Organic Amendments and Sampling Date Influences on Soil Bacterial Community Composition and Their Predictive Functional Profiles in an Olive Grove Ecosystem
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design and Edaphoclimatic Conditions
2.2. Soil Sampling
2.3. Molecular Analyses of Soil Bacteria
2.4. Predictive Metagenomics Profiling
2.5. Statistical and Diversity Analysis
3. Results
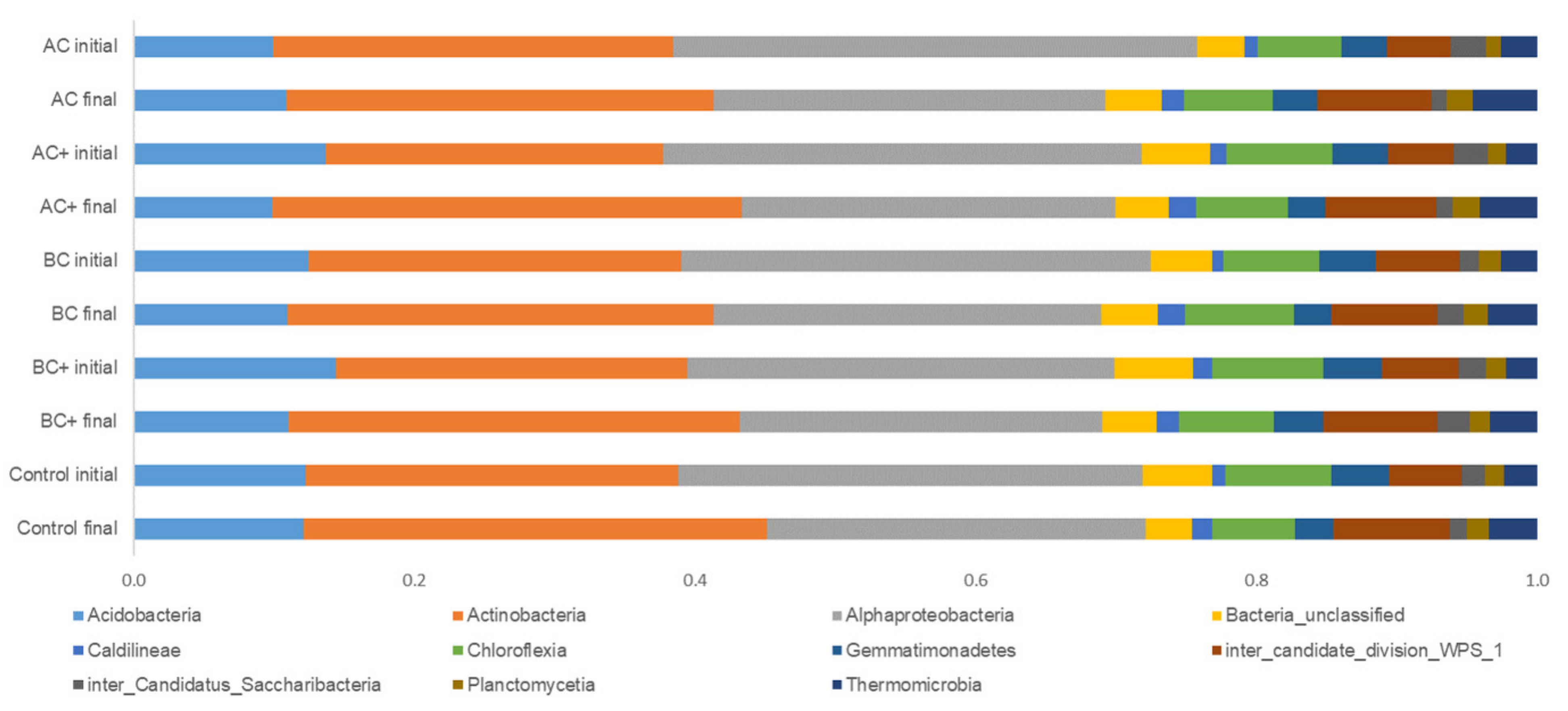
3.1. Bacterial Community Abundance
3.2. Bacterial Communities
| Shannon–Wiener Diversity Index | Species Richness (S) | Simpson Diversity Index | Evenness | |||||
|---|---|---|---|---|---|---|---|---|
| INITIAL | FINAL | INITIAL | FINAL | INITIAL | FINAL | INITIAL | FINAL | |
| AC | 5.84 ± 0.13 | 6.09 ± 0.03 | 2854 ± 246 | 2323 ± 34 | 0.013 ± 0.000a | 0.008 ± 0.000b | 0.735 ± 0.008b | 0.786 ± 0.003a |
| AC+ | 6.05 ± 0.01 | 6.01 ± 0.02 | 3327 ± 30a | 2470 ± 82b | 0.013 ± 0.000 | 0.010 ± 0.001 | 0.747 ± 0.000 | 0.769 ± 0.004 |
| BC | 6.04 ± 0.07 | 6.19 ± 0.04 | 3435 ± 262 | 2637 ± 36 | 0.012 ± 0.002a | 0.008 ± 0.001b | 0.743 ± 0.002b | 0.786 ± 0.003a |
| BC+ | 6.19 ± 0.03 | 6.11 ± 0.05 | 3625 ± 153 | 2645 ± 120 | 0.010 ± 0.000 | 0.009 ± 0.001 | 0.756 ± 0.000 | 0.776 ± 0.006 |
| Control | 6.14 ± 0.07 | 5.92 ± 0.13 | 3497 ± 146a | 2318 ± 126b | 0.012 ± 0.001 | 0.011 ± 0.002 | 0.753 ± 0.012 | 0.765 ± 0.013 |
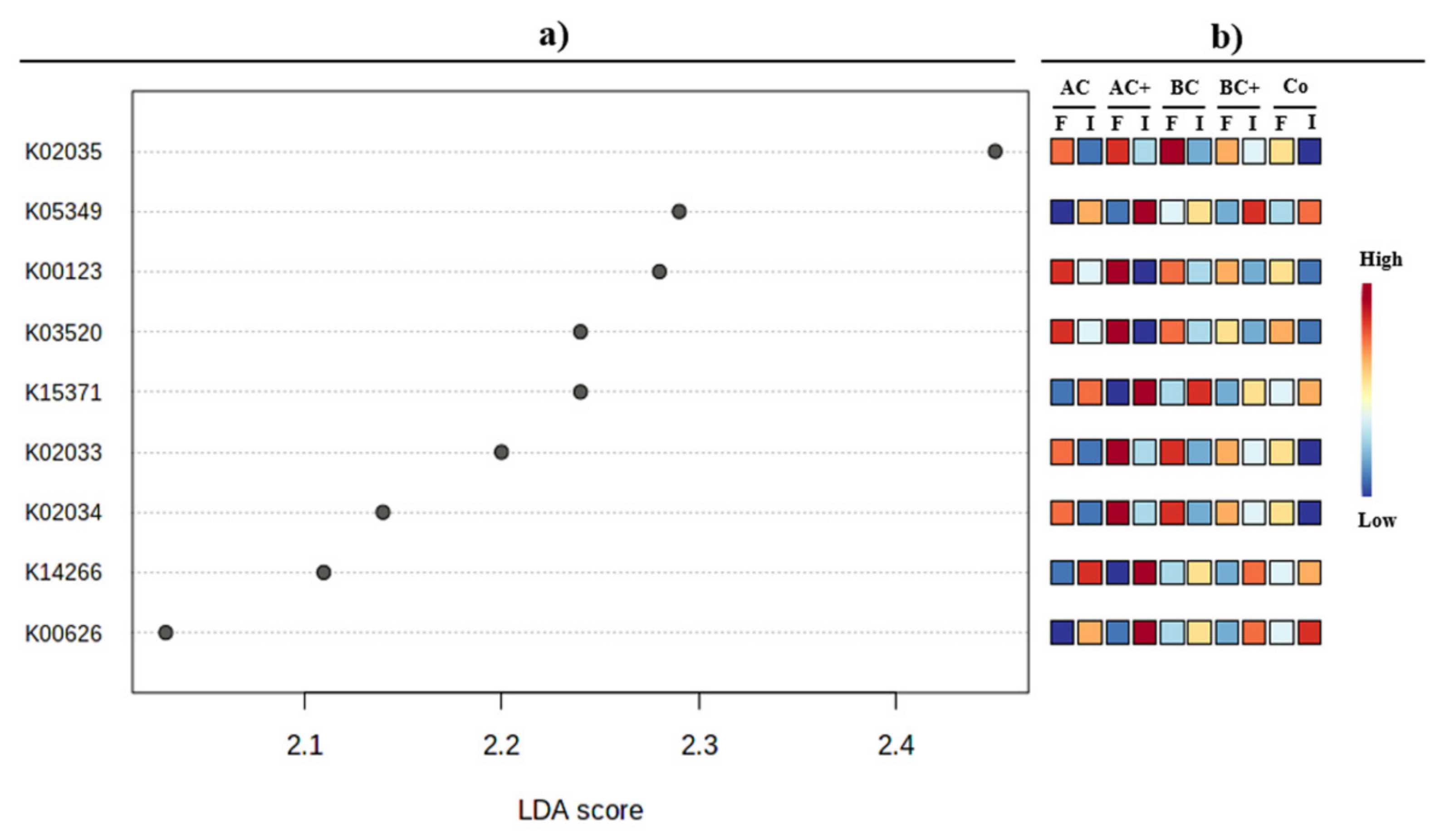
3.3. Bacterial Community Functional Analysis
4. Discussion
4.1. Bacterial Communities Composition and Abundance
