The Immense Functional Attributes of Maize Rhizosphere Microbiome: A Shotgun Sequencing Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Description of Study Area and Soil Sampling
2.2. Chemical Analysis of Soil
2.3. DNA Extraction, Metagenomic Sequencing, and Downstream Analysis
2.4. Statistical Analysis
3. Results
3.1. Chemical Properties of Maize Rhizosphere Soil and Control Samples
3.2. Sequence Information and Processing Output
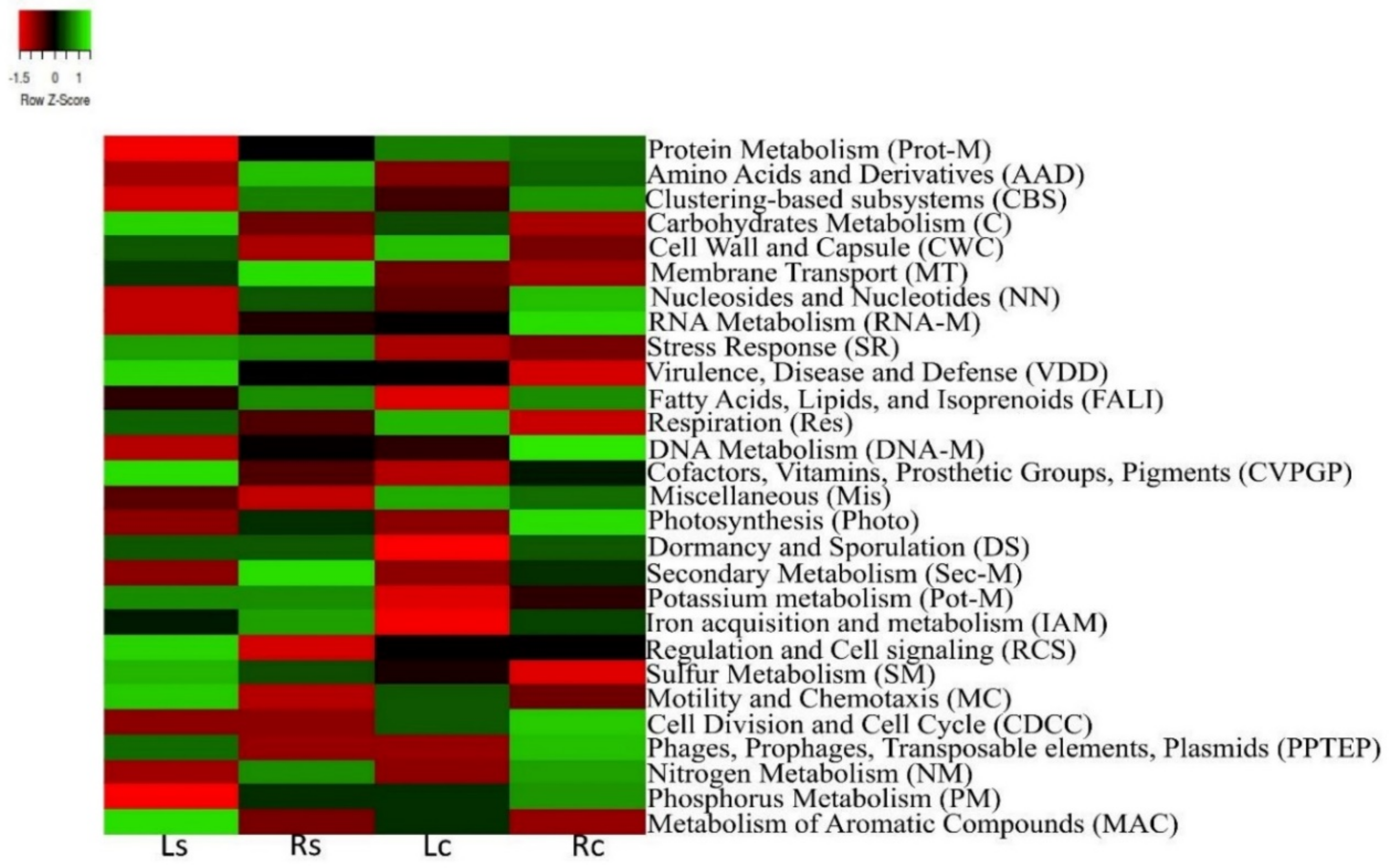
