Abstract
Self-incompatibility is an important evolutionary feature in angiosperms and has major implications for breeding strategies in horticultural crops. In citrus, when coupled with parthenocarpy, it enables the production of seedless fruits in a mono-varietal orchard. A gametophytic incompatibility system with one S locus was proposed for citrus, but its molecular mechanisms remain the subject of debate. The objective of this work was to locate the S locus by the analyzing segregation distortion in reciprocal crosses of two self-incompatible citrus sharing one self-incompatible allele and to compare this location with previously published models. High density genetic maps of ‘Fortune’ mandarin and ‘Ellendale tangor’ with, respectively, 2164 SNP and 1467 SNP markers, were constructed using genotyping by sequencing data. They are highly syntenic and collinear with the clementine genome. Complete rejection of one allele was only observed in male segregation in the two parents and in only one genomic area, at the beginning of chromosome 7 of the clementine reference genome. Haplotype data in the area surrounding the theoretical S locus were in agreement with previously proposed S genotypes. Overall, our results are in full agreement with the recently proposed gametophytic S-RNase system with the S locus at the beginning of chromosome 7 of the clementine reference genome.
1. Introduction
Seedlessness is a major citrus breeding objective for the fresh fruit market. Strategies for breeding seedless varieties are based on the association of parthenocarpy and mechanisms that prevent fertilization of the ovules by pollen or results in embryo degeneration. Gametic sterility can result from sterility genes, such as the nucleocytoplasmic male sterility in Satsuma [1] or from ploidy manipulation to create triploid hybrids with unbalanced meiosis [2,3]. Self-incompatibility (SI), the inability for a male and female fertile plant to produce seeds from self-fertilization, is also an efficient way to select for seedless cultivars. In citrus, SI was first described in pummelos [4], but the seedlessness of some small citrus cultivars such as ‘Ellendale’, ‘Fortune’, ‘Nadorcott’, ‘Nova’, and most clementine varieties, if grown in solid blocks, results from the association of parthenocarpy and SI [5,6,7,8].
SI is widespread in angiosperm species. It is an important evolutive reproductive biological feature, favoring cross pollination, preventing inbreeding, and conserving high diversity within natural populations [9]. It is found in many horticultural crops. In legumes, the Solanaceae and Brassicaceae families were important models to decipher the molecular mechanisms of SI [10,11,12]. In fruit tree species, SI has been described in most species including the family Rosaceae (apple, pear, plum, and apricot [13,14,15,16]) and other families such as Malvaceae (cocoa [17]), Oleaceae (olive tree [18]) and Rutaceae (citrus species; Zhang et al. [19] for a review).
In annual crops, SI is considered as an advantage for the production of F1 hybrid seed [20]. As already mentioned for citrus and also described in pineapple [21], when coupled with parthenocarpy, SI enables the producing seedless fruits in mono-varietal context. Conversely, for non-parthenocarpic species or varieties, SI is a constraint, and implies mixing inter-compatible varieties with overlapping flowering periods in the same orchard [22]. SI has also important implications for breeding strategies and schemes and its determinism has been widely studied in several crop families.
Two main forms of SI have been described: sporophytic SI (SSI) and gametophytic SI (GSI). In SSI, the pollen-pistil interaction is determined by the diploid genotype of the parents. In contrast, in GSI, the compatibility phenotype for pollen is directly related to the pollen haplotype. Several incompatibility systems have been reported within the SSI and GSI categories, but only three of them have been characterized at molecular level: the Brassicaceae system for SSI [23], and two for GSI. In the latter, one system was described in Solanaceae, shared with Plantaginaceae and Rosaceae, and the other in Papaveraceae [24,25,26]. The GSI system is controlled by a single polymorphic locus called the S locus that is expressed in several combinations in both the pistil and the pollen grain. In the Solanaceae system, the stylar S locus codes for ribonuclease glycoproteins (S-RNases) that are taken up by the pollen tube. In compatible pollen, S-RNases are degraded, whereas in incompatible pollen, they remain active, causing the degradation of pollen RNA and blocking growth of the pollen tube [27,28,29,30].
Using controlled pollination data, Soost [4,31] proposed a gametophytic SI system for citrus with one self-fertility allele and at least eight S incompatibility alleles that resulted in pollen tube arrest in self-pollinated pistils. More recent pollination studies established that many citrus accessions are self-incompatible [32,33,34,35]. Based on distorted segregation of the isozyme gene Got-3, assumed to be linked to S gene in the progenies, Ngo et al. [35] also estimated the existence of eight alleles for the S gene. Wakana et al. [36] successfully produced S1 seeds homozygous for different S alleles in several monoembryonic SI cultivars, including self-incompatible clementine. Used as male parents, these S homozygotic lines provided basic resources for the analysis of the distribution of SI alleles in the citrus germplasm, particularly for S1 and S2 [34], S4 and S5 [37], S9 and S10 [32], and S3 and S11 [38] alleles. These studies provided important information for the identification of the S-determinants and the study of molecular mechanism underlying the SI response in citrus.
Molecular and transcriptomic studies revealed that different molecules and gene families may be involved in SI mechanisms in citrus. Among the factors involved in the SI mechanism, a role for transglutaminases and polyamines and particularly spermidine was proposed [39,40,41]. Over the last 15 years, comparative transcriptome approaches have been applied to identify the genes and proteins involved in the SI response. Several pairs of self-incompatible and self-compatible (SC) mutants were used for the purpose. For clementine, analyzing the SI ‘Commune’ and the SC ‘Monreal’ cultivars, Distefano et al. [7] demonstrated that the self-compatibility in ‘Monreal’ clementine was due to changes in pistil function but not in pollen function. Caruso et al. [42] identified differentially expressed genes in laser-microdissected stylar canal cells in the SI ‘Commune’ and in the SC mutant ‘Monreal’, particularly three up-regulated Asp-rich protein genes and the DELLA gene located on chromosome 9 of the clementine genome. Recent work revealed potential involvement of S-RNase genes. Indeed, an S-RNase gene (homolog to those of Rosaceae and Solanaceae) has been identified in pummelo, whose product was able to inhibit pollen tube growth [43]. The proposed S-RNase system for GSI in citrus was in agreement with the previous transcriptomic studies of Miao et al. [44] in mandarin and by Zhang et al. [45] in limon. Recently, Liang et al. [46] identified several polymorphic pistil-expressed S-RNases in pummelo and showed that they segregate with S haplotypes. These authors provided strong evidence that the S-RNase based SI system was prevalent in citrus and that S-RNases functioned as pistil S determinants, inhibiting pollen in a S-specific manner. They located the corresponding SI locus that also included F-box genes, at the beginning of the pseudo-chromosome 7 of the clementine reference genome. The involvement of T2/S-RNase in self-incompatibility of citrus was also proposed by Honsho et al. [47] on the basis of transcriptomic, phylogenetic and genetic approaches. Both studies [46,47] analyzed the segregation of markers of the S-RNase genes on controlled progenies and found, for some of them, segregations in agreement with the gametophytic model for self-incompatibility. However no genetic studies based on whole genome segregation analysis definitively validated the location of the SI locus and its unicity. Under the GSI system, in compatible crosses between parents sharing one incompatibility allele, in male gametes, fully skewed segregations are expected at the SI locus, with the rejection of the shared haplotype, and decreasing segregation distortion directly linked to the genetic distance of the markers from the SI locus. Such a skewed segregation pattern associated with a SI locus has been described in Cocoa [26]. In the present work, we provide additional evidence for a gametophytic SI system and its location, based on the analysis of male and female segregation distortions all along the genome in reciprocal crosses between two self-incompatible small citrus cultivars that share one incompatibility allele: ‘Fortune’ mandarin and ‘Ellendale’ tangor. Previous studies of pollination of ‘Ellendale’ and clementine with homozygous lines for the S locus have shown that clementine and ‘Ellendale’ share the same S3–S11 genotype [38]. As ‘Fortune’ is a self-incompatible direct hybrid of clementine [48] it is expected to share one SI allele with ‘Ellendale’. The present study was based on high density genetic mapping established from genotyping by sequencing data (GBS [49]). Local and chromosome haplotyping of the two parents was performed using phase markers and provides new insights into the origin of these two varieties and S haplotype diversity.
2. Materials and Methods
2.1. Plant Material
Two populations of diploid hybrids were created and grown at the Inra-Cirad San Giuliano research station (Corsica, France) from two reciprocal crosses between the diploids ‘Fortune’ mandarin and ‘Ellendale’ tangor, both used as female and male genitors. For a while, ‘Fortune’ mandarin was considered to be a hybrid between clementine and ‘Dancy’ mandarin [50,51]. However, molecular studies [48] suggested that it rather resulted from a cross between clementine and ‘Orlando’ tangelo. ‘Orlando’ being itself a hybrid of ‘Duncan’ grapefruit and ‘Dancy’ mandarin. Based on its phenotype, ‘Ellendale’ is considered as a tangor (C. reticulata Ten. × C. sinensis L. hybrid) but its precise origin remains unknown. Flow cytometry analysis was performed, as described in Aleza et al. [2] to discard triploids resulting from 2n gametes. In the following, ForEl stands for ‘Fortune’ mandarin × ‘Ellendale’ tangor hybrids, and ElFor stands for ‘Ellendale’ tangor × ‘Fortune’ mandarin hybrids.
2.2. Plant Genotyping
A total of 167 diploid mandarin hybrids (79 ForEl and 88 ElFor) and replicates of the two parents were subjected to genotyping by sequencing (GBS). Genomic DNA was isolated using the Plant DNAeasy® kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The concentration of genomic DNA was adjusted to 20 ng/μL, and ApekI GBS libraries were prepared following the protocol described by Eslhire et al. [49]. DNA of each sample (200 ng) was digested with the ApekI enzyme (New England Biolabs, Hitchin, UK). Digestion took place at 75 °C for 2 h and then at 65 °C for 20 min to inactivate the enzyme. The ligation reaction was completed in the same plate as the digestion, again using T4 DNA ligase enzyme (New England Biolabs, Hitchin, UK) at 22 °C for 1 h and the ligase was inactivated prior to pooling the samples by holding it at 65 °C for 20 min. For each library, ligated samples were pooled (i.e., two multiplex libraries of 96 samples) and PCR-amplified in a single tube. Complexity was further reduced using PCR primers with one selective base (A) as described by Sonah et al. [52]. Single-end sequencing was performed on a single lane of an Illumina HiSeq4000 at the Genoscope facilities (Paris, France) with two runs for each library. Keygene N.V. (Keygene, Wageningen, The Netherlands) owns the patents and patent applications protecting its Sequence Based Genotyping technologies.
SNP genotype calling was performed from the DNA sequence reads with the Tassel 4.0 pipeline [53] to identify good quality, unique, sequence reads with barcodes. These sequences were aligned with the C. clementina 1.0 reference genome (available at https://phytozome.jgi.doe.gov, accessed on 21 April 2021) using Bowtie2 v2.2.672. For genotype calling, positions with less than five reads were considered as missing data. Next, polymorphic positions were filtered for diallelic SNPs and minor allele frequency (MAF) over 0.05.
2.3. Linkage Analysis and Genetic Mapping
The two-way pseudo-testcross mapping strategy implemented for genetic mapping from progenies resulting from crosses between two heterozygous parents (Ritter et al., 1990) and used in previous high density mapping studies in citrus [54,55,56,57] was applied to establish ‘Fortune’ and ‘Ellendale’ genetic maps. For each map, SNP markers were selected according to their respective heterozygosity for the mapped parent and homozygosity for the other one. Each set of data for the 167 hybrids was filtered to retain markers and hybrids with less than 15% of missing data.
Linkage analysis and genetic mapping were then performed using JoinMap5 (https://www.kyazma.nl/index.php/JoinMap/; accessed on 21 April 2021). Linkage mapping was performed in the « Hap » option for both ‘Fortune’ mandarin and ‘Ellendale’ tangor. Markers were grouped using the independence LOD score. Phases (coupling and repulsion) of the linked marker loci were automatically detected by the software. Map distances were estimated in centiMorgan (cM) using the regression mapping algorithm. After a first mapping round, singletons, i.e., an individual genotype that suggests recombination with its two flanking markers, were identified. On the high-density map, the probability of having two successive cross-overs within a small genomic area is very low, while genotyping errors strongly affect the estimation of genetic distances that erroneously expand the genetic linkage groups. Therefore, as recommended by Van Os et al. [58], we replaced singletons with missing data using a homemade excel page routine and performed a second mapping round. At the same time, a few individuals displaying an aberrant number of recombinations, set by examining the global recombination distribution, were removed as we considered their genotype calling quality was insufficient. The synteny and collinearity of both ‘Fortune’ and ‘Ellendale’ genetic maps with the reference clementine genome were visualized using Circos [59]; http://circos.ca; accessed on 21 April 2021 in Galaxy [60]. Marey maps were drawn using Excel to visualize changes in the recombination rate along the genome.