4.2. Bacterial Community Functional Potential
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saccá, M.L.; Barra Caracciolo, A.; Di Lenola, M.; Grenni, P. Ecosystem Services Provided by Soil Microorganisms. In Soil Biological Communities and Ecosystem Resilience. Sustainability in Plant and Crop Protection; Lukac, M., Grenni, P., Gamboni, M., Eds.; Springer: Cham, Switzerland, 2017; pp. 9–24. [Google Scholar]
- Marco, D.E.; Abram, F. Using genomics, metagenomics and other “Omics” to assess valuable microbial ecosystem services and novel biotechnological applications. Front. Microbiol. 2019, 10, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Palacios, P.; Crowther, T.W.; Dacal, M.; Hartley, I.P.; Reinsch, S.; Rinnan, R.; Rousk, J.; van del Hoogen, J.; Ye, J.S.; Bradford, M.A. Evidence for large microbial-mediated losses of soil carbon under anthropogenic warming. Nat. Rev. Earth Environ. 2021, 2, 507–517. [Google Scholar] [CrossRef]
- Panettieri, M.; de Sosa, L.L.; Domínguez, M.T.; Madejón, E. Long-term impacts of conservation tillage on Mediterranean agricultural soils: Shifts in microbial communities despite limited effects on chemical properties. Agric. Ecosyst. Environ. 2020, 304, 107144. [Google Scholar] [CrossRef]
- Panettieri, M.; Guigue, J.; Chemidlin Prevost-Bouré, N.; Thévenot, M.; Lévêque, J.; Le Guillou, C.; Maron, P.A.; Santoni, A.L.; Ranjard, L.; Mounier, S.; et al. Grassland-cropland rotation cycles in crop-livestock farming systems regulate priming effect potential in soils through modulation of microbial communities, composition of soil organic matter and abiotic soil properties. Agric. Ecosyst. Environ. 2020, 299, 106973. [Google Scholar] [CrossRef]