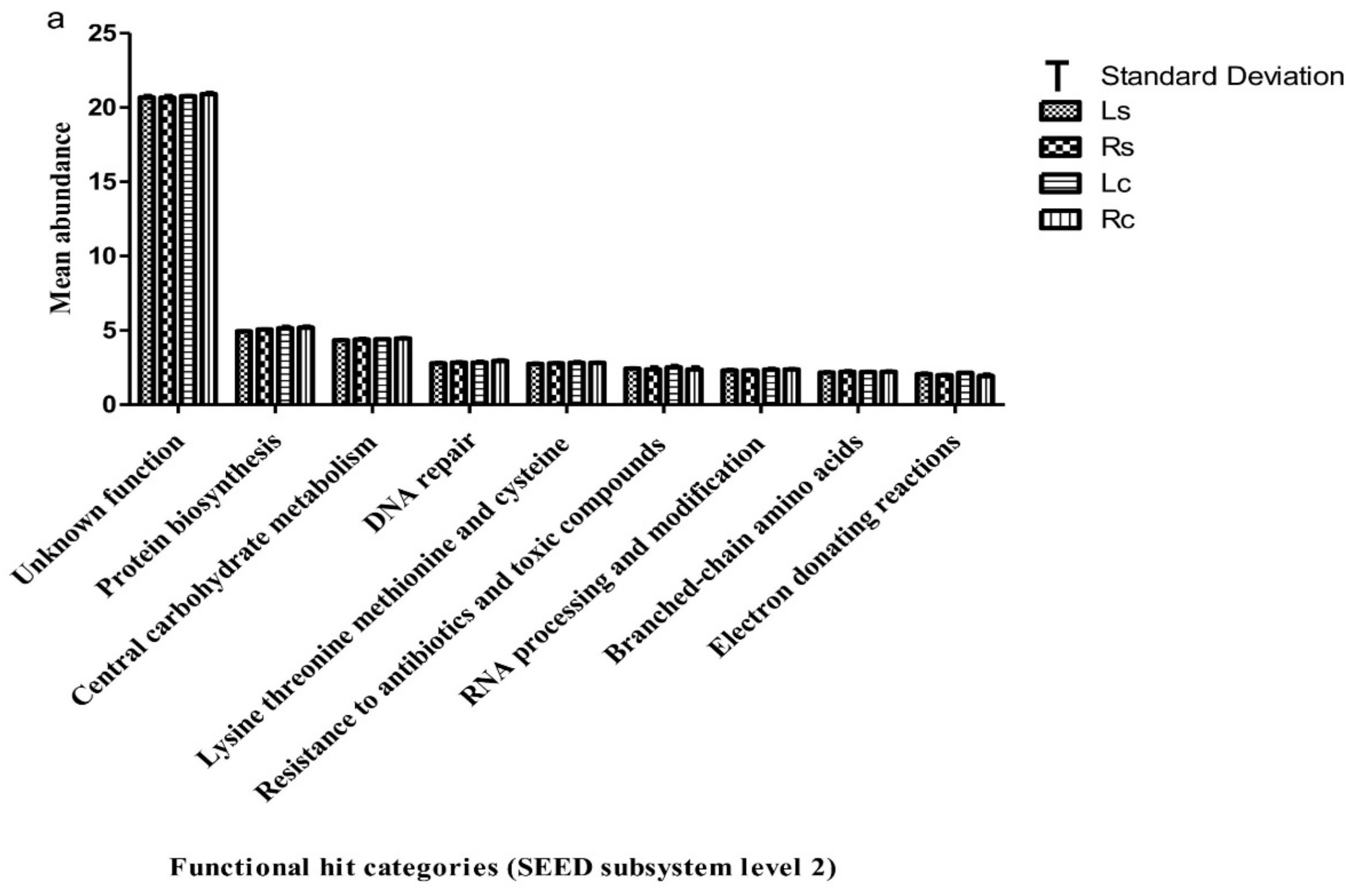
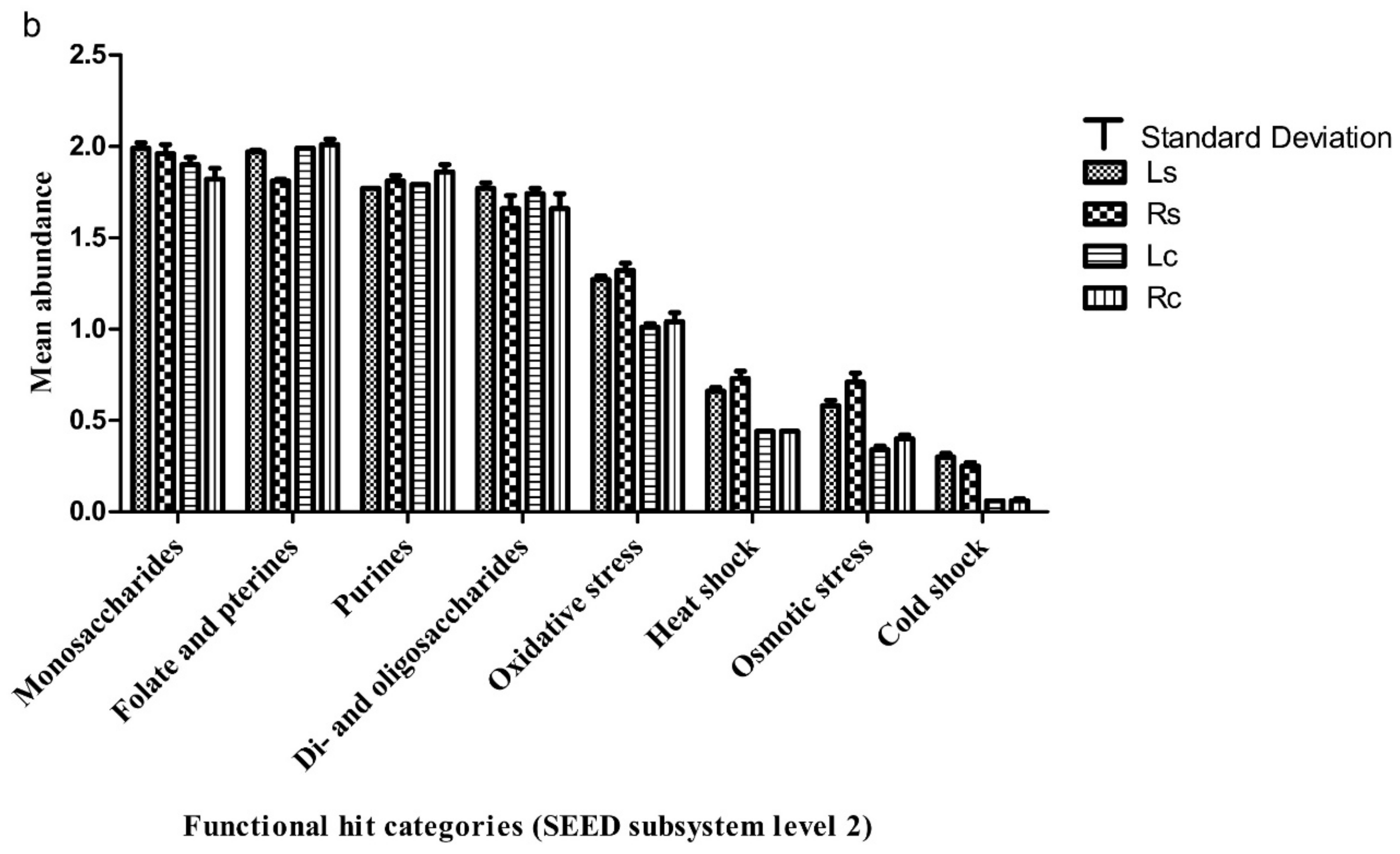
3.3. Functional Attributes Associated with the Rhizosphere Samples and Their Controls
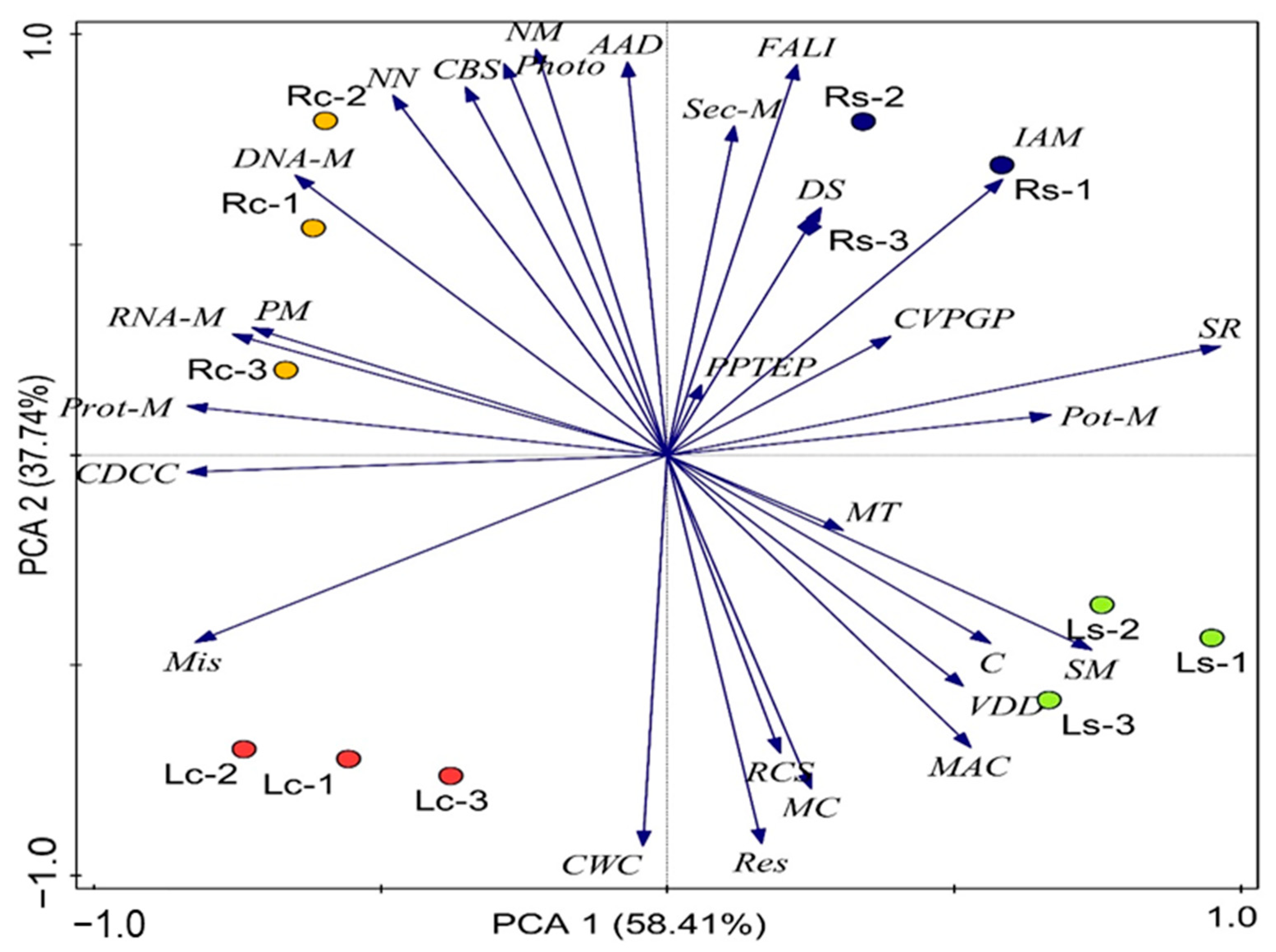
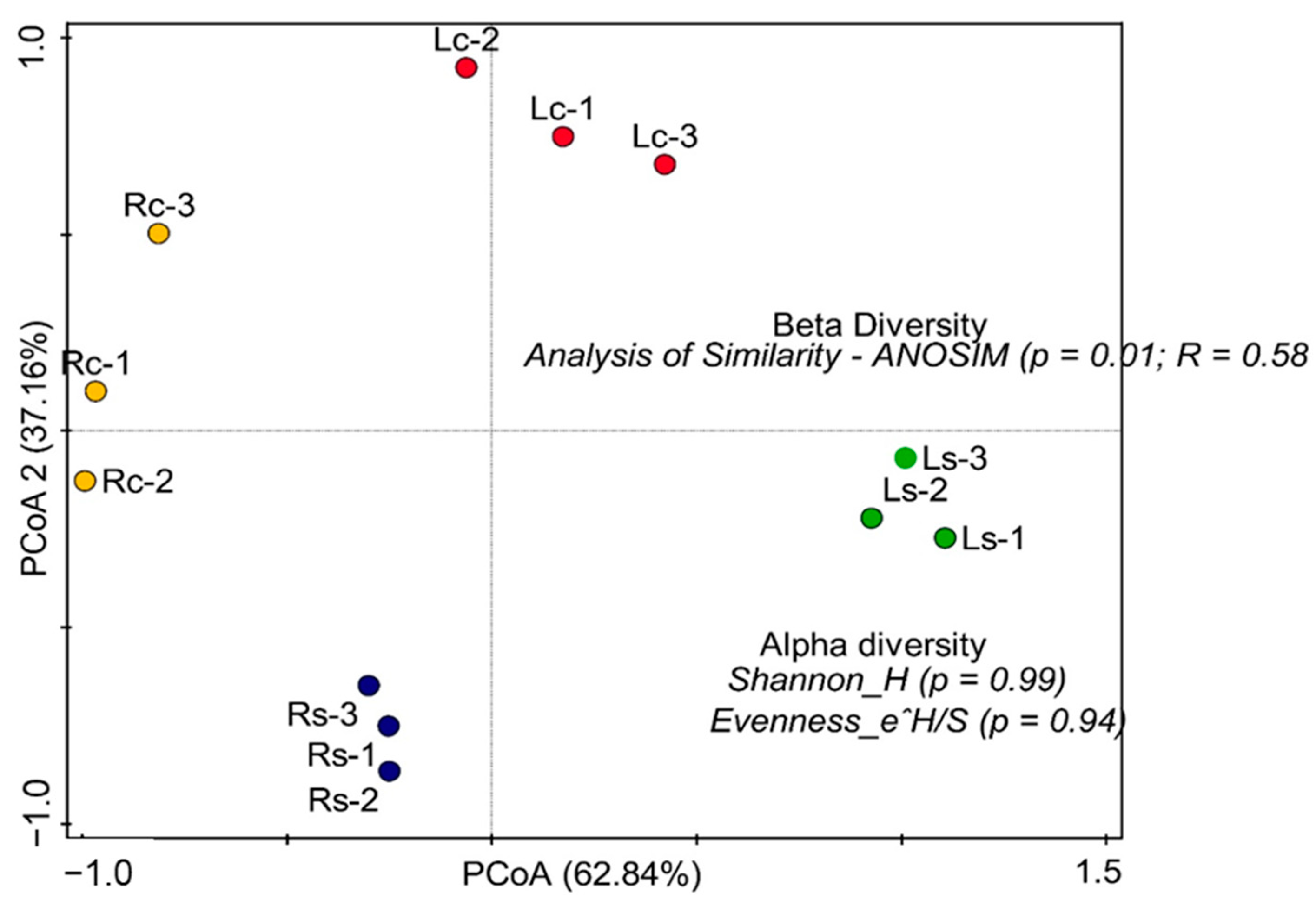
3.4. Alpha and Beta Diversity of Assessed Functional Hits across Soil Samples
3.5. Impact of Environmental Factors on Rhizobiome Functional Categories
4. Discussion
5. Conclusions
- Differences in the functional attributes were observed in the metagenomics study of maize rhizosphere and bulk soil.
- The presence of enormous functions conferring response to soil stressors in the rhizosphere samples could highlight the presence of novel organisms with biotechnological importance.
