Effect of Red Orange and Lemon Extract-Enriched Diet in Suckling Lambs’ Fecal Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Management and Feeding
2.2. Composition of Red Orange and Lemon Extract
2.3. Fecal Sampling and Microbiome Analysis
2.4. Statistical Analysis
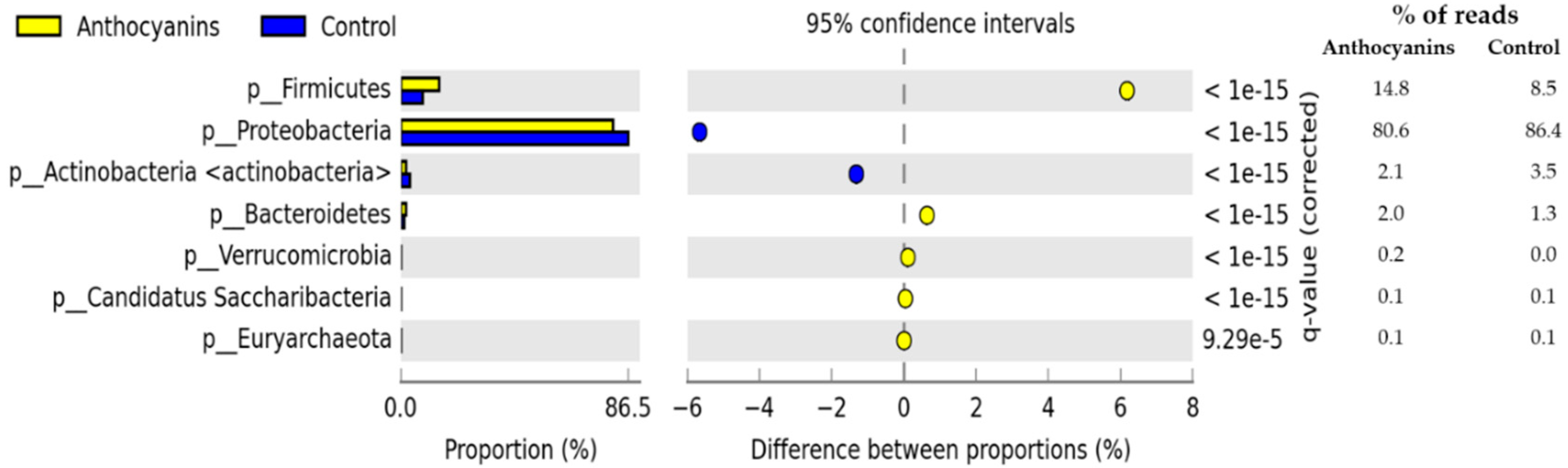
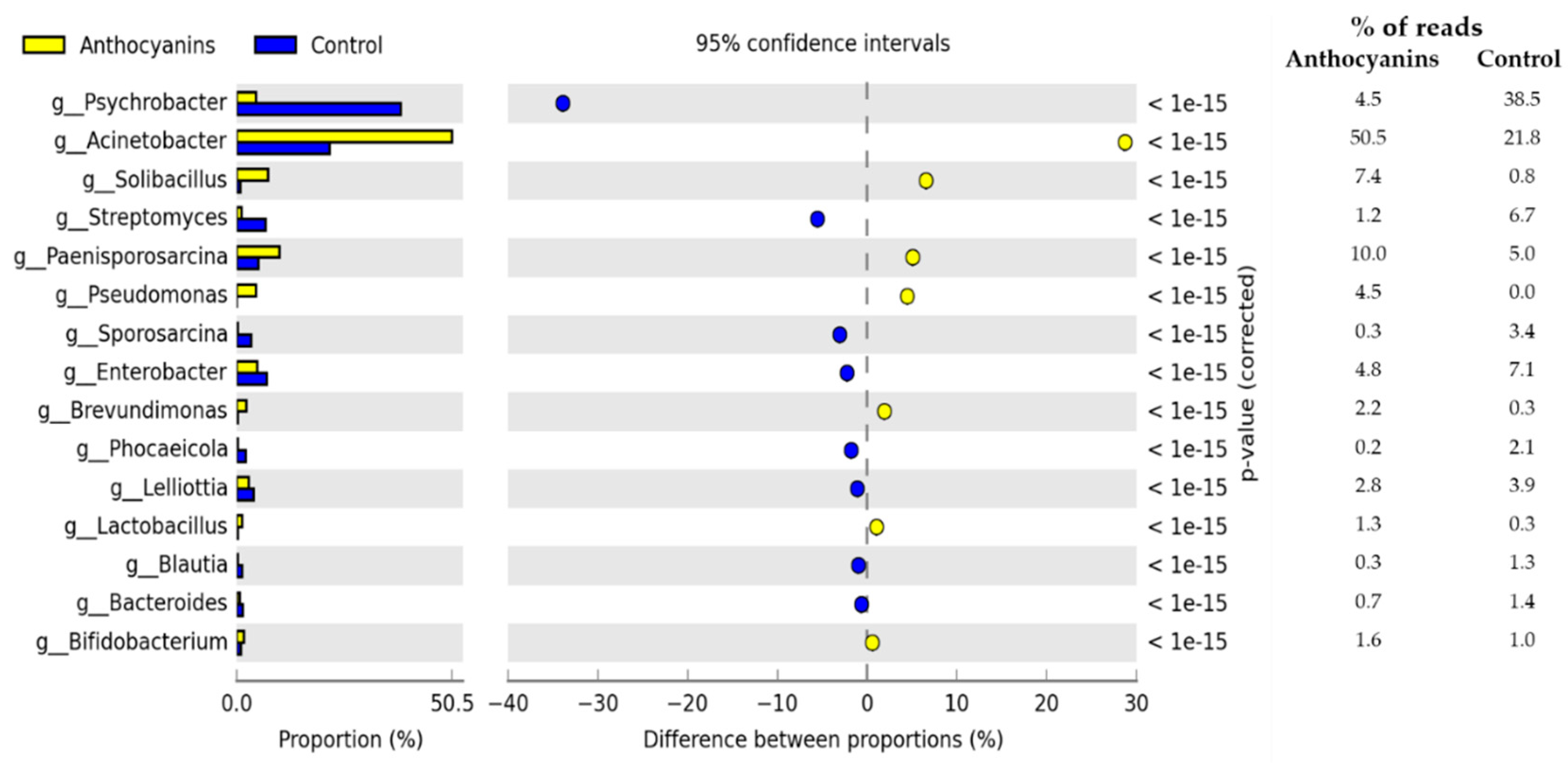
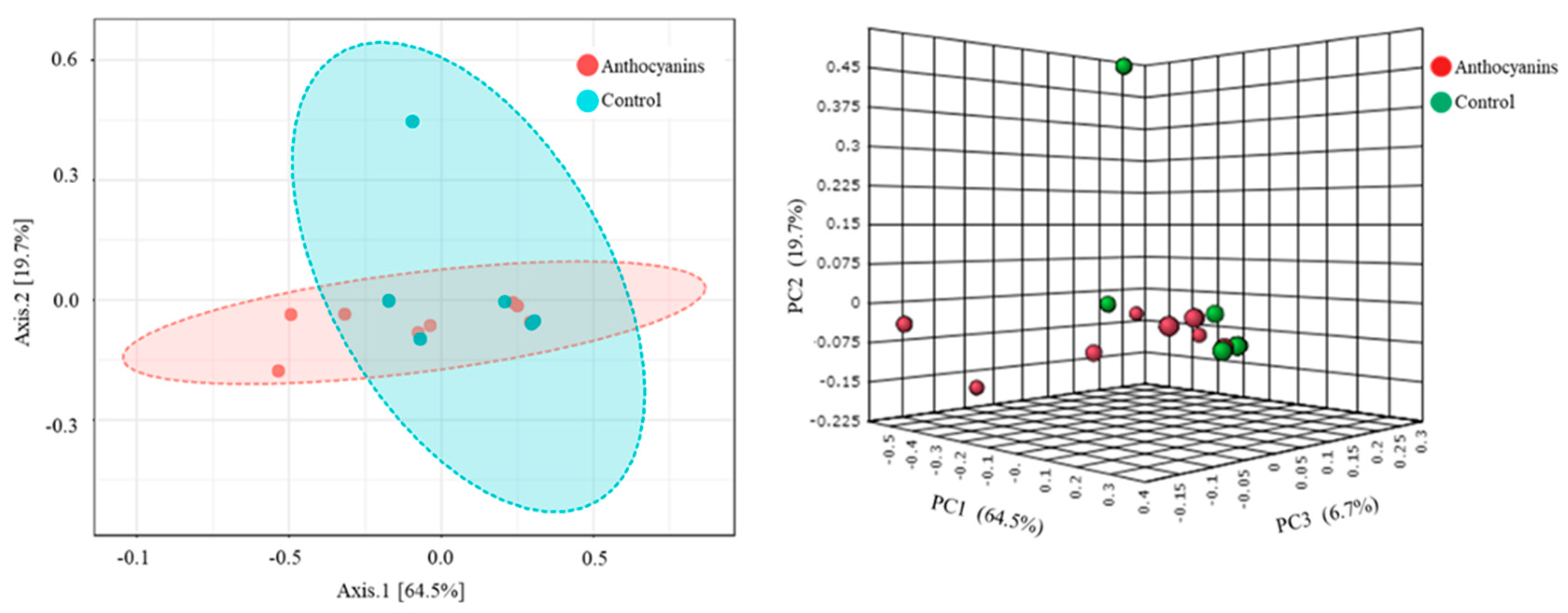
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Zhang, H.; Zhu, L.; Xu, Y.; Liu, N.; Sun, X.; Hu, L.; Huang, H.; Wei, K.; Zhu, R. Dynamic Distribution of Gut Microbiota in Goats at Different Ages and Health States. Front. Microbiol. 2018, 9, 2509. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ. Microbiol. 2007, 9, 1101–1111. [Google Scholar] [CrossRef]
- Kuhn, K.A.; Stappenbeck, T.S. Peripheral education of the immune system by the colonic microbiota. Semin. Immunol. 2013, 25, 364–369. [Google Scholar] [CrossRef] [Green Version]
- Russell, J.B.; Rychlik, J.L. Factors That Alter Rumen Microbial Ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Bäckhed, F. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Kittelmann, S.; Miri, V.; Zethof, M.; Noel, S.; Waghorn, G.; Janssen, P. Effect of DNA Extraction Methods and Sampling Techniques on the Apparent Structure of Cow and Sheep Rumen Microbial Communities. PLoS ONE 2013, 8, e74787. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Yang, Y.; Qin, S.; Lv, S.; Jin, T.; Li, K.; Han, Z.; Li, Y. Microbiome analysis reveals gut microbiota alteration of early-weaned Yimeng black goats with the effect of milk replacer and age. Microb. Cell Factories 2021, 20, 78. [Google Scholar] [CrossRef]