2.4. Analysis of Segregation Distortion
The matrix of phased data resulting from each previous genetic map analysis was used to study the skewed segregation all along the genome for each parent, globally and when used as male or female parent. The p-values for the Chi2 test according to a 0.5 theoretical frequency for each allele were computed with Excel and we used the approach proposed by Benjamini and Hochberg [61] to limit the false discovery rate (FDR) in multiple testing; the approach was performed according to the method of Storey [62] with a q value threshold of 0.05. The results were visualized in a Circos plot.
2.5. Haplotype Analysis
SNP phase markers in ‘Fortune’ and ‘Ellendale’ were identified with the CP option of JoinMap 5 using all segregating markers in the ForEl and ElFor progenies and allowing 15% of missing data. Then we selected the set of SNP markers shared with the ones used in the diversity study based on GBS with the same methodology published by Oueslati et al. [63]. The parentage of ‘Fortune’ mandarin was analyzed by examining the compatibility of its haplotypes with different potential parents: clementine, ‘Orlando’ tangelo and ‘Dancy’ mandarin. ‘Ellendale’ tangor haplotypes were also analyzed in relation with those of sweet orange. We took advantage of the haploid sequence of clementine and the parenthood network with the other accession to infer their haplotypes, as described in Amaral et al. [64].
Local haplotyping in the genomic region surrounding the S locus was performed using the same approach for accessions included in the clementine parenthood network. The relationships between haplotypes were then analyzed by neighbor-joining using DARwin software version 6.0 (https://darwin.cirad.fr/; accessed on 21 April 2021). The analyses were based on the Manhattan index:
where i and j are the two individuals, k is the locus, K is the total number of loci and xik is the frequency of the alternative allele at locus k for the individual i.
3. Results
3.1. SNP Calling
Tassel software identified 23,875 polymorphic positions. Among them, we filtered positions where all replicate of the parents were identical, with a least one of the parents heterozygous, and with less than 15% of missing data. This resulted in the selection of 8458 SNPs.
3.2. Genetic Linkage Maps of ‘Fortune’ Mandarin and ‘Ellendale’ Tangor
The SNP matrix containing 167 individuals was filtered for markers heterozygous for ‘Fortune’ mandarin and homozygous for ‘Ellendale’ tangor that had less than 15% of missing data and segregations in agreement with the parental genotypes. Linkage mapping of the ‘Fortune’ mandarin was then performed using a matrix of 2184 segregating and 167 individuals. Five individuals displaying an abnormal number of recombinations during the first mapping round were discarded before the final mapping. A total of 2164 out of the 2184 SNPs were assigned to one of the nine resulting linkage groups (LG) which corresponds to the number of haploid chromosomes in citrus (Table 1; Supplementary Table S1). The number of markers was unequally distributed among the linkage groups. LG8 included only 55 SNPs while 370 SNPs were attributed to LG7. The small number of markers in LG8 is due to high homozygosity of ‘Fortune’ mandarin in a large part of the corresponding chromosome. LG8 displayed the lowest genetic size (75.937 cM). LG3, gathering 352 SNPs, displayed the largest genetic size (276.43 cM) (Table 1). The whole map spanned 1508.4 cM, with an average inter-locus distance of 0.7 cM. 95.1% of SNPs had an inter-locus gap of less than 3 cM and only 0.51% had a gap of more than 10 cM. A total of 1577 markers were located in genes, with 1523 genes marked (Table 1; Table S1).

Table 1.
Summary of ‘Fortune’ mandarin mapping data.
After filtering the markers heterozygous for Ellendale and homozygous for Fortune with less than 15% of missing data and with segregation in agreement with the parental genotypes, 1503 segregating SNPs genotyped in 167 hybrids were used to construct the genetic map of the ‘Ellendale’ tangor. Nine linkage groups including 1467 markers were generated. The number of markers ranged from 59 for LG6 to 268 for LG5 (Table 2; Supplementary Table S2). The total size of the genetic map was 1034.3 cM with an average inter-locus distance of 0.71 cM. The smallest linkage group was LG6 with 90.18 cM, while LG3 was the largest (164.85 cM). The inter-locus gap of 94.68% of the SNPs was less than 3 cM, while the genetic distance was more than 10 cM in only 0.68% of them. A total of 1040 markers were located on genes and 1000 genes had at least one mapped marker (Table 2; Supplementary Table S2).

Table 2.
Summary of ‘Ellendale’ tangor mapping data.
3.3. Synteny and Collinearity with the Reference Genome of Clementine
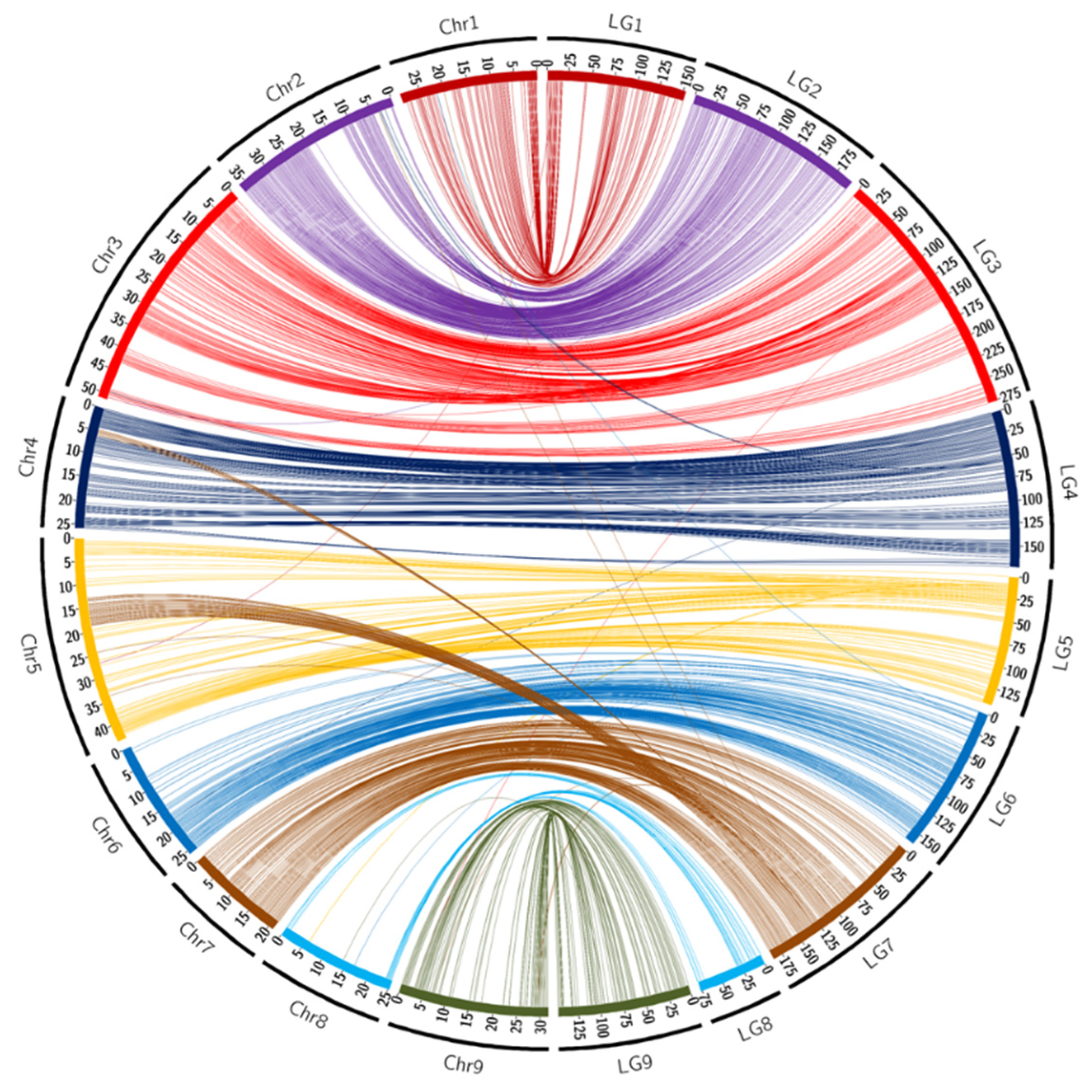
In ‘Fortune’ mandarin, the majority of the linkage groups were composed of SNPs mapped onto syntenic pseudo-chromosomes of the clementine reference genome (Table 1) and the genetic map displayed high global synteny (95.6%). LG7 stood out, with linkage mapping of 12 markers physically located on chromosome 4 and 63 on chromosome 5. The map also counted five unassigned markers that were not previously positioned on the nine pseudo-chromosomes of the clementine reference genome. These five SNPs belong to the same scaffold (Scaff 10), indicating that Scaff 10 may be joined to pseudochromosome 7. The circos representations (Figure 1) and the Marey map (Supplementary Figure S1A) between the genetic positions and physical locations over the clementine genome allowed us to identify a cluster of 78 markers displaying clear incongruency between the genetic map and the physical positions on chromosome 3. This cluster encompasses a genomic region located between 29 Mb and 34 Mb. The other markers display global high collinearity between the ‘Fortune’ genetic map and the Clementine reference genome. The evolution of the recombination rate along the chromosome (Supplementary Figure S1A) is very similar to evolution observed in clementine (Wu et al., 2014), that is directly linked with the density of genes and repeat elements along the genome.
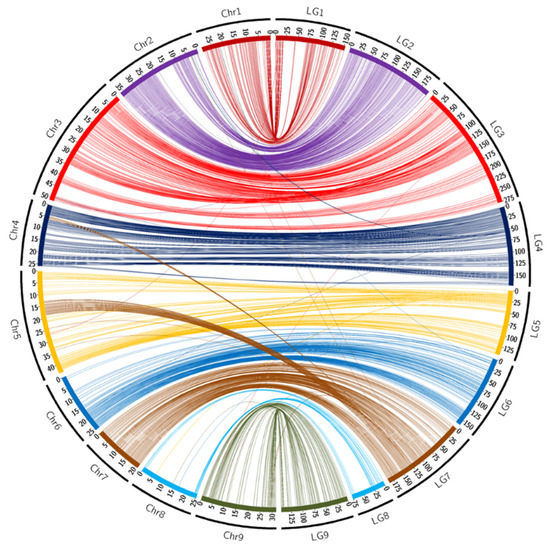
Figure 1.
Links between the position of markers on the ‘Fortune’ genetic map and on the chromosome assembly of clementine genome [65]. Scales are in cM for LGs and in Mb for chromosomes.