- Environmental variables viz. N-NO3, sulfate, and pH had great impact on soil rhizobiome functioning.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Enebe, M.C.; Babalola, O.O. The impact of microbes in the orchestration of plants’ resistance to biotic stress: A disease management approach. J. Appl. Microbiol. Biotechnol. 2019, 103, 9–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alawiye, T.T.; Babalola, O.O. Bacterial diversity and community structure in typical plant rhizosphere. Diversity 2019, 11, 179. [Google Scholar] [CrossRef] [Green Version]
- Gougoulias, C.; Clark, J.M.; Shaw, L.J. The role of soil microbes in the global carbon cycle: Tracking the below-ground microbial processing of plant-derived carbon for manipulating carbon dynamics in agricultural systems. J. Sci. Food Agric. 2014, 94, 2362–2371. [Google Scholar] [CrossRef] [PubMed]
- Mhete, M.; Eze, P.N.; Rahube, T.O.; Akinyemi, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. J. Sci. Afr. 2020, 7, e00246. [Google Scholar] [CrossRef]
- Igiehon, N.O.; Babalola, O.O. Rhizosphere microbiome modulators: Contributions of nitrogen fixing bacteria towards sustainable agriculture. Int. J. Environ. Res. Pub. Health 2018, 15, 574. [Google Scholar] [CrossRef] [Green Version]
- Jéan du Plessis, D.D.V.B.; Justinus, M.; Apie, P.; Thinus, P. The Department of Agriculture and Obtainable from Resource Centre Directorate Agricultural Information Services Private Bag X144, Pretoria, 0001; The Department of Agriculture: Pretoria, South Africa, 2003; pp. 1–35.
- Gravelet-Blondin, R. Prospectus on the South African Maize Industry; The Maize Trust: Pretoria, South Africa, 2013; Available online: https://sacota.co.za/wp-content/uploads/Prospectus-on-the-South-African-Maize-Industry.pdf (accessed on 15 May 2020).
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Banach, A.; Izak, D.; Stępniewska, Z.; Błaszczyk, M. Metagenomic analysis of some potential nitrogen-fixing bacteria in arable soils at different formation processes. J. Microb. Ecol. 2017, 73, 162–176. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Y.; Hu, J.; Xia, Q.; Zhang, S.; Li, X.; Pan, X.; Zhao, R.; Wang, R.; Yan, W.; Shangguan, Z. Soil microbial mechanisms promoting ultrahigh rice yield. J. Soil Biol. Biochem. 2020, 143, 107741. [Google Scholar] [CrossRef]
- Olsen, S.; Sommers, L. Phosphorus. In Methods of Soil Analysis Part 2; Page, A.L., Miller, R.H., Keneey, D.R., Eds.; American Society of Agronomy: Madison, WI, USA, 1982; pp. 403–430. [Google Scholar]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. J. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Deke, A.L.; Adugna, W.T.; Fite, A.T. Soil physico-chemical properties in termite mounds and adjacent control soil in Miyo and Yabello Districts of Borana Zone, Southern Ethiopia. Am. J. Agric. For. 2016, 4, 69. [Google Scholar]
- Shi, J.-Y.; Yuan, X.-F.; Lin, H.-R.; Yang, Y.-Q.; Li, Z.-Y. Differences in soil properties and bacterial communities between the rhizosphere and bulk soil and among different production areas of the medicinal plant Fritillaria thunbergii. Int. J. Mol. Sci. 2011, 12, 3770–3785. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [Green Version]
- Kent, W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilke, A.; Harrison, T.; Wilkening, J.; Field, D.; Glass, E.M.; Kyrpides, N.; Mavrommatis, K.; Meyer, F. The M5nr: A novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. J. BMC Bioinform. 2012, 13, 141. [Google Scholar] [CrossRef] [Green Version]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. J. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Amoo, A.E.; Babalola, O.O. Impact of land use on bacterial diversity and community structure in temperate pine and indigenous forest soils. Diversity 2019, 11, 217. [Google Scholar] [CrossRef] [Green Version]