- Morgavi, D.P.; Rathahao-Paris, E.; Popova, M.; Boccard, J.; Nielsen, K.F.; Boudra, H. Rumen microbial communities influence metabolic phenotypes in lambs. Microb. Cell Factories 2015, 20, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Benson, A.K.; Kelly, S.A.; Legge, R.; Ma, F.; Low, S.J.; Kim, J.; Zhang, M.; Oh, P.L.; Nehrenberg, D.; Hua, K.; et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18933–18938. [Google Scholar] [CrossRef] [Green Version]
- Lane, M.A.; Baldwin, R.L., IV; Jesse, B.W. Developmental changes in ketogenic enzyme gene expression during sheep rumen development1. J. Anim. Sci. 2002, 80, 1538–1544. [Google Scholar] [CrossRef]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing Global Ruminant Agricultural Challenges Through Understanding the Rumen Microbiome: Past, Present, and Future. J. Anim. Sci. 2018, 80, 1538–1544. [Google Scholar] [CrossRef]
- Palomba, A.; Tanca, A.; Fraumene, C.; Abbondio, M.; Fancello, F.; Atzori, A.S.; Uzzau, S. Multi-Omic Biogeography of the Gastrointestinal Microbiota of a Pre-Weaned Lamb. Front. Microbiol. 2017, 5, 36. [Google Scholar] [CrossRef] [Green Version]
- Meale, S.J.; Li, S.; Azevedo, P.; Derakhshani, H.; Plaizier, J.C.; Khafipour, E.; Steele, M.A. Development of Ruminal and Fecal Microbiomes Are Affected by Weaning But Not Weaning Strategy in Dairy Calves. Front. Microbiol. 2016, 7, 582. [Google Scholar] [CrossRef] [Green Version]
- Lyons, T.; Boland, T.; Storey, S.; Doyle, E. Linseed Oil Supplementation of Lambs’ Diet in Early Life Leads to Persistent Changes in Rumen Microbiome Structure. Front. Microbiol. 2017, 8, 1656. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Holman, D.B.; Alexander, T.; Hu, T.; Jin, L.; Xu, Z.; McAllister, T.A.; Acharya, S.; Zhao, G.; Wang, Y. Fecal microbiota of lambs fed purple prairie clover (Dalea purpurea Vent.) and alfalfa (Medicago sativa). Arch. Microbiol. 2018, 200, 137–145. [Google Scholar] [CrossRef]