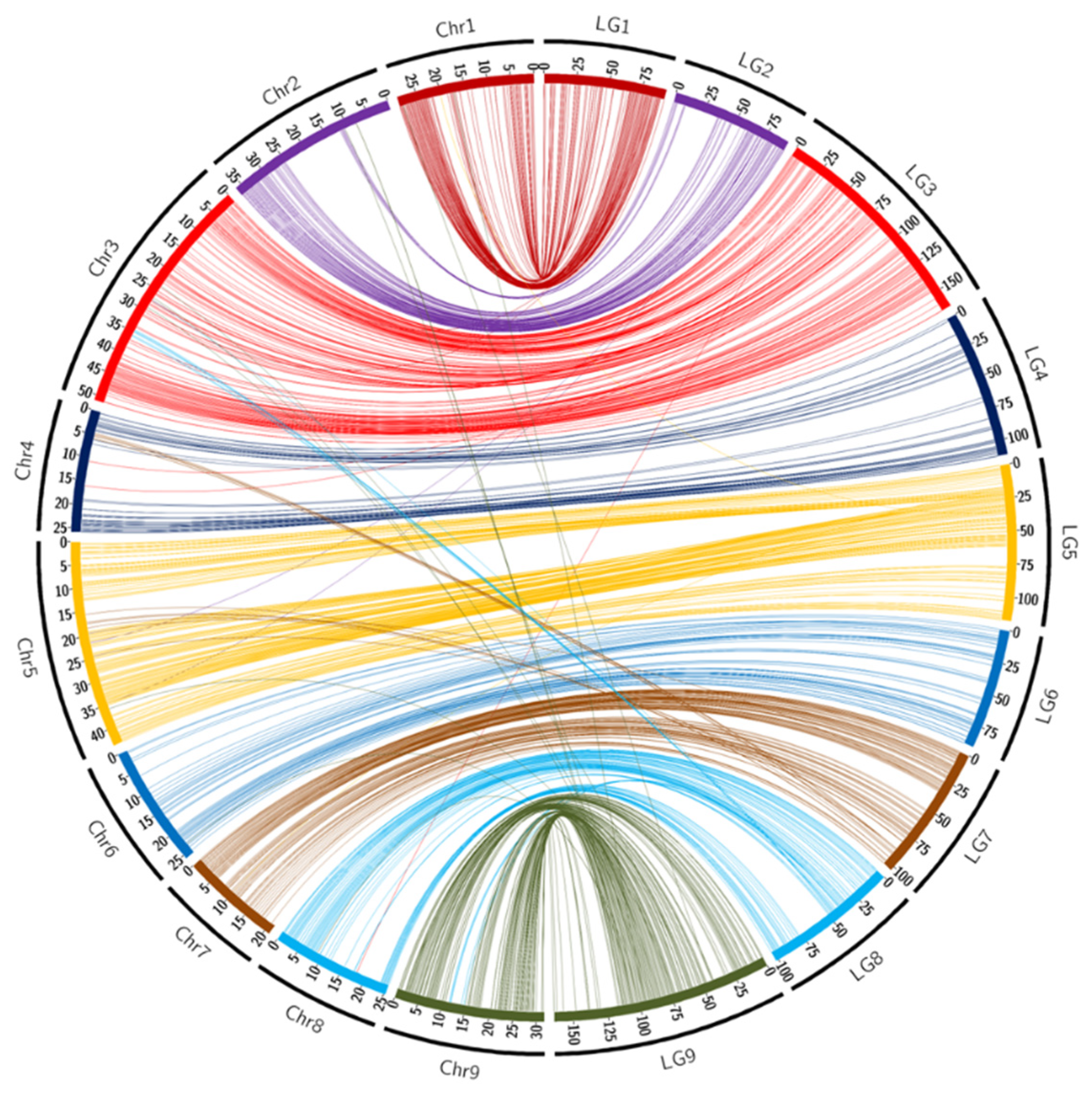
Overall synteny was also high (96.8%) between the Ellendale genetic map and the Clementine reference genome (Table 2 and Figure 2). LG4 and LG6 displayed full synteny with the reference genome. As already observed in the ‘Fortune’ mandarin, some SNPs located on pseudochromosomes 4 and 5 were linked to LG7 (six markers for each pseudo-chromosome). LG8 had more SNPs that were not mapped on the corresponding pseudo-chromosome with respectively eight and seven SNPs (out of a total of 120) located on the physical assembly of pseudo-chromosomes 3 and 9.
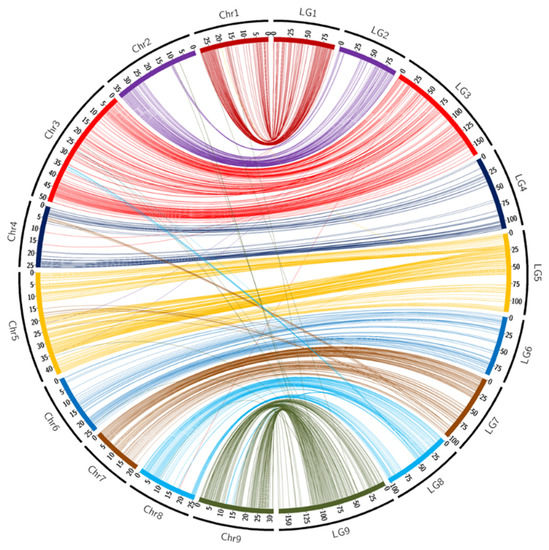
Figure 2.
Links between the position of markers on the ‘Ellendale’ genetic map (LGs) and on the chromosome assembly (Chr) of the clementine genome [65]. Scales are in cM for LGs and in Mb for chromosomes.
Like ‘Fortune mandarin’, the Circos representation (Figure 2) and the Marey map (Supplementary Figure S1B) highlighted a genomic region located between 29 Mb and 33 Mb of chromosome 3 that differed from the genetic positions. The collinearity of the other markers was high. Very few markers presented inversions in the middle and at the end of LG1 and at the beginning of LG6. The evolution of the recombination rate along the chromosome (Supplementary Figure S1B) was very similar to the one observed in ‘Fortune’.
3.4. Segregation Distortion
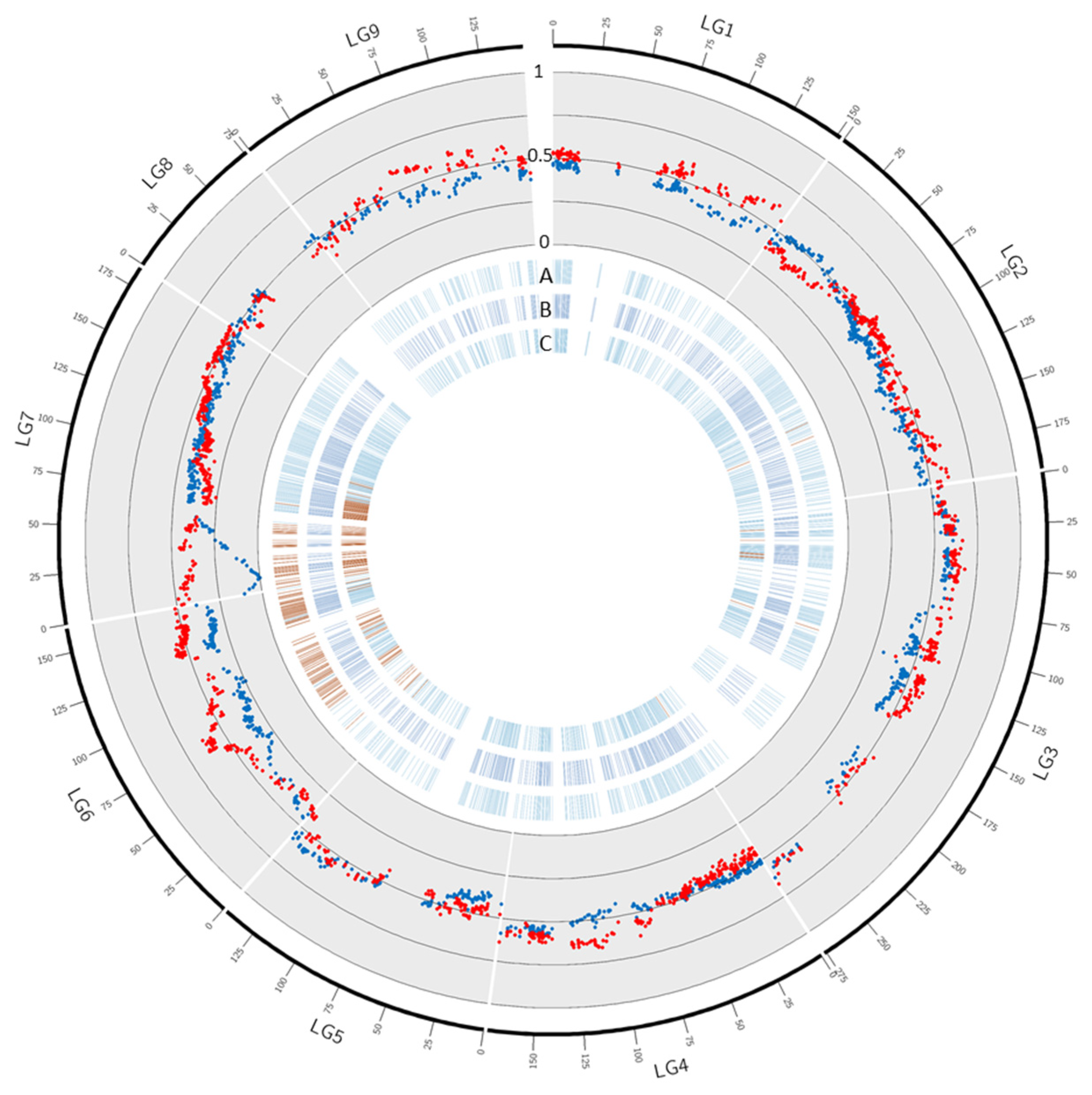
Among the 2164 SNPs assigned on the ‘Fortune’ mandarin genetic map, 202 showed significant segregation distortions according to the X2 test adjusted to the q value for multiple hypothesis testing (Table 3; Supplementary File S1). No significant distortion was found in LG 1, 4, 5, and 8. Distortion concerned only one marker in LG 2 (0.3%) and LG9 (0.9%) and 10 markers (2.8%) in LG3. LG6 and LG7 displayed the highest numbers and rates of skewed markers (69–22.6% and 121–32.7% respectively). Segregation distortion was also investigated in efficient male and female ‘Fortune’ gametes (the ones that contributed to the progenies) (Table 3; Figure 3; Supplementary Table S1). No significant distortion was found in efficient female ‘Fortune’ mandarin gametes while 313 markers displayed significant distortion in efficient male gametes. The skewed markers were concentrated in LG6 (255) and LG7 (54). Even if the number of significant skewed markers was higher in LG6, the level of the distortion was much higher in a cluster of markers at the beginning of LG7. Indeed, in LG7, it reached the maximum segregation distortion value with complete elimination of one of the male alleles for two markers located at 11.4 cM, corresponding to positions 1,296,255 and 1,310,473 on chromosome 7 of the reference clementine genome. The level of distortion segregation decreased, in high correlation with genetic distance on both sides of this genetic position and remained significant from 0 to 55.8 cM.

Table 3.
Number of markers with significant segregation distortions for all efficient male and female gametes of ‘Fortune’ mandarin and ‘Ellendale’ tangor.
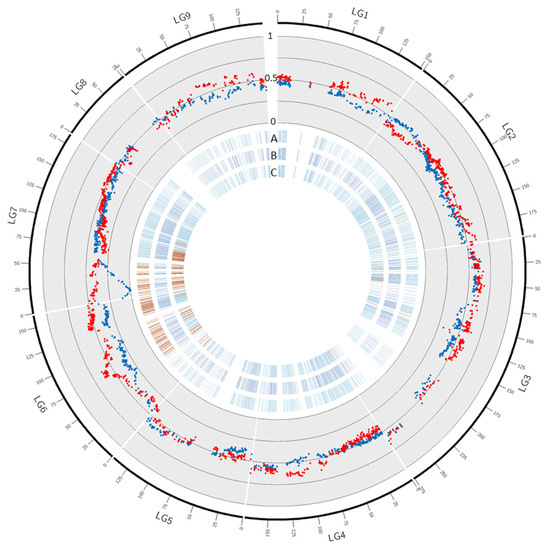
Figure 3.
Segregation distortion in ‘Fortune’. The blue and red dots in the scatter plot represent respectively, male, and female gamete frequency in ‘Fortune’ haplotype 1. A, B, and C: blue and red dashes: markers with non-significant and significant distortion in (A): male gametes, (B): female gametes and (C): all gametes.