- Tale, K.S.; Ingole, S. A review on role of physico-chemical properties in soil quality. J. Chem. Sci. Rev. Lett. 2015, 4, 57–66. [Google Scholar]
- Salam, L.B.; Obayori, O.S. Structural and functional metagenomic analyses of a tropical agricultural soil. Span. J. Soil Sci. 2019, 9. [Google Scholar] [CrossRef]
- Salam, L.B.; Obayori, S.O.; Nwaokorie, F.O.; Suleiman, A.; Mustapha, R. Metagenomic insights into effects of spent engine oil perturbation on the microbial community composition and function in a tropical agricultural soil. J. Environ. Sci. Pollut. Res. 2017, 24, 7139–7159. [Google Scholar] [CrossRef]
- Dinsdale, E.A.; Edwards, R.A.; Hall, D.; Angly, F.; Breitbart, M.; Brulc, J.M.; Furlan, M.; Desnues, C.; Haynes, M.; Li, L. Functional metagenomic profiling of nine biomes. Nature 2008, 452, 629–632. [Google Scholar] [CrossRef]
- García-Salamanca, A.; Molina-Henares, M.A.; van Dillewijn, P.; Solano, J.; Pizarro-Tobías, P.; Roca, A.; Duque, E.; Ramos, J.L. Bacterial diversity in the rhizosphere of maize and the surrounding carbonate-rich bulk soil. Microb. Biotechnol. 2013, 6, 36–44. [Google Scholar] [CrossRef]
- Uroz, S.; Buée, M.; Murat, C.; Frey-Klett, P.; Martin, F. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2010, 2, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Chukwuneme, C.F.; Babalola, O.O.; Kutu, F.R.; Ojuederie, O.B. Characterization of actinomycetes isolates for plant growth promoting traits and their effects on drought tolerance in maize. J. Plant Interact. 2020, 15, 93–105. [Google Scholar] [CrossRef]
- Kumar, P.; Dubey, R.; Maheshwari, D. Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol. Res. 2012, 167, 493–499. [Google Scholar] [CrossRef]
- Madhaiyan, M.; Saravanan, V.S.; Blom, J.; Smits, T.H.; Rezzonico, F.; Kim, S.J.; Weon, H.Y.; Kwon, S.W.; Whitman, W.B.; Ji, L. Phytobacter palmae sp. nov., a novel endophytic, N2 fixing, plant growth promoting Gammaproteobacterium isolated from oil palm (Elaeis guineensis Jacq.). Int. J. Syst. Evol. Microbiol. 2020, 70, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, R.; Lang, E.; Midha, S.; Patil, P.B.; Rameshkumar, N. Isolation and characterization of a novel 1-aminocyclopropane-1-carboxylate (ACC) deaminase producing plant growth promoting marine Gammaproteobacteria from crops grown in brackish environments. Proposal for Pokkaliibacter plantistimulans gen. nov., sp. nov., Balneatrichaceae fam. nov. in the order Oceanospirillales and an emended description of the genus Balneatrix. Syst. Appl. Microbiol. 2018, 41, 570–580. [Google Scholar]
- Chukwuneme, C.F.; Ayangbenro, A.S.; Babalola, O.O. Functional diversity of microbial communities in two contrasting maize rhizosphere soils. Rhizosphere 2020, 17, 100282. [Google Scholar] [CrossRef]
- Akinola, S.A.; Babalola, O.O. The importance of adverse soil microbiomes in the light of omics: Implications for food safety. Plant. Soil Environ. 2020, 66, 421–430. [Google Scholar] [CrossRef]
- Fan, K.; Weisenhorn, P.; Gilbert, J.A.; Chu, H. Wheat rhizosphere harbors a less complex and more stable microbial co-occurrence pattern than bulk soil. Soil Biol. Biochem. 2018, 125, 251–260. [Google Scholar] [CrossRef]