- Bauer, J.T.; Blumenthal, N.; Miller, A.J.; Ferguson, J.K.; Reynolds, H.L. Effects of between-site variation in soil microbial communities and plant-soil feedbacks on the productivity and composition of plant communities. J. Appl. Ecol. 2017, 54, 1028–1039. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.L.; Ding, J.; Zhu, Y.G.; He, J.Z.; Hu, H.W. Soil bacterial taxonomic diversity is critical to maintaining the plant productivity. Environ. Int. 2020, 140, 105766. [Google Scholar] [CrossRef]
- Simon, C.; Daniel, R. Metagenomic Analyses: Past and Future Trends. Appl. Environ. Microbiol. 2011, 77, 1153–1161. [Google Scholar] [CrossRef] [Green Version]
- Smulders, L.; Benítez, E.; Moreno, B.; López-García, Á.; Pozo, M.J.; Ferrero, V.; de la Peña, E.; Alcalá Herrera, R. Tomato Domestication Affects Potential Functional Molecular Pathways of Root-Associated Soil Bacteria. Plants 2021, 10, 1942. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher-Jenull, J.; Ceccherini, M.T.; Pietramellara, G.; Renella, G.; Schloter, M. Beyond microbial diversity for predicting soil functions: A mini review. Pedosphere 2020, 30, 5–17. [Google Scholar] [CrossRef]
- Agriculture and Rural Development. In Soil Matters for Our Future. Available online: https://ec.europa.eu/info/news/soil-matters-our-future-2019-dec-05_en (accessed on 5 June 2020).