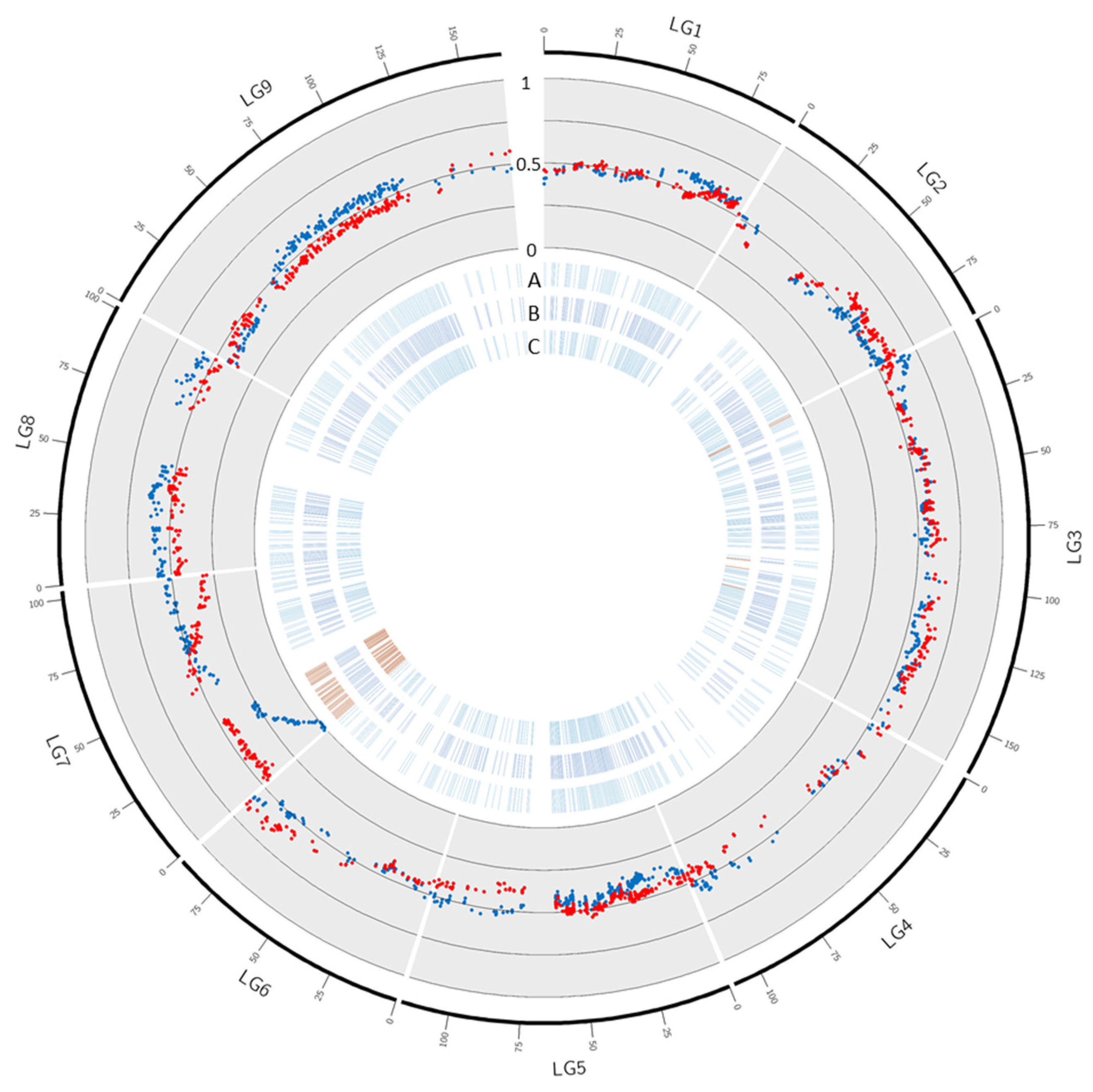
In ‘Ellendale’, 103 out of the 1467 mapped markers displayed a significant deviation from the expected genotypic proportions (Table 3; Figure 4; Supplementary Table S2). No significant distortion was observed in LGs 1, 2, 4, 5, 8, and 9. Only two (3.4%) and eight (3.5%) markers were skewed in LG6 and LG3, respectively. Ninety-three markers (58.9%) were significantly skewed in LG7. No significant distortion was observed in efficient female ‘Ellendale’ tangor gametes while 96 markers displayed skewed segregation in efficient male gametes. Three skewed markers were located in LG3 and the remaining 93 were located in the first part of LG7. The distortions in LG7 reached complete elimination of one allele in 20 markers located between 1.3 and 5.5 cM on the genetic map and 0.083 and 1.549 MB of pseudo-chromosome 7. The level of distortion segregation decreased in high correlation with genetic distance and remained significant from 0 to 34.0 cM (there was then a gap in the genetic map between the marker at 34.0 cM and the following one at 50.9 cM).
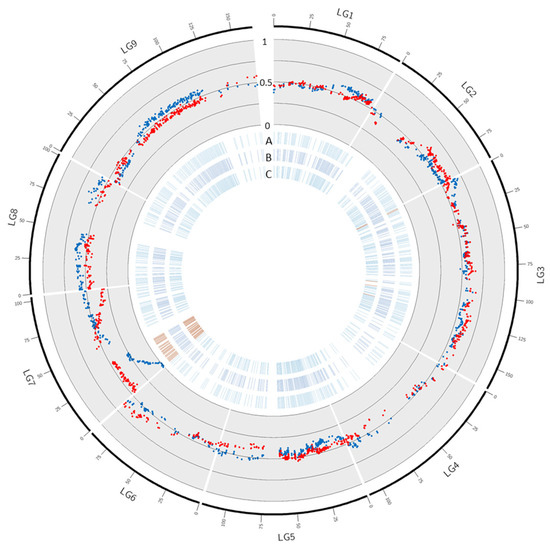
Figure 4.
Segregation distortion in ‘Ellendale’. Blue and red dots in the scatter plot represent respectively male and female gamete frequency in ‘Ellendale’ haplotype 1. A, B, and C: blue and red dashes represent markers with non-significant and significant distortion in (A): male gametes, (B): female gametes and (C): all gametes.
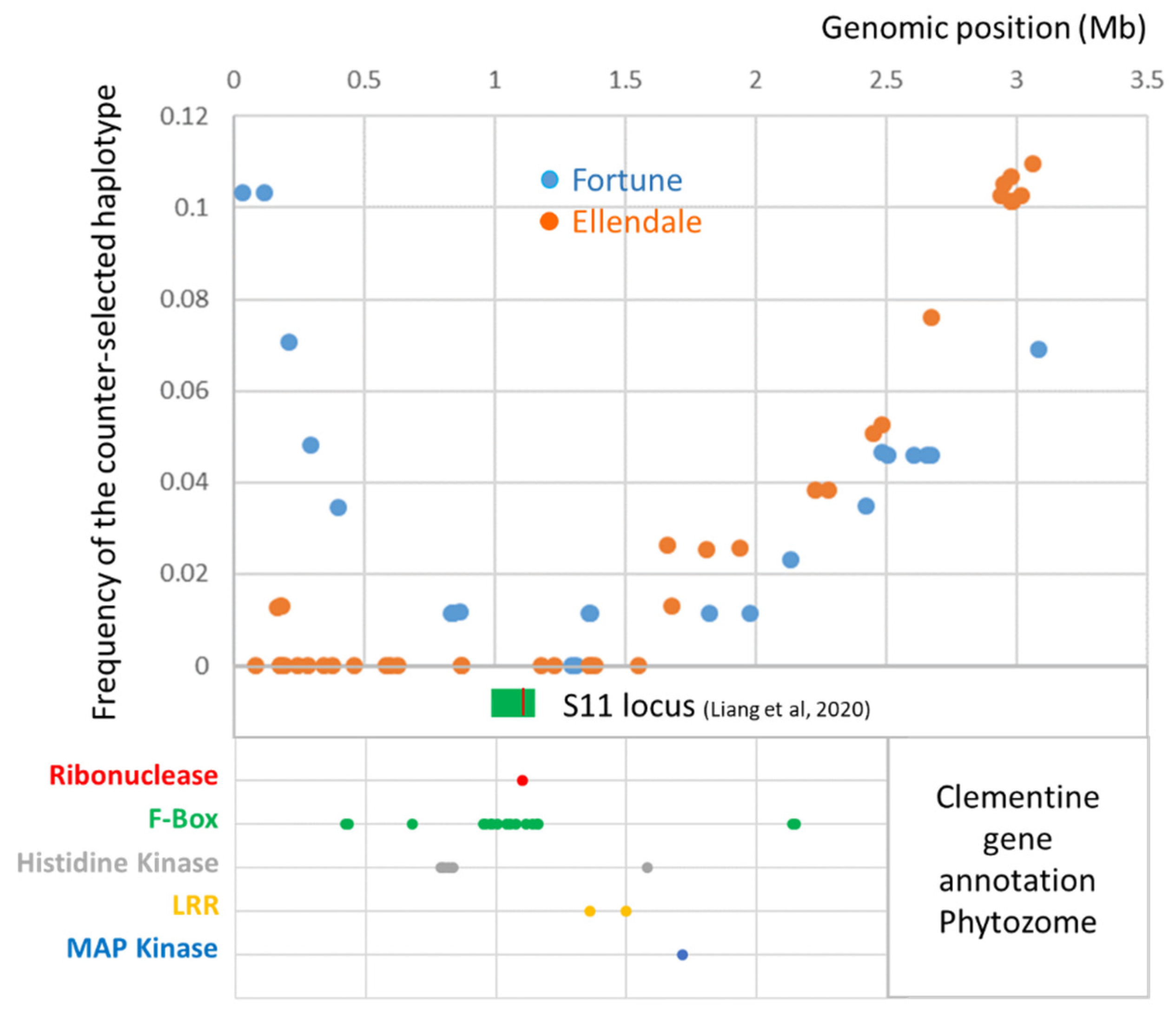
3.5. Gene Annotation in the Fully Skewed Region of Chromosome 7 in Male Parents
Gene annotation of the clementine reference genome (Wu et al. [65]), in the genomic region where the counter-selected haplotype frequency was less than 0.04 in the two parents (0.35–2.2 Mb), revealed 252 genes with 26 genes related to families reported to be involved in SI: 17 F-Box, 5 histidine kinase, 2 Leucine reach repeat (LRR), 1 map kinase and 1 ribonuclease (Figure 5). Six of these F-Box genes and the ribonuclease (Ciclev10027322m.g.) form the S11 locus (position 0.98–1.20 Mb on chromosome 7) identified as being responsible for SI in clementine by Liang et al. [46]. The S11 locus is located where skewed segregation for the two parents is most marked.
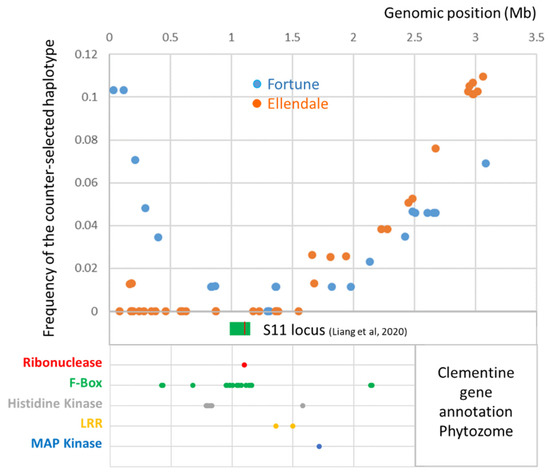
Figure 5.
Gene annotation in the fully skewed genomic region of chromosome 7 for the male parents. The green box indicates the position of the S11 locus and the red line in this box the position of the ribonuclease gene involved in SI, identified by Liang et al. [46].
3.6. Haplotype Structure and Origin of ‘Fortune’ Mandarin and ‘Ellendale’ Tangor
We established the phase between markers and hence the chromosome haplotypes of ‘Fortune’ and ‘Ellendale’ with JoinMap-5 using the CP scheme for two-way pseudo-testcross mapping. We used the 8458 SNPs of the initial vcf file filtered at a rate of 15% for missing data. A total of 8415 SNPs were assigned to nine linkage groups. Among the 8044 syntenic markers, 6693 of these phased markers were the same as the ones in the GBS diversity analysis performed by Oueslati et al. [63] and were used to study the origin of ‘Fortune’ and ‘Ellendale’ varieties. Considering the high collinearity between the genetic maps of ‘Fortune’ and ‘Ellendale’ and the clementine reference genome, we used the positions on the clementine reference genome to analyze the haplotypic similarities between individuals along the genome using windows of 10 successive markers.
In the first step, we identified the two haplotypes of clementine: one inherited from the ‘Commune’ mandarin (mother of clementine) gamete ClHMc and the other from sweet orange (father of clementine) gamete ClHOr that generated the clementine [65,66]. For this purpose, taking advantage of the fact that the clementine reference genome was established using a haploid clementine, we analyzed the compatibility of the haploid sequence and its complementary sequence (to obtain the diploid clementine genotype), using the ‘Commune’ mandarin and the sweet orange genotypic data all along the genome.
The two clementine haplotypes were reconstructed from this information. We then compared the two clementine haplotypes with the two haplotypes deduced from the linkage analysis based on the hybrid progenies between ‘Fortune’ and ‘Ellendale’. The comparison revealed that for each chromosome, haplotype 1 from ‘Fortune’ (FH1) was the one inherited from clementine (Table 4; Supplementary Figure S2A).

Table 4.
Compatibility of each haplotype chromosome of, respectively, ‘Fortune’ (FH1, FH2) and ‘Ellendale’ (EH1, EH2) with the haplotypes of clementine and sweet orange based on the average similarity of ten marker windows (average of the best percentage of similarity per window for each ‘Fortune’ and ‘Ellendale haplotype). Rec FH1: number of recombinations in the FHI haplotype.
The average similarity values of the 10 marker windows along each chromosome ranged from 98.6% for chromosome 6–100% for chromosomes 2 and 8 (with a total average similarity of 99.5%) while the average similarity for the ‘Fortune’ haplotype 2 (FH2) ranged between 37.6% for chromosome 7 and 86.4% for chromosome 8 (global average: 67.9). The analysis of similarities between FH1 and the two clementine haplotypes based on sliding windows of 10 markers (Supplementary Figure S2A) revealed nine recombination events between the ClHMc and ClHOr chromosome (Table 4) during the formation of the gamete that generated the ‘Fortune’ mandarin. Given that for each chromosome, FH1 was inherited from clementine (the female parent of ‘Fortune’), we tested the compatibility of FH2 with three potential male parents: ‘Orlando’ tangelo, grapefruit, and ‘Dancy’ mandarin (Table 5). For ‘Orlando’ tangelo the average compatibility over the whole genome was very high (98.43%) with little variation between chromosomes (97.1% on chromosome 7 to 99.0% on chromosome 4). The average values were lower for grapefruit and ‘Dancy’ mandarin (87.4% and 75.8%, respectively) with very low values for some chromosomes (55.6% for grapefruit chromosome 9 and 49.4% for ‘Dancy’ chromosome 7). Considering ‘Orlando’ as the male parent of ‘Fortune’, we identified 14 recombination events between the ‘Dancy’ mandarin and grapefruit genomes that constituted ‘Orlando’ tangelo during the genesis of the male gamete that generated ‘Fortune’. Supplementary Figure S3 give a schematic diagram of the parentage of ‘Fortune’ mandarin based on this analysis and locates the different recombination points that took place on the clementine and tangelo gametes.