- Nkuekam, G.K.; Cowan, D.A.; Valverde, A. Arable agriculture changes soil microbial communities in the South African Grassland Biome. S. Afr. J. Sci. 2018, 114, 1–7. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Bickford, W.A.; Zak, D.R.; Kowalski, K.P.; Goldberg, D.E. Differences in rhizosphere microbial communities between native and non-native Phragmites australis may depend on stand density. Ecol. Evol. 2020, 10, 11739–11751. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, P.; de Miera, L.E.S.; Ansola, G. Influence of environmental variables on the structure and composition of soil bacterial communities in natural and constructed wetlands. J. Sci. Total Environ. 2015, 506, 380–390. [Google Scholar] [CrossRef] [PubMed]

| Sample Locations → | Ls | Rs | Lc | Rc | p-Value | |
|---|---|---|---|---|---|---|
| Physicochemical parameters | pH (H2O) | 5.62 ± 0.09 a | 6.76 ± 0.28 b | 5.87 ± 0.22 a | 6.73 ± 0.26 b | <0.000 |
| P (mgkg−1) | 50.98 ± 1.77 a | 257.14 ± 35.32 b | 65.86 ± 13.71 a | 206.54 ± 81.73 b | 0.001 | |
| K (mgkg−1) | 240.00 ± 2.94 a | 167.00 ± 11.63 b | 243.00 ± 0.82 a | 148.50 ± 34.95 b | <0.000 | |
| Sulfate (mgkg−1) | 1.60 ± 1.68 a | 2.56 ± 2.66 a | 0.44 ± 0.36 a | 2.32 ± 2.75 a | 0.623 | |
| Total C (%) | 0.90 ± 0.05 a | 1.34 ± 0.24 a | 0.90 ± 0.01 a | 0.85 ± 0.50 a | 0.187 | |
| Org C (%) | 0.61 ± 0.02 a | 1.09 ± 0.09 b | 0.60 ± 0.01 a | 0.87 ± 0.15 c | <0.000 | |
| Org M (%) | 3.40 ± 0.16 a | 3.43 ± 0.39 a | 3.25 ± 0.03 a | 2.95 ± 0.85 a | 0.609 | |
| N-NO3(mgkg−1) | 16.29 ± 2.25 a | 8.52 ± 2.68 b | 16.24 ± 0.59 a | 7.38 ± 2.46 b | 0.001 | |
| N-NH4 (mgkg−1) | 3.61 ± 0.29 a,b | 2.91 ± 1.12 a | 2.42 ± 0.19 a | 4.75 ± 1.21 b | 0.044 | |
| Sample Sites | Ls | Rs | Lc | Rc |
|---|---|---|---|---|
| Uploaded Information | ||||
| bp Count | 2,863,587,272 | 2,237,924,006 | 2,269,959,337 | 2,113,440,642 |
| Sequences Count | 19,276,118 | 14,928,201 | 14,988,818 | 14,053,905 |
| Mean sequence length (bp) | 149 ± 51 | 150 ± 48 | 152 ± 47 | 151 ± 48 |
| Mean G + C content (%) | 64 ± 11 | 65 ± 11 | 65 ± 10 | 65 ± 11 |
| Post Quality Control Information | ||||
| bp count | 2,687,455,368 | 2,115,280,833 | 2,147,410,521 | 1,994,176,095 |
| Sequence count | 17,596,177 | 13,823,192 | 13,925,537 | 13,006,005 |
| Mean sequence length (bp) | 153 ± 47 | 154 ± 45 | 154 ± 44 | 154 ± 45 |
| Mean G + C content (%) | 65 ± 9 | 65 ± 9 | 65 ± 9 | 65 ± 9 |
| Processed Sequences | ||||
| Predicted protein features | 15,344,917 | 12,427,664 | 12,428,891 | 11,695,150 |
| Predicted rRNA features | 35,945 | 31,594 | 27,292 | 27,927 |
| Aligned Sequences | ||||
| Identified protein features | 5,959,395 | 4,732,504 | 4,654,996 | 4,507,871 |
| Identified rRNA features | 8225 | 7129 | 6347 | 7240 |
| Environmental Variable | Contribution% | Pseudo-F | p |
|---|---|---|---|
| N-NO3 | 45.5 | 1.7 | 0.65 |
| Sulfate | 46.4 | 5.7 | 0.53 |
| pH | 8.1 | <0.1 | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akinola, S.A.; Ayangbenro, A.S.; Babalola, O.O. The Immense Functional Attributes of Maize Rhizosphere Microbiome: A Shotgun Sequencing Approach. Agriculture 2021, 11, 118. https://doi.org/10.3390/agriculture11020118
Akinola SA, Ayangbenro AS, Babalola OO. The Immense Functional Attributes of Maize Rhizosphere Microbiome: A Shotgun Sequencing Approach. Agriculture. 2021; 11(2):118. https://doi.org/10.3390/agriculture11020118
Chicago/Turabian StyleAkinola, Saheed Adekunle, Ayansina Segun Ayangbenro, and Olubukola Oluranti Babalola. 2021. "The Immense Functional Attributes of Maize Rhizosphere Microbiome: A Shotgun Sequencing Approach" Agriculture 11, no. 2: 118. https://doi.org/10.3390/agriculture11020118
APA StyleAkinola, S. A., Ayangbenro, A. S., & Babalola, O. O. (2021). The Immense Functional Attributes of Maize Rhizosphere Microbiome: A Shotgun Sequencing Approach. Agriculture, 11(2), 118. https://doi.org/10.3390/agriculture11020118