- Regni, L.; Nasini, L.; Ilarioni, L.; Brunori, A.; Massaccesi, L.; Agnelli, A.; Proietti, P. Long term amendment with fresh and composted solid olive mill waste on olive grove affects carbon sequestration by prunings, fruits and soil. Front. Plant Sci. 2017, 7, 2042. [Google Scholar] [CrossRef]
- Bechara, E.; Papafilippaki, A.; Doupis, G.; Sofo, A.; Koubouris, G. Nutrient dynamics, soil properties and microbiological aspects in an irrigated olive orchard managed with five different management systems involving soil tillage, cover crops and compost. J. Water Clim. Chang. 2018, 9, 736–747. [Google Scholar] [CrossRef]
- Benítez, E.; Paredes, D.; Rodríguez, E.; Aldana, D.; González, M.; Nogales, R.; Campos, M.; Moreno, B. Bottom-up effects on herbivore-induced plant defences: A case study based on compositional patterns of rhizosphere microbial communities. Sci. Rep. 2017, 7, 6251. [Google Scholar] [CrossRef] [Green Version]
- Maron, P.A.; Sarr, A.; Kaisermann, A.; Lvêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Benard, L.; Dequiedt, S.; Terat, S.; et al. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 2018, 84, e02738-17. [Google Scholar] [CrossRef] [Green Version]
- Schloter, M.; Nannipieri, P.; Sørensen, S.J.; van Elsas, J.D. Microbial indicators for soil quality. Biol. Fertil. Soils 2018, 54, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Llimós, M.; Segarra, G.; Sancho-Adamson, M.; Trillas, M.I.; Romanyà, J. Impact of Olive Saplings and Organic Amendments on Soil Microbial Communities and Effects of Mineral Fertilization. Front. Microbiol. 2021, 12, 653027. [Google Scholar] [CrossRef]
- Rajhi, H.; Sanz, J.L.; Abichou, M.; Morato, A.; Jradi, R.; Zaouari, R.; Bousnina, H.; Rojas, P. The Impact of Different Techniques of Soil Management on Soil Fertility and the Associated Bacterial Communities in Semi-arid Olive Tree Fields. J. Soil Sci. Plant Nutr. 2021, 21, 547–558. [Google Scholar] [CrossRef]
- Bevivino, A.; Paganin, P.; Bacci, G.; Florio, A.; Pellicer, M.S.; Papaleo, M.C.; Mengoni, A.; Ledda, L.; Fani, R.; Benedetti, A.; et al. Soil Bacterial Community Response to Differences in Agricultural Management along with Seasonal Changes in a Mediterranean Region. PLoS ONE 2014, 9, e105515. [Google Scholar] [CrossRef] [Green Version]
- Yuste, J.C.; Fernandez-Gonzalez, A.J.; Fernandez-Lopez, M.; Ogaya, R.; Penuelas, J.; Sardans, J.; Lloret, F. Strong functional stability of soil microbial communities under semiarid Mediterranean conditions and subjected to long-term shifts in baseline precipitation. Soil Biol. Biochem. 2014, 69, 223–233. [Google Scholar] [CrossRef]
- L de Sosa, L.; Benítez, E.; Girón, I.; Madejón, E. Agro-Industrial and Urban Compost as an Alternative of Inorganic Fertilizers in Traditional Rainfed Olive Grove under Mediterranean Conditions. Agronomy 2021, 11, 1223. [Google Scholar] [CrossRef]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microb. 1993, 59, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Cañizares, R.; Moreno, B.; Benítez, E. Biochemical characterization with detection and expression of bacterial β-glucosidase encoding genes of a Mediterranean soil under different long-term management practices. Biol. Fertil. Soils 2012, 48, 651–663. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundberg, D.S.; Yourstone, S.; Mieczkowski, P.; Jones, C.D.; Dangl, J.L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 2013, 10, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Větrovský, T.; Baldrian, P.; Morais, D. SEED 2: A user-friendly platform for amplicon high-throughput sequencing data analyses. Bioinformatics 2018, 34, 2292–2294. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE, highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur, open-source; platform-independent; community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project, improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical; functional; and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst, a web-based tool for comprehensive statistical; visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, 180–188. [Google Scholar] [CrossRef]
- Aßhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun, predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, 457–462. [Google Scholar] [CrossRef] [Green Version]
- Hammer, O.; Harper, D.; Ryan, P. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https:www.R-project.org/ (accessed on 10 June 2021).
- RStudio Team. RStudio, Integrated Development Environment for R. 2016. Available online: http://www.rstudio.com/ (accessed on 10 June 2021).
- Parham, J.A.; Deng, S.P.; Da, H.N.; Sun, H.Y.; Raun, W.R. Long-term cattle manure application in soil. II. Effect on soil microbial populations and community structure. Biol. Fertil. Soils 2003, 38, 209–215. [Google Scholar] [CrossRef]
- Dangi, S.; Gao, S.; Duan, Y.; Wang, D. Soil microbial community structure affected by biochar and fertilizer sources. Appl. Soil Ecol. 2020, 150, 103452. [Google Scholar] [CrossRef]
- Lazcano, C.; Gómez-Brandón, M.; Revilla, P.; Domínguez, J. Short-term effects of organic and inorganic fertilizers on soil microbial community structure and function. Biol. Fertil. Soils 2013, 49, 723–733. [Google Scholar] [CrossRef]
- Teutscherova, N.; Vazquez, E.; Santana, D.; Navas, M.; Masaguer, A.; Benito, M. Influence of pruning waste compost maturity and biochar on carbon dynamics in acid soil: Incubation study. Eur. J. Soil Biol. 2017, 78, 66–74. [Google Scholar] [CrossRef]
- Knapp, B.A.; Ros, M.; Insam, H. Do composts affect the soil microbial community? In Microbes at Work; Insam, H., Franke-Whittle, I., Goberna, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 271–291. [Google Scholar]
- Li, N.; Lei, W.; Long, J.; Han, X. Restoration of Chemical Structure of Soil Organic Matter Under Different Agricultural Practices from a Severely Degraded Mollisol. J. Soil Sci. Plant Nutr. 2021, 1–14. [Google Scholar] [CrossRef]
- Ho, A.; Di Lonardo, D.P.; Bodelier, P.L.E. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Männistö, M.; Ganzert, L.; Tiirola, M.; Häggblom, M.; Stark, S. Do shifts in life strategies explain microbial community responses to increasing nitrogen in tundra soil? Soil Biol. Biochem. 2016, 96, 216–228. [Google Scholar] [CrossRef] [Green Version]
- Plaza, C.; Courtier-Murias, D.; Fernández, J.M.; Polo, A.; Simpson, A.J. Physical, chemical, and biochemical mechanisms of soil organic matter stabilization under conservation tillage systems: A central role for microbes and microbial by-products in C sequestration. Soil Biol. Biochem. 2013, 57, 124–134. [Google Scholar] [CrossRef] [Green Version]
- Tyler, H.L. Shifts in bacterial community in response to conservation management practices within a soybean production system. Biol. Fertil. Soils 2021, 57, 575–586. [Google Scholar] [CrossRef]