Table 5.
Compatibility of the FH2 haplotype with three potential parents of ‘Fortune’.
We also tested ‘Ellendale’ haplotype inheritance from sweet orange (Table 4). One of the haplotypes considered for sweet orange (OrHCl) was the one deduced previously from the identification of the clementine haplotype (ClHOr) originating from sweet orange and the secondone (OrH2) was complementary to obtain the diploid sweet orange genotype. We then analyzed the similarity between the two haplotypes of sweet orange and the two for Ellendale (EH1 and EH2) all along the genome (Supplementary Figure S2B). On each chromosome, one of the ‘Ellendale’ haplotypes displayed high local similarity with at least one sweet orange haplotype. Indeed, the average similarity value of the set of 10 marker windows along each chromosome ranged between 97.2 and 99.7% for EH2 on chromosomes 1–6 and 9 and between 97.5 and 99.5% for EH1 on chromosomes 7 and 8, respectively. It is therefore highly probable that ‘Ellendale’ is a direct hybrid of sweet orange. Considering the clear difference in similarity for EH1 and EH2 haplotypes with sweet orange on each chromosome, we can assign the ‘Ellendale’ haplotype inherited from sweet orange: EH2 for chromosomes 1–6 and 9 and EH1 for chromosome 7 and 8.
3.7. Haplotypic Structure around the SI Locus
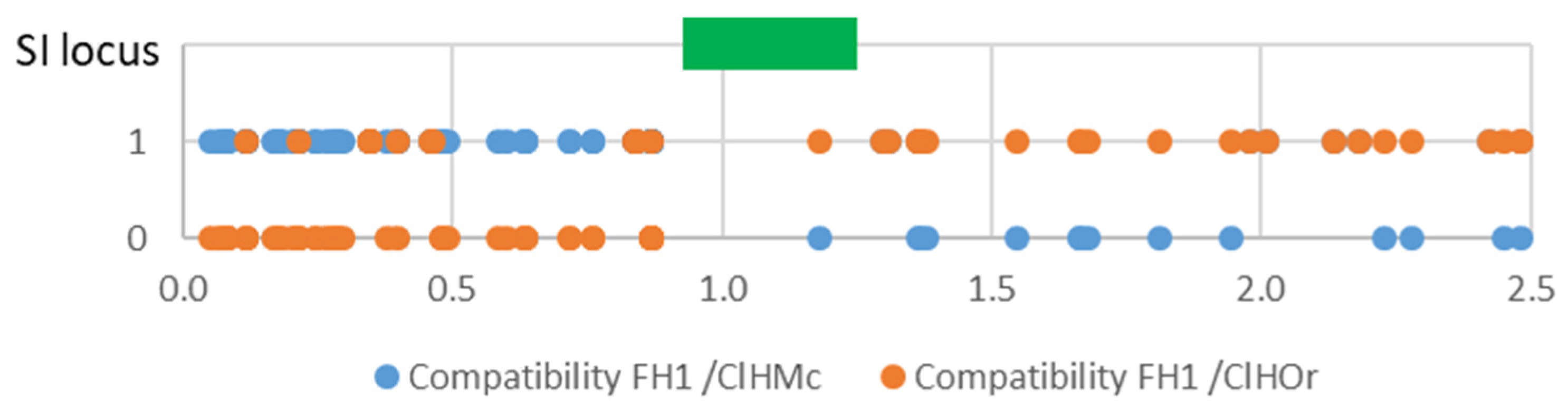
A more detailed analysis of the area surrounding the SI region (as defined by Liang et al. 2020), revealed that a recombination occurred during clementine meiosis in the ‘Fortune’ haplotype originating from Clementine (FH1). Indeed (Figure 6) FH1 conforms well with the clementine haplotype inherited from ‘Commune’ mandarin (ClHMc) from the start of the chromosome up to position 871,872. Then, from 1,180,894 to at least 2.5 Mb, it conforms with the sweet orange haplotype of clementine (ClHOr). Data for the S07_1180894 marker indicate that, at this position, the FH1 haplotype was inherited from its sweet-orange grandfather. Unfortunately, only the S07_1180894 marker provided information within the SI region. Accordingly, the recombination should have occurred before or within the SI locus.
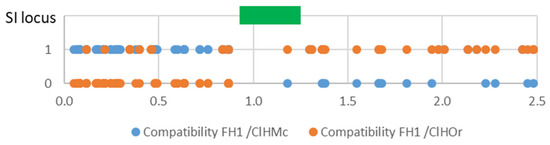
Figure 6.
Compatibility of the FH1 haplotype of ‘Fortune’ mandarin with the mandarin haplotype (ClHMc) and sweet orange haplotype (ClHOr) of clementine for SNP markers near the SI locus (green rectangle); x axis: position in the chromosome 7 in Mb.
Analysis of the genotype of the ElFor segregating progeny for five loci heterozygous in ‘Fortune’ and homozygous in ‘Ellendale’ surrounding the SI locus (from position 869,201 to 1,369,079) revealed that the FH1 haplotype inherited from clementine was the one strongly counter-selected (Supplementary Table S3A) in this region. For ‘Ellendale’, 14 markers between position 601,330 and 1,381,530 were available for a similar analysis in ForEl progenies and revealed that the EH1 haplotype inherited from sweet orange was the one strongly counter-selected in the SI area (Supplementary Table S3B).
According to Kim et al. [38], clementine and ‘Ellendale’ share the same S3–S11 genotype at the SI locus and we therefore expected that ‘Ellendale’ and ‘Fortune’ would share one of the clementine alleles and that this common allele would be counter-selected. We had previously identified the counter-selected haplotypes in ‘Fortune’ and ‘Ellendale’ pollen as respectively FH1 (inherited from clementine) and EH1 (inherited from sweet orange). These results confirmed our hypothesis and suggest that ‘Fortune’ possesses the functional SI alleles of sweet orange, inherited through clementine, and shared with ‘Ellendale’. This in turn implies that the recombination in the clementine gamete that produced ‘Fortune’ occurred before the functional SI genes with a switch from ‘Commune’ mandarin haplotype, at the beginning of the chromosome, to the sweet orange haplotype.
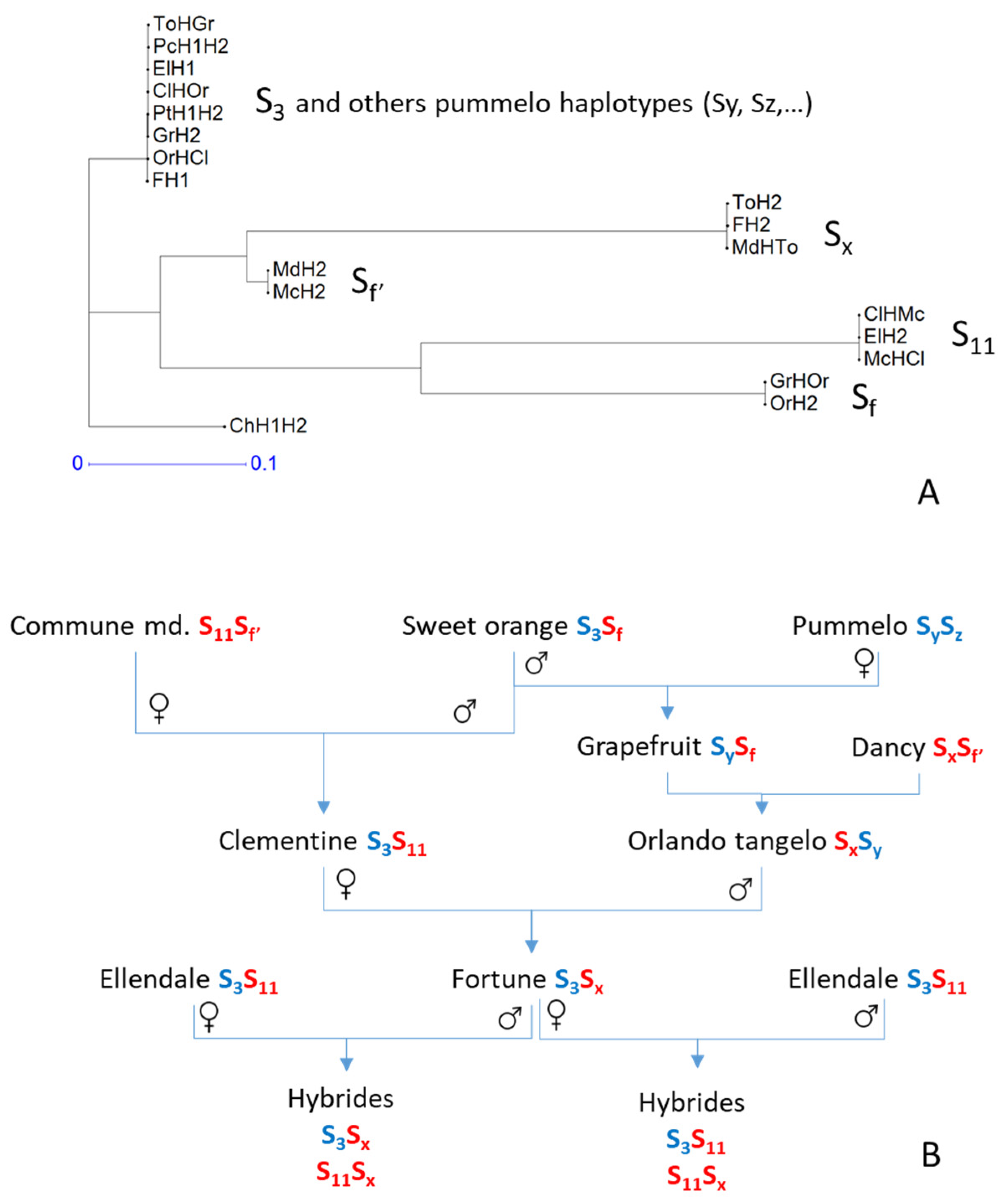
To check this hypothesis, we performed neighbor-joining analyses using two sets of markers located in the vicinity of the SI locus; one with markers in the area where clementine’s contribution to ‘Fortune’ comes from the ‘Commune’ mandarin (position 601,330–869,997; with 13 markers; Supplementary Figure S4) and a second with markers where the clementine haplotype of ‘Fortune’ (FH1) concerned was inherited from sweet orange (position 1,180,894–1,381,530; with eight markers; Figure 7A). We included in the analysis the inferred haplotypes of ‘Fortune’, ‘Ellendale’, ‘Nules’ clementine, ‘Commune’ mandarin and sweet orange. We expanded the inference of haplotype genotypes in this genomic region to grapefruit, ‘Orlando’ tangelo, and ‘Dancy’ mandarin, using the previously inferred haplotypes for sweet orange and the known parental relationships: grapefruit = sweet orange x pummelo and ‘Orlando’ tangelo = ‘Dancy’ mandarin x grapefruit. We also added the haplotypes of ‘Hupang’ citron, ‘Chandler’ and ‘Timor’ pummelos taking advantage of the full homozygosity of these varieties for the markers concerned.
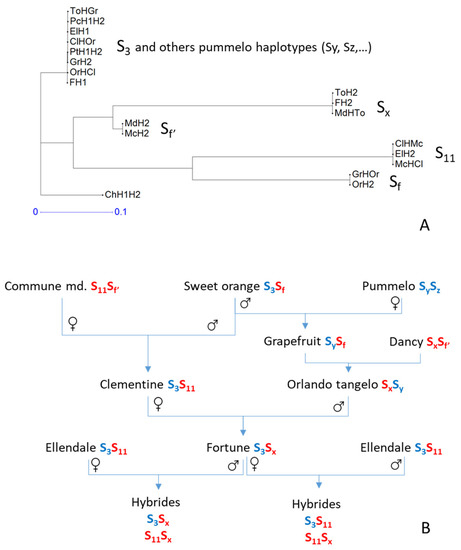
Figure 7.
