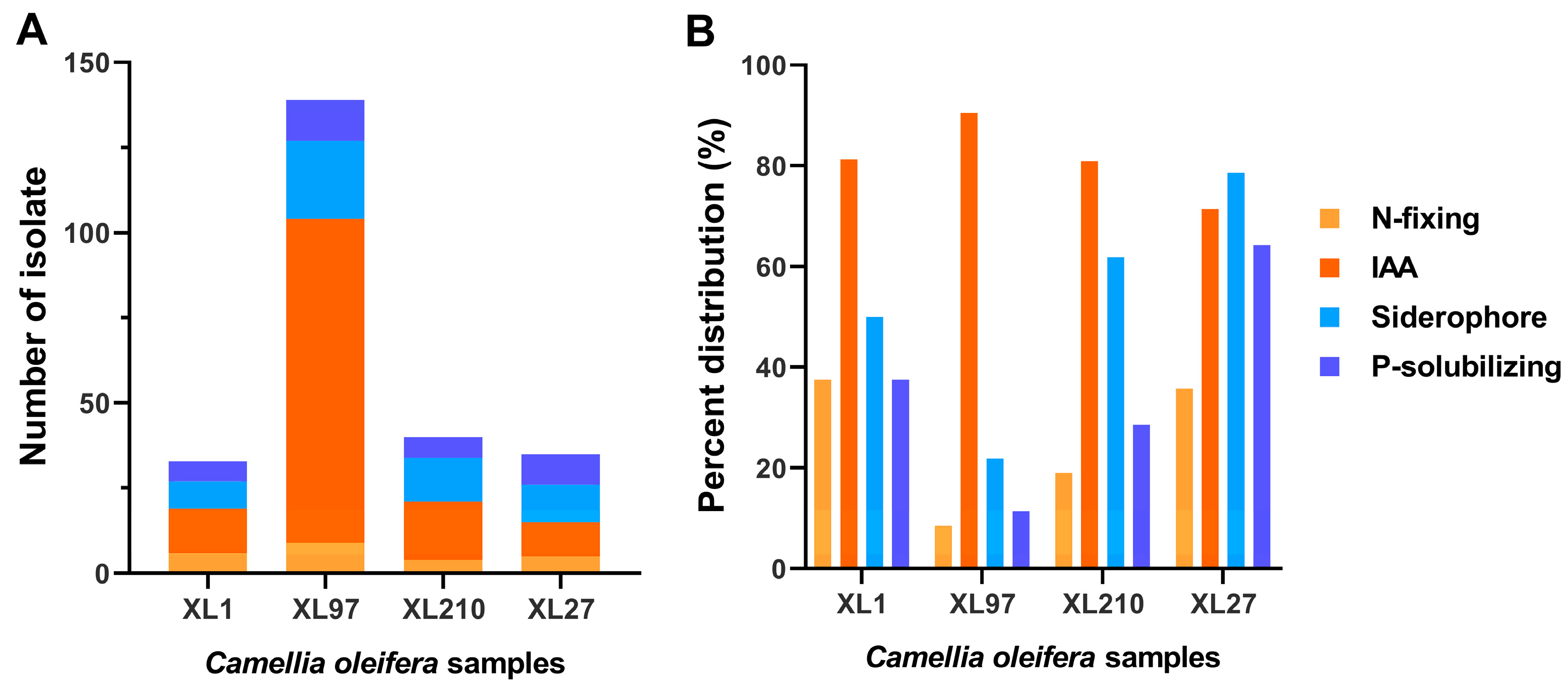

| XL1 | XL97 | XL210 | XL27 | |
|---|---|---|---|---|
| Fruit diameter/mm | 30–44 | 26–37 | 33–48 | 30–45 |
| Fruit number per 500 g | 15–30 | 21–40 | 15–30 | 15–28 |
| Fresh seed rate/% | 46.8 | 43.4–46.4 | 44.8 | 50–56 |
| Seed kernel oil rate/% | 35.0 | 50.5 | 36.4 | 34.7 |
| Fruit oil rate/% | 8.9 | 10.9 | 5.8 | 10.7 |
| Oil production/kg ha−1 | 772.5 | 901.5 | 618.8 | 995.4 |
| XL1 | XL97 | XL210 | XL27 | |
|---|---|---|---|---|
| pH | 5.71 ± 0.03 a | 5.05 ± 0.03 b | 5.07 ± 0.24 b | 4.73 ± 0.02 c |
| Organic matter (g kg−1) | 34.86 ± 1.48 c | 57.33 ± 2.24 a | 55.37 ± 3.11 a | 41.01 ± 2.52 b |
| Total nitrogen (g kg−1) | 2.68 ± 0.01 b | 3.41 ± 0.27 a | 2.63 ± 0.04 b | 2.63 ± 0.23 b |
| Total phosphorus (g kg−1) | 0.61 ± 0.13 b | 0.59 ± 0.05 b | 0.42 ± 0.04 b | 1.16 ± 0.14 a |
| Total potassium (g kg−1) | 5.24 ± 0.14 b | 4.53 ± 0.05 bc | 4.11 ± 0.08 c | 7.69 ± 0.52 a |
| Available nitrogen (mg kg−1) | 170.67 ± 15.73 b | 171.88 ± 7.02 b | 124.16 ± 15.44 c | 215.37 ± 13.24 a |
| Available phosphorus (mg kg−1) | 11.81 ± 0.74 bc | 12.96 ± 0.72 b | 10.90 ± 0.24 c | 15.21 ± 2.49 a |
| Available potassium (mg kg−1) | 107.74 ± 2.03 b | 86.76 ± 4.67 c | 88.39 ± 6.81 c | 133.88 ± 3.78 a |
| Calcium (g kg−1) | 0.53 ± 0.06 b | 0.74 ± 0.09 a | 0.40 ± 0.05 c | 0.77 ± 0.09 a |
| Iron (g kg−1) | 14.87 ± 0.56 d | 23.23 ± 0.73 b | 18.23 ± 1.53 c | 30.25 ± 1.31 a |
| Aluminum (g kg−1) | 1.10 ± 0.17 a | 1.11 ± 0.13 a | 0.96 ± 0.04 a | 1.12 ± 0.16 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, T.; Cui, K.; Chen, J.; Wang, R.; Wang, X.; Chen, L.; Zhang, Z.; He, Z.; Liu, C.; Tang, W.; et al. Biodiversity of Culturable Endophytic Actinobacteria Isolated from High Yield Camellia oleifera and Their Plant Growth Promotion Potential. Agriculture 2021, 11, 1150. https://doi.org/10.3390/agriculture11111150
Xu T, Cui K, Chen J, Wang R, Wang X, Chen L, Zhang Z, He Z, Liu C, Tang W, et al. Biodiversity of Culturable Endophytic Actinobacteria Isolated from High Yield Camellia oleifera and Their Plant Growth Promotion Potential. Agriculture. 2021; 11(11):1150. https://doi.org/10.3390/agriculture11111150
Chicago/Turabian StyleXu, Ting, Kunpeng Cui, Jiawei Chen, Rui Wang, Xiangnan Wang, Longsheng Chen, Zhen Zhang, Zhilong He, Caixia Liu, Wei Tang, and et al. 2021. "Biodiversity of Culturable Endophytic Actinobacteria Isolated from High Yield Camellia oleifera and Their Plant Growth Promotion Potential" Agriculture 11, no. 11: 1150. https://doi.org/10.3390/agriculture11111150