- Saison, C.; Degrange, V.; Oliver, R.; Millard, P.; Commeaux, C.; Montange, D.; Le Roux, X. Alteration and resilience of the soil microbial community following compost amendment: Effects of compost level and compost-borne microbial community. Environ. Microbiol. 2006, 8, 247–257. [Google Scholar] [CrossRef]
- Tian, W.; Zhang, Z.; Hu, X.; Tian, R.; Zhang, J.; Xiao, X.; Xi, Y. Short-term changes in total heavy metal concentration and bacterial community composition after replicated and heavy application of pig manure-based compost in an organic vegetable production system. Biol. Fertil. Soils 2015, 51, 593–603. [Google Scholar] [CrossRef]
- Biggs, C.R.; Yeager, L.A.; Bolser, D.G.; Bonsell, C.; Dichiera, A.M.; Hou, Z.; Keyser, S.R.; Khursigara, A.J.; Lu, K.; Muth, A.F.; et al. Does functional redundancy affect ecological stability and resilience? A review and meta-analysis. Ecosphere 2020, 11, e03184. [Google Scholar] [CrossRef]
- Stevenson, F.J. Humus Chemistry, Genesis, Composition, Reactions, 2nd ed.; John Wiley and Sons: New York, NY, USA, 1994. [Google Scholar]
- Liu, C.; Li, L.; Xie, J.; Coulter, J.A.; Zhang, R.; Luo, Z.; Cai, L.; Wang, L.; Gopalakrishnan, S. Soil Bacterial Diversity and Potential Functions Are Regulated by Long-Term Conservation Tillage and Straw Mulching. Microorganisms 2020, 8, 836. [Google Scholar] [CrossRef]
- Gryta, A.; Frąc, M.; Oszust, K. Genetic and Metabolic Diversity of Soil Microbiome in Response to Exogenous Organic Matter Amendments. Agronomy 2020, 10, 546. [Google Scholar] [CrossRef] [Green Version]
- Spaccini, R.; Piccolo, A. Amendments with humified compost effectively sequester organic carbon in agricultural soils. Land Degrad. Dev. 2020, 31, 1206–1216. [Google Scholar] [CrossRef]
- Kang, S.; Ma, W.; Li, F.Y.; Zhang, Q.; Niu, J.; Ding, Y.; Han, F.; Sun, X. Functional Redundancy Instead of Species Redundancy Determines Community Stability in a Typical Steppe of Inner Mongolia. PLoS ONE 2015, 10, e0145605. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sosa, L.L.d.; Moreno, B.; Herrera, R.A.; Panettieri, M.; Madejón, E.; Benítez, E. Organic Amendments and Sampling Date Influences on Soil Bacterial Community Composition and Their Predictive Functional Profiles in an Olive Grove Ecosystem. Agriculture 2021, 11, 1178. https://doi.org/10.3390/agriculture11111178
Sosa LLd, Moreno B, Herrera RA, Panettieri M, Madejón E, Benítez E. Organic Amendments and Sampling Date Influences on Soil Bacterial Community Composition and Their Predictive Functional Profiles in an Olive Grove Ecosystem. Agriculture. 2021; 11(11):1178. https://doi.org/10.3390/agriculture11111178
Chicago/Turabian StyleSosa, Laura L. de, Beatriz Moreno, Rafael Alcalá Herrera, Marco Panettieri, Engracia Madejón, and Emilio Benítez. 2021. "Organic Amendments and Sampling Date Influences on Soil Bacterial Community Composition and Their Predictive Functional Profiles in an Olive Grove Ecosystem" Agriculture 11, no. 11: 1178. https://doi.org/10.3390/agriculture11111178
APA StyleSosa, L. L. d., Moreno, B., Herrera, R. A., Panettieri, M., Madejón, E., & Benítez, E. (2021). Organic Amendments and Sampling Date Influences on Soil Bacterial Community Composition and Their Predictive Functional Profiles in an Olive Grove Ecosystem. Agriculture, 11(11), 1178. https://doi.org/10.3390/agriculture11111178