Analysis of potential SI haplotypes according to the flanking sequences of the SI locus and published SI alleles from cross compatibility analysis (Kim et al., 2020) [38]. (A): Neighbor-joining analysis of haplotypes for markers located after the recombination event in the ‘Fortune’ FH1 maternal gamete; (B): proposed inheritance of the SI alleles in the related varieties and hybrids analyzed. Blue represents C. maxima and red represents C. reticulata haplotype ancestry, according to Oueslati et al. (2017).
If we consider the flanking haplotypes to be informative (due to linkage disequilibrium) for the SI locus haplotypes, it appears that the similarity of the FH1 haplotype with the Ellendale haplotypes before the cross-over (inherited from ClHMc) is not compatible with the observed rejection of FH1 and EH1 in the SI region for the reciprocal crosses (implying identity of FH1 and EH1 for the SI locus). Indeed, before the CO involved in the genesis of the FH1 gamete, FH1 is identical to ELH2 (and identical to ClHMc = McHCl), while ELH1 is shared with OrHcl = ClHor, the two Chandler pummelo haplotypes and one haplotype of Orlando (ToHGr) and the ‘Chandler’ pummelo haplotypes (Supplementary Figure S4).
Conversely, in the region after the recombination identified in the FH1 haplotype (Figure 7A), the identities of the ‘Fortune’ and ‘Ellendale’ haplotypes agree with what would be expected for the SI locus according to ELH1 and FH1 rejection. ElH1 and FH1 are identical and logically identical to the haplotype of clementine inherited from sweet orange (ClHOr = OrHCl). The same genotype for the concerned markers is also shared with the ‘Chandler’ and ‘Timor’ pummelo haplotype and one grapefruit haplotype (GrH2). The FH2 haplotype inherited from ‘Orlando’ tangelo (ToH2) is the haplotype coming from the ‘Dancy’ mandarin (MdHTo) while ElH2 is identical to McHCl (=ClHMc) for the considered markers. Considering that the haplotype information in this part of the genome is indicative of the one for the SI locus for related varieties, we schematized the SI allele inheritance along related varieties (Figure 7B) according to the nomenclature of Kim et al. [38] for clementine and ‘Ellendale’. We added the information on the phylogenomic structure in the area surrounding the SI locus resulting from the analysis performed by Oueslati et al. [63]. The SI allele shared by ELH1 and FH1 should then be the S3 allele identified by Kim et al. [38]. Furthermore, the marker haplotypes suggest that the origin of the compatibility allele Sf’ found in ‘Commune’ and ‘Dancy’ mandarin differs from that of the Sf allele in sweet orange. Both Sf and Sf’ should have originated in the C. reticulata gene pool.
4. Discussion
4.1. High Density Genetic Maps of ‘Fortune’ Mandarin and Ellendale ‘Tangor’ Were Produced
‘Fortune’ mandarin and ‘Ellendale’ tangor are important progenitors for mandarin breeding. It was therefore essential to establish molecular resources such as saturated genetic maps to optimize their exploitation in breeding schemes. Two genetic maps of ‘Fortune’ mandarin were previously published. The first one constructed using a reciprocal cross between ‘Fortune’ and ‘Chandler’ pummelo spanned 577 cM with 95 markers, mostly SSRs, defined in 13 linkage groups [67]. More recently, another ‘Fortune’ genetic map was constructed from an F1 population derived from ‘Fortune’ x ‘Murcott’ mandarin [68]. The map spanned 681.07 cM and consists of 189 SNP markers distributed along nine linkage groups. In the present study, a high-density genetic map of ‘Fortune’ mandarin was built for the first time. It consists of 2164 SNP markers spread among nine linkage groups, corresponding to the nine citrus chromosomes, with a total size of 1508.4 cM. All chromosomes except chromosome 8 were regularly covered. The very partial coverage of chromosome 8 is due to high homozygosity resulting from the inheritance of the same haplotype region of sweet orange genome from the two parents of ‘Fortune’ mandarin. Indeed, both parents share parentage with sweet orange: ‘clementine’ = mandarin × sweet orange and ‘Orlando’ tangelo = (mandarin × (pummelo × sweet orange). No ‘Ellendale’ tangor genetic map has been published to date. The one we implemented includes 1467 SNPs defined in nine linkage groups. It spans a total of 1034.3 cM. Most chromosomes display good and homogeneous coverage. However, markers are lacking in the first part of chromosome 2 and the middle of chromosome 4. These gaps result from high homozygosity probably due to inbreeding in the origin of Ellendale.
Synteny is high and the linear orders of the markers are highly conserved in the two genetic maps and the clementine reference genome [65]. This is consistent with previous studies concluding on high synteny and collinearity between Citrus species [54,67,68] and even between Citrus species and Poncirus trifoliata [57,69,70]. However, a few discrepancies were observed between our two genetic maps and the clementine reference genome. On LG7, both ‘Fortune’ and ‘Ellendale’ displayed two sets of SNP markers located on chromosomes 4 and 5 of the clementine reference genome. Similar results were previously reported in the ‘Fortune’ genetic map [68] but also on sweet orange and trifoliate orange genetic maps [57,70]. The analysis of collinearity between LG3 and chromosome 3 evidenced a misplaced and probably inverted genomic region, particularly visible in ‘Fortune’, located between 29 and 34 Mb. The same genomic area was also identified as misplaced in the high-density genetic maps of sweet orange and trifoliate orange [57] and even in the reference genetic map of Clementine [54]. It is therefore probable that most of the apparent non-syntenic or non-colinear markers are rather due to minor errors in the clementine genome assembly than to real structural variations between ‘Fortune’ or ‘Ellendale’ and clementine.
4.2. The Origins of ‘Fortune’ Mandarin and ‘Ellendale’ Tangor Were Assessed through Analysis of Chromosome Haplotypes
‘Fortune’ mandarin is a late high-quality mandarin widely used as the female parent in mandarin breeding programs [2] due to its self-incompatibility and non-apomictic reproductive behavior. It was presented by its plant breeders as a hybrid between clementine and ‘Dancy’ mandarin [50,51]. However genotyping data with 17 SSR markers discarded ‘Dancy’ as direct parent and suggested that ‘Orlando’ tangelo, a hybrid between ‘Duncan’ tangelo and ‘Dancy’ mandarin, was the male parent of ‘Fortune’ [48]. In our study, we established the chromosome haplotypes of ‘Fortune’ from phased data of 6693 markers, and for each chromosome, we identified the ‘Fortune’ haplotype inherited from clementine and checked the compatibility of the remaining haplotypes with three potential male parents: ‘Orlando’ tangelo, grapefruit, and ‘Dancy’ mandarin. ‘Orlando’ tangelo was validated as male parent with 98.5% compatibility over the whole genome. The contributions of the genomes of the four grandparents of ‘Fortune (‘Commune’ mandarin and sweet orange inherited from clementine; grapefruit and ‘Dancy’ mandarin inherited from ‘Orlando’ tangelo) were analyzed all along the genome and revealed respectively, nine and 14 recombination events in the clementine and the ‘Orlando’ tangelo gametes that generated ‘Fortune’ mandarin. Interestingly, one of the cross-over in the clementine gamete occurred near the SI locus identified by Liang et al. [46] at the beginning of chromosome 7.
The ‘Ellendale’ variety originated in Queensland as a chance seedling at the end of the 19th century and became an important variety in Australia and a standard parent for mandarin breeding due to its self-incompatibility and non-apomictic reproductive behavior. Bowman [71] considered it a natural tangor (mandarin x sweet orange hybrid) based on its fruit attributes. However, until now no concrete proof has been provided for this origin. Comparison of the chromosome haplotypes of ‘Ellendale’ tangor and sweet orange allowed us to identify, for each chromosome, one of the two ‘Ellendale’ haplotypes that fit the sweet orange ones and the potential breaking points between the two sweet orange haplotypes needed to reconstitute the ‘Ellendale’ haplotype. Overall compatibility was 98.2% and we can therefore assume that sweet orange is one of the direct parents of ‘Ellendale’. The identification of the second parent will need additional studies based on high throughput genotyping of mandarins and mandarin hybrid germplasm.
4.3. Segregation Distortion in the Male Parent Revealed a Genomic Region Involved in Self-Incompatibility
Significant segregation distortions were observed in 9.3% and 7.0% of the markers on the ‘Fortune’ and ‘Ellendale’ genetic maps, respectively. LG6 and LG7 had the highest percentages of distorted molecular markers in ‘Fortune’, while in ‘Ellendale’, distortion mostly concerned LG7. Interestingly, no skewed segregation was found to be significant in female gametes whereas skewed segregation reached 14.4% and 6.5% in male gametes of ‘Fortune’ and ‘Ellendale’, respectively. In citrus, segregation distortion has been described in many previous mapping studies and male parent markers have often been reported to display higher distortion than female ones [54,67,68,72], probably due to pollen competition [54]. However, none of the previous studies evidenced complete counter-selection of one male gamete allele, as would be expected for markers located near the self-incompatibility locus in the GSI system. In the present study, such a situation was observed in both ‘Fortune’ and ‘Ellendale’, for markers located at the beginning of LG7, whereas no significant distortion was observed in female gametes. Both ‘Fortune’ and ‘Ellendale’ are self-incompatibles with one assumed SI allele in common and this area of LG7 is the only area in the whole genome where such a distortion pattern was observed on the saturated genetic maps. Therefore, it is highly probable that the distortion at the beginning of LG7 reveals the presence of a major pollen gene for self-incompatibility. The annotation of the clementine genome revealed the presence of 17 F-box genes and one ribonuclease (Ciclev10027322m.g.) gene classically involved in gametophytic SI systems. Our observations are consistent with previous description for several SI varieties of pollen tube rejection after growing through the top one-third of the style, indicative of gametophytic rather than sporophytic control [7,33,43]. Our results are also in agreement with the differential expression of several F-box genes between ‘Wuzishatangju’ (SI) and ‘Shatangju’ (SC) mandarin pollens observed by Miao et al. [44] and the identification of a S-RNase gene implied in pummelo SI [43]. Above all, our results concerning skewed segregation are in full agreement with the conclusions of Liang et al. [46]. Indeed, these authors identified a SI locus for GSI system (including six of the F-Box genes and the ribonuclease we identified in the clementine annotation) located between 0.98 and 1.20 Mb of chromosome 7 of the clementine reference genome, where the skewed segregations were most marked for the two male parents of our progenies.
Moreover, the SI genotypes inferred from the haplotypes of the surrounding region of the SI locus for ‘Fortune’, ‘Ellendale’ and their progenitors provided a logical pattern, in agreement with SI phenotypes (sweet orange, grapefruit, ‘Commune’ and ‘Dancy’ mandarin being self-compatible and clementine, ‘Orlando’ tangelo ‘Fortune’ and ‘Ellendale’ self-incompatible) as well as with the genotypes at SI locus for ‘Ellendale’ and clementine proposed by Kim et al. [38]. Considering the flanking sequence, the S11 haplotype appears to have a C. reticulata origin according to the phylogenomic study of Oueslati et al. [63] and the Sf’ self-compatible allele shared by ‘Commune’ and ‘Dancy’ mandarin could have a different origin from the one identified by Liang et al. [46] in sweet oranges.
The conclusion concerning a gametophytic SI system in citrus with the SI locus located at the beginning of chromosome 7 of the clementine reference genome proposed by Liang et al. [46] is therefore strongly confirmed by our study. However, the influence of environmental conditions on pollen–pistil interactions has already been documented [41,73,74] and different transglutaminase features and polyamine pattern were recently described depending on the prevailing temperature during pollination [41]. Additional studies are needed to gain a full understanding of the pollen-pistil interaction in different citrus species under different environments.
5. Conclusions
Two high-density genetic maps of ‘Fortune’ mandarin and ‘Ellendale’ tangor were constructed, for the first time, thanks to GBS analysis of two populations resulting from reciprocal crosses. These two maps consisted, respectively, of 2164 and 1467 markers, grouped in nine linkage groups corresponding to the nine pseudo-chromosomes of the clementine reference genome. These two genetic maps were characterized by high synteny and collinearity compared to the clementine reference genome. The inference of ‘Fortune’ and ‘Ellendale’ chromosomal haplotypes based on phase marker information, and their comparison with genotypic and haplotypic data of potential parents allowed us to decipher their origins. ‘Fortune’ mandarin results from clementine x ‘Orlando’ tangelo hybridization while ‘Ellendale’ tangor has sweet orange as a direct parent. The analysis of skewed segregation of male and female parents revealed a complete counter-selection of one haplotype for each male parent in the same region at the beginning of chromosome 7. These skewed segregations concerned a shared haplotypic region that includes a SI candidate locus for a gametophytic S-RNase system, recently identified by deep genomic analysis. The S alleles deduced for ‘Fortune’ mandarin, ‘Ellendale’ and their progenitors, from flanking haplotypic sequences, are consistent with their phenotypes for self-incompatibility. The new high-density genetic maps for two non-apomictic and self-incompatible varieties and the confirmation of a gametophytic S-RNase system, with the SI locus located at the beginning of chromosome 7 of the clementine reference genome, pave the way for more efficient use of self-incompatibility in breeding projects aimed at creating new seedless mandarin cultivars at diploid level.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/agriculture11050379/s1: Figure S1: Marey map plot of the nine linkage groups of ‘Fortune’ mandarin (A) and ‘Ellendale’ tangor (B) compared with the clementine reference genome (Wu et al., 2014), Figure S2: Similarity of haplotypes along the genome; (A): ‘Fortune’ mandarin with clementine; (B): ‘Ellendale’ tangor with sweet orange, Figure S3: Parentage of ‘Fortune’ mandarin and contribution of its grandparent genomes ‘Commune’ mandarin, sweet orange, ‘Dancy’ mandarin and grapefruit) to its genomic structure, Figure S4: Analysis of potential SI haplotypes according to the flanking sequences of the SI locus; Neighbor-joining analysis of haplotypes for markers located before the recombination event in the ‘Fortune’ FH1 maternal gamete, Table S1: Detail of the ‘Fortune’ genetic map including information on physical position of the markers (clementine reference genome), reference and alternative alleles, genetic position, segregation distortions and test of significance for all gametes, male gametes and female gametes, and the gene on which the marker is located (if any), Table S2: Detail of the ‘Ellendale’ genetic map including information on physical position of the markers (clementine reference genome), reference and alternative alleles, genetic position, segregation distortions, and test of significance for all gametes, male gametes and female gametes, and the gene on which the marker is located (if any), Table S3: Genotypic frequencies of the ‘Ellendale’ × ‘Fortune’ (EllFor) and ‘Fortune x Ellendale’ (ForEll) hybrids in the vicinity of the SI locus. (A): genotypic frequencies of ‘EllFor’ progenies for markers homozygous in ‘Ellendale’ and heterozygous in ‘Fortune’; (B): genotypic frequencies of ‘ForEll’ progenies for markers homozygous in ‘Fortune’ and heterozygous in ‘Ellendale’.
Author Contributions
Conceptualization, P.O. and Y.F.; methodology, P.M., C.C. and A.P.; formal analysis, P.O. and D.A.; investigation, P.O., D.A. and J.-C.E.; data curation, G.C.; writing—original draft preparation, P.O.; writing—review and editing, Y.F.; project administration, Y.F.; funding acquisition, Y.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work received financial support from the European Regional Development Fund under the framework PO FEDER-FSE Corse 2014–2020 number 247SAEUFEDER1A, project called Innov’Agrumes (ARR-18/517 CE, synergie number: CO 0009083). GBS analyses were funded by the project France Genomique “Dynamo”. We thank also the Collectivité de Corse for the grant of DA (number ARR-15.036680.SR).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Illumina Hiseq4000 sequencing raw data are available in the NCBI SRA (Sequence Read Archive), under the BioProject number PRJNA705866.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Goto, S.; Yoshioka, T.; Ohta, S.; Kita, M.; Hamada, H.; Shimizu, T. QTL Mapping of Male Sterility and Transmission Pattern in Progeny of Satsuma Mandarin. PLoS ONE 2018, 13, e0200844. [Google Scholar] [CrossRef] [PubMed]
- Aleza, P.; Juarez, J.; Cuenca, J.; Ollitrault, P.; Navarro, L. Recovery of Citrus Triploid Hybrids by Embryo Rescue and Flow Cytometry from 2x x 2x Sexual Hybridisation and Its Application to Extensive Breeding Programs. Plant Cell Rep. 2010, 29, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.; Smith, M.W.; Froelicher, Y.; Russo, G.; Gmitter, F.G. Chapter 7—Traditional breeding. In The Genus Citrus; Talon, M., Caruso, M., Gmitter, F.G., Eds.; Woodhead Publishing: Cambridge, UK, 2020; pp. 129–148. ISBN 978-0-12-812163-4. [Google Scholar]
- Soost, R.K. The Incompatibility Gene System in Citrus. Proc. First Int. Citrus Symp. 1968, 1, 189–190. [Google Scholar]
- Vardi, A.; Neumann, H.; Frydman-Shani, A.; Yaniv, Y.; Spiegel-Roy, P. Tentative Model on the Inheritance of Juvenility, Self-Incompatibility and Parthenocarpy. Acta Hortic. 2000, 199–206. [Google Scholar] [CrossRef]
- Raveh, E.; Goldenberg, L.; Porat, R.; Carmi, N.; Gentile, A.; La Malfa, S. Conventional Breeding of Cultivated Citrus Varieties. In The Citrus Genome; Gentile, A., La Malfa, S., Deng, Z., Eds.; Compendium of Plant Genomes; Springer International Publishing: Cham, Switzerland, 2020; pp. 33–48. ISBN 978-3-030-15308-3. [Google Scholar]
- Distefano, G.; Casas, G.L.; Malfa, S.L.; Gentile, A.; Tribulato, E.; Herrero, M. Pollen Tube Behavior in Different Mandarin Hybrids. J. Am. Soc. Hortic. Sci. 2009, 134, 583–588. [Google Scholar] [CrossRef]
- Gambetta, G.; Gravina, A.; Fasiolo, C.; Fornero, C.; Galiger, S.; Inzaurralde, C.; Rey, F. Self-Incompatibility, Parthenocarpy and Reduction of Seed Presence in ‘Afourer’ Mandarin. Sci. Hortic. 2013, 164, 183–188. [Google Scholar] [CrossRef]
- Ferrer, M.M.; Good, S.V. Self-Sterility in Flowering Plants: Preventing Self-Fertilization Increases Family Diversification Rates. Ann. Bot. 2012, 110, 535–553. [Google Scholar] [CrossRef]
- McClure, B. S-RNase and SLF Determine S-Haplotype–Specific Pollen Recognition and Rejection. Plant Cell 2004, 16, 2840–2847. [Google Scholar] [CrossRef]
- Yamamoto, M.; Nishio, T. Commonalities and Differences between Brassica and Arabidopsis Self-Incompatibility. Hortic. Res. 2014, 1, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, N.; Singh, S. Progress on Deciphering the Molecular Aspects of Cell-to-Cell Communication in Brassica Self-Incompatibility Response. 3 Biotech 2018, 8, 347. [Google Scholar] [CrossRef]
- Maliepaard, C.; Alston, F.H.; van Arkel, G.; Brown, L.M.; Chevreau, E.; Dunemann, F.; Evans, K.M.; Gardiner, S.; Guilford, P.; van Heusden, A.W.; et al. Aligning Male and Female Linkage Maps of Apple (Malus Pumila mill.) Using Multi-Allelic Markers. Theor. Appl. Genet. 1998, 97, 60–73. [Google Scholar] [CrossRef]
- Claessen, H.; Keulemans, W.; Van de Poel, B.; De Storme, N. Finding a Compatible Partner: Self-Incompatibility in European Pear (Pyrus Communis); Molecular Control, Genetic Determination, and Impact on Fertilization and Fruit Set. Front. Plant Sci. 2019, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, D.; Baraket, G.; Perez, V.; Ben Mustapha, S.; Salhi-Hannachi, A.; Hormaza, J.I. Analysis of Self-Incompatibility and Genetic Diversity in Diploid and Hexaploid Plum Genotypes. Front. Plant Sci. 2019, 10, 896. [Google Scholar] [CrossRef]
- Brancher, T.L.; Hawerroth, M.C.; Kvitschal, M.V.; Manenti, D.C.; Guidolin, A.F.; Brancher, T.L.; Hawerroth, M.C.; Kvitschal, M.V.; Manenti, D.C.; Guidolin, A.F. Self-Incompatibility Alleles in Important Genotypes for Apple Breeding in Brazil. Crop Breed. Appl. Biotechnol. 2020, 20. [Google Scholar] [CrossRef]
- Knight, R.; Rogers, H.H. Sterility in Theobroma cacao L. Nature 1953, 172, 164. [Google Scholar] [CrossRef] [PubMed]
- Alagna, F.; Caceres, M.E.; Pandolfi, S.; Collani, S.; Mousavi, S.; Mariotti, R.; Cultrera, N.G.M.; Baldoni, L.; Barcaccia, G. The Paradox of Self-Fertile Varieties in the Context of Self-Incompatible Genotypes in Olive. Front. Plant Sci. 2019, 10, 725. [Google Scholar] [CrossRef]
- Zhang, S.; Liang, M.; Wang, N.; Xu, Q.; Deng, X.; Chai, L. Reproduction in Woody Perennial Citrus: An Update on Nucellar Embryony and Self-Incompatibility. Plant Reprod. 2018, 31, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Denna, D.W. The Potential Use of Self-Incompatibility for Breeding F1 Hybrids of Naturally Self-Pollinated Vegetable Crops. Euphytica 1971, 20, 542–548. [Google Scholar] [CrossRef]
- Brewbaker, J.L.; Gorrez, D.D. Genetics of Self-Incompatibility in the Monocot Genera, Ananas (Pineapple) and Gasteria. Am. J. Bot. 1967, 54, 611–616. [Google Scholar] [CrossRef]
- Goldway, M.; Stern, R.; Zisovich, A.; Raz, A.; Sapir, G.; Schnieder, D.; Nyska, R. The Self-Incompatibility Fertilization System in Rosaceae: Agricultural and Genetic Aspects. Acta Hortic. 2012, 77–82. [Google Scholar] [CrossRef]
- Durand, E.; Chantreau, M.; Veve, A.L.; Stetsenko, R.; Dubin, M.; Genete, M.; Llaurens, V.; Poux, C.; Roux, C.; Billiard, S.; et al. Evolution of Self-Incompatibility in the Brassicaceae: Lessons from a Textbook Example of Natural Selection. Evol. Appl. 2020, 13, 1279–1297. [Google Scholar] [CrossRef]
- Higashiyama, T. Peptide Signaling in Pollen–Pistil Interactions. Plant Cell Physiol. 2010, 51, 177–189. [Google Scholar] [CrossRef] [PubMed]
- McClure, B.; Cruz-García, F.; Romero, C. Compatibility and Incompatibility in S-RNase-Based Systems. Ann. Bot. 2011, 108, 647–658. [Google Scholar] [CrossRef]
- Lanaud, C.; Fouet, O.; Legavre, T.; Lopes, U.; Sounigo, O.; Eyango, M.C.; Mermaz, B.; Da Silva, M.R.; Loor Solorzano, R.G.; Argout, X.; et al. Deciphering the Theobroma Cacao Self-Incompatibility System: From Genomics to Diagnostic Markers for Self-Compatibility. J. Exp. Bot. 2017, 68, 4775–4790. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.G.; Huang, S.; Li, S.; Staiger, C.J.; Franklin-Tong, V.E. Actin Depolymerization Is Sufficient to Induce Programmed Cell Death in Self-Incompatible Pollen. J. Cell Biol. 2006, 174, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, K.A.; Poulter, N.S.; Franklin-Tong, V.E. Taking One for the Team: Self-Recognition and Cell Suicide in Pollen. J. Exp. Bot. 2014, 65, 1331–1342. [Google Scholar] [CrossRef]
- Wang, C.-L.; Wu, J.; Xu, G.-H.; Gao, Y.-B.; Chen, G.; Wu, J.-Y.; Wu, H.-Q.; Zhang, S.-L. S-RNase Disrupts Tip-Localized Reactive Oxygen Species and Induces Nuclear DNA Degradation in Incompatible Pollen Tubes of Pyrus Pyrifolia. J. Cell Sci. 2010, 123, 4301–4309. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Meng, D.; Gu, Z.; Yang, Q.; Yuan, H.; Li, Y.; Chen, Q.; Yu, J.; Liu, C.; Li, T. Apple S-RNase Triggers Inhibition of TRNA Aminoacylation by Interacting with a Soluble Inorganic Pyrophosphatase in Growing Self-Pollen Tubes in Vitro. New Phytol. 2018, 218, 579–593. [Google Scholar] [CrossRef]
- Soost, R.K. Incompatibility Alleles in the Genus Citrus. Proc. Amer. Soc. Hort. Sci. 1965, 87, 176–180. [Google Scholar]
- JungHee, K.; Mori, T.; Wakana, A.; BinhXuan, N.; Masuda, J.; Sakai, K.; Kajiwara, K. Production of Homozygous S1 Seedlings for S Gene in “Hirado Buntan” Pummelo (Citrus Grandis osbeck) and Determination of the S Alleles (S9 and S10) by Pollination with the S1 Seedlings to Citrus Cultivars. J. Fac. Agric. Kyushu Univ. 2010, 55, 239–245. [Google Scholar]
- Yamamoto, M.; Kubo, T.; Tominaga, S. Self- and Cross-Incompatibility of Various Citrus Accessions. J. Jpn. Soc. Hortic. Sci. 2006, 75, 372–378. [Google Scholar] [CrossRef]
- Kim, J.-H.; Mori, T.; Wakana, A.; Ngo, B.X.; Sakai, K.; Kajiwara, K. Determination of Self-Incompatible Citrus Cultivars with S1 and/or S2 Alleles by Pollination with Homozygous S1 Seedlings (S1S1 or S2S2) of ‘Banpeiyu’ Pummelo. J. Jpn. Soc. Hortic. Sci. 2011, 80, 404–413. [Google Scholar] [CrossRef]
- Ngo, B.X.; Kim, J.-H.; Wakana, A.; Isshiki, S.; Mori, T. Estimation of Self-Incompatibility Genotypes of Citrus Cultivars with Got-3 Allozyme Markers. J. Jpn. Soc. Hortic. Sci. 2011, 80, 284–294. [Google Scholar] [CrossRef]
- Wakana, A.; Ngo, B.; Fukudome, I.; Kajiwara, K. Estimation of the Degree of Self-Incompatibility Reaction during Flower Bud Development and Production of Self-Fertilized Seeds by Bud Pollination in Self-Incompatible Citrus Cultivars. J. Fac. Agric. Kyushu Univ. 2004, 49, 307–320. [Google Scholar] [CrossRef]
- Zhou, X.-H.; Kim, J.-H.; Wakana, A.; Sakai, K.; Kajiwara, K.; Mizunoe, Y. Distribution and Evolution of Citrus with S4and/or S5gene Alleles for Self-Incompatibility with Special Focus on the Origin of Satsuma Mandarin (Citrus Unshiu marc.; SfS4). Genet. Resour. Crop Evol. 2018, 65, 1013–1033. [Google Scholar] [CrossRef]
- Kim, J.H.; Handayani, E.; Wakana, A.; Sato, M.; Miyamoto, M.; Miyazaki, R.; Zhou, X.; Sakai, K.; Mizunoe, Y.; Shigyo, M.; et al. Distribution and Evolution of Citrus Accessions with S3 and/or S11 Alleles for Self-Incompatibility with an Emphasis on Sweet Orange [Citrus Sinensis (L.) Osbeck; Sf S3 or Sf S3sm]. Genet. Resour. Crop Evol. 2020, 67, 2101–2117. [Google Scholar] [CrossRef]
- Gentile, A.; Antognoni, F.; Iorio, R.A.; Distefano, G.; Las Casas, G.; La Malfa, S.; Serafini-Fracassini, D.; Del Duca, S. Polyamines and Transglutaminase Activity Are Involved in Compatible and Self-Incompatible Pollination of Citrus Grandis. Amino Acids 2012, 42, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Uchida, A.; Abe, A.; Yamamoto, A.; Hirano, T.; Kunitake, H. Effects of Polyamines on Self-Incompatibility-like Responses in Pollen Tubes of Citrus Cultivars, Banpeiyu and Hyuganatsu. J. Am. Soc. Hortic. Sci. 2015, 140, 183–190. [Google Scholar] [CrossRef]
- Aloisi, I.; Distefano, G.; Antognoni, F.; Potente, G.; Parrotta, L.; Faleri, C.; Gentile, A.; Bennici, S.; Mareri, L.; Cai, G.; et al. Temperature-Dependent Compatible and Incompatible Pollen-Style Interactions in Citrus Clementina hort. Ex Tan. Show Different Transglutaminase Features and Polyamine Pattern. Front. Plant Sci. 2020, 11, 1018. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.; Merelo, P.; Distefano, G.; La Malfa, S.; Lo Piero, A.R.; Tadeo, F.R.; Talon, M.; Gentile, A. Comparative Transcriptome Analysis of Stylar Canal Cells Identifies Novel Candidate Genes Implicated in the Self-Incompatibility Response of Citrus Clementina. BMC Plant Biol. 2012, 12, 20. [Google Scholar] [CrossRef]
- Liang, M.; Yang, W.; Su, S.; Fu, L.; Yi, H.; Chen, C.; Deng, X.; Chai, L. Genome-Wide Identification and Functional Analysis of S-RNase Involved in the Self-Incompatibility of Citrus. Mol. Genet. Genom. MGG 2017, 292, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Ye, Z.; da Silva, J.A.T.; Qin, Y.; Hu, G. Identifying Differentially Expressed Genes in Pollen from Self-Incompatible “Wuzishatangju” and Self-Compatible “Shatangju” Mandarins. Int. J. Mol. Sci. 2013, 14, 8538–8555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ding, F.; He, X.; Luo, C.; Huang, G.; Hu, Y. Characterization of the ‘Xiangshui’ Lemon Transcriptome by de Novo Assembly to Discover Genes Associated with Self-Incompatibility. Mol. Genet. Genom. 2015, 290, 365–375. [Google Scholar] [CrossRef]
- Liang, M.; Cao, Z.; Zhu, A.; Liu, Y.; Tao, M.; Yang, H.; Xu, Q.; Wang, S.; Liu, J.; Li, Y.; et al. Evolution of Self-Compatibility by a Mutant S m -RNase in Citrus. Nat. Plants 2020, 6, 131–142. [Google Scholar] [CrossRef]
- Honsho, C.; Ushijima, K.; Anraku, M.; Ishimura, S.; Yu, Q.; Gmitter, F.G.J.; Tetsumura, T. Association of T2/S-RNase With Self-Incompatibility of Japanese Citrus Accessions Examined by Transcriptomic, Phylogenetic, and Genetic Approaches. Front. Plant Sci. 2021, 12, 121. [Google Scholar] [CrossRef]
- Barry, G.H.; Gmitter, F.G., Jr.; Chen, C.; Roose, M.L.; Federici, C.T.; McCollum, G.T. Investigating the Parentage of “Orri” and “Fortune” Mandarin Hybrids. Acta Hortic. 2015, 449–456. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Furr, J.R. New Tangerines for the Desert. Calif. Citrogr. 1964, 49, 266–276. [Google Scholar]
- Hodgson, R.W. Horticultural varieties of citrus. In The Citrus Industry; UC Press: Berkeley, CA, USA; Los Angeles, CA, USA, 1967; Volume 1, pp. 431–591. [Google Scholar]
- Sonah, H.; Bastien, M.; Iquira, E.; Tardivel, A.; Légaré, G.; Boyle, B.; Normandeau, É.; Laroche, J.; Larose, S.; Jean, M.; et al. An Improved Genotyping by Sequencing (GBS) Approach Offering Increased Versatility and Efficiency of SNP Discovery and Genotyping. PLoS ONE 2013, 8, e54603. [Google Scholar] [CrossRef]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef]
- Ollitrault, P.; Terol, J.; Chen, C.; Federici, C.T.; Lotfy, S.; Hippolyte, I.; Ollitrault, F.; Berard, A.; Chauveau, A.; Cuenca, J.; et al. A Reference Genetic Map of C. Clementina Hort. Ex Tan.; Citrus Evolution Inferences from Comparative Mapping. BMC Genom. 2012, 13, 1–20. [Google Scholar] [CrossRef]
- Guo, F.; Yu, H.; Tang, Z.; Jiang, X.; Wang, L.; Wang, X.; Xu, Q.; Deng, X. Construction of a SNP-Based High-Density Genetic Map for Pummelo Using RAD Sequencing. Tree Genet. Genomes 2015, 11, 2. [Google Scholar] [CrossRef]
- Curtolo, M.; Cristofani-Yaly, M.; Gazaffi, R.; Takita, M.A.; Figueira, A.; Machado, M.A. QTL Mapping for Fruit Quality in Citrus Using DArTseq Markers. BMC Genom. 2017, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Roose, M.L.; Yu, Q.; Du, D.; Yu, Y.; Zhang, Y.; Deng, Z.; Stover, E.; Gmitter, F.G.J. Construction of High-Density Genetic Maps and Detection of QTLs Associated With Huanglongbing Tolerance in Citrus. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- van Os, H.; Stam, P.; Visser, R.G.; van Eck, H.J. SMOOTH: A Statistical Method for Successful Removal of Genotyping Errors from High-Density Genetic Linkage Data. TAG Theoretical. Appl. Genet. Angew. Genet. 2005, 112, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Rasche, H.; Hiltemann, S. Galactic Circos: User-Friendly Circos Plots within the Galaxy Platform. GigaScience 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Storey, J.D. A Direct Approach to False Discovery Rates. J. R. Stat. Soc. Ser. B Stat. Methodol. 2002, 64, 479–498. [Google Scholar] [CrossRef]
- Oueslati, A.; Salhi-Hannachi, A.; Luro, F.; Vignes, H.; Mournet, P.; Ollitrault, P. Genotyping by Sequencing Reveals the Interspecific C. Maxima/C. Reticulata Admixture along the Genomes of Modern Citrus Varieties of Mandarins, Tangors, Tangelos, Orangelos and Grapefruits. PLoS ONE 2017, 12, e0185618. [Google Scholar] [CrossRef]
- do Amaral, M.; Barbosa de Paula, M.F.; Ollitrault, F.; Rivallan, R.; de Andrade Silva, E.M.; da Silva Gesteira, A.; Luro, F.; Garcia, D.; Ollitrault, P.; Micheli, F. Phylogenetic Origin of Primary and Secondary Metabolic Pathway Genes Revealed by C. Maxima and C. Reticulata Diagnostic SNPs. Front. Plant Sci. 2019, 10, 1128. [Google Scholar] [CrossRef]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of Diverse Mandarin, Pummelo and Orange Genomes Reveals Complex History of Admixture during Citrus Domestication. Nat. Biotechnol. 2014, 32, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Ollitrault, P.; Terol, J.; Garcia-Lor, A.; Bérard, A.; Chauveau, A.; Froelicher, Y.; Belzile, C.; Morillon, R.; Navarro, L.; Brunel, D.; et al. SNP Mining in C. Clementina BAC End Sequences; Transferability in the Citrus Genus (Rutaceae), Phylogenetic Inferences and Perspectives for Genetic Mapping. BMC Genom. 2012, 13, 13. [Google Scholar] [CrossRef] [PubMed]
- Bernet, G.; Fernandez-Ribacoba, J.; Carbonell, E.; Asins, M. Comparative Genome-Wide Segregation Analysis and Map Construction Using a Reciprocal Cross Design to Facilitate Citrus Germplasm Utilization. Mol. Breed. 2010, 659–673. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, C.; Gmitter, F.G. QTL Mapping of Mandarin (Citrus Reticulata) Fruit Characters Using High-Throughput SNP Markers. Tree Genet. Genomes 2016, 12, 77. [Google Scholar] [CrossRef]
- Chen, C.X.; Bowman, K.D.; Choi, Y.A.; Dang, P.M.; Rao, M.N.; Huang, S.; Soneji, J.R.; McCollum, T.G.; Gmitter, F.G., Jr. EST-SSR Genetic Maps for Citrus Sinensis and Poncirus Trifoliata. Tree Genet. Genomes 2008, 4, 1–10. [Google Scholar] [CrossRef]
- Calvez, L.; Dereeper, A.; Mournet, P.; Froelicher, Y.; Bruyère, S.; Morillon, R.; Ollitrault, P. Intermediate Inheritance with Disomic Tendency in Tetraploid Intergeneric Citrus × Poncirus Hybrids Enhances the Efficiency of Citrus Rootstock Breeding. Agronomy 2020, 10, 1961. [Google Scholar] [CrossRef]
- Bowman, F.T. Citrus Growing in Australia; Halstead Press: Sydney, New South Wales, Australia, 1956; p. 311. [Google Scholar]
- Carlos de Oliveira, A.; Bastianel, M.; Cristofani-Yaly, M.; Morais do Amaral, A.; Machado, M.A. Development of Genetic Maps of the Citrus Varieties “Murcott” Tangor and “Pera” Sweet Orange by Using Fluorescent AFLP Markers. J. Appl. Genet. 2007, 48, 219–231. [Google Scholar] [CrossRef]
- Distefano, G.; Hedhly, A.; Las Casas, G.; La Malfa, S.; Herrero, M.; Gentile, A. Male–Female Interaction and Temperature Variation Affect Pollen Performance in Citrus. Sci. Hortic. 2012, 140, 1–7. [Google Scholar] [CrossRef]
- Distefano, G.; Gentile, A.; Hedhly, A.; La Malfa, S. Temperatures during Flower Bud Development Affect Pollen Germination, Self-Incompatibility Reaction and Early Fruit Development of Clementine (Citrus Clementina hort. Ex Tan.). Plant Biol. Stuttg. Ger. 2018, 20, 191–198. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).