- Boto-Ordóñez, M.; Urpi-Sarda, M.; Queipo-Ortuño, M.I.; Tulipani, S.; Tinahones, F.J.; Andres-Lacueva, C. High levels of Bifidobacteria are associated with increased levels of anthocyanin microbial metabolites: A randomized clinical trial. Food Funct. 2014, 5, 1932–1938. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Ding, S.; Fei, Y.; Liu, G.; Jang, H.; Fang, J. Antimicrobial activity of anthocyanins and catechins against foodborne pathogens Escherichia coli and Salmonella. Food Control. 2019, 106, 106712. [Google Scholar] [CrossRef]
- Anhê, F.F.; Roy, D.; Pilon, G.; Dudonné, S.; Matamoros, S.; Varin, T.V.; Garofalo, C.; Moine, Q.; Desjardins, Y.; Levy, E.; et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 2015, 64, 872–883. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Wu, T.; Chu, X.; Tang, S.; Cao, W.; Liang, F.; Fang, Y.; Pan, S.; Xu, X. Fermented blueberry pomace with antioxidant properties improves fecal microbiota community structure and short chain fatty acids production in an in vitro mode. LWT Food Sci. Tehnol. 2020, 125, 109260. [Google Scholar] [CrossRef]
- Puupponen-Pimiä, R.; Nohynek, L.; Hartmann-Schmidlin, S.; Kähkönen, M.; Heinonen, M.; Määttä-Riihinen, K.; Oksman-Caldentey, K.-M. Berry phenolics selectively inhibit the growth of intestinal pathogens. J. Appl. Microbiol. 2005, 98, 991–1000. [Google Scholar] [CrossRef]
- Choy, Y.Y.; Quifer-Rada, P.; Holstege, D.M.; Frese, S.A.; Calvert, C.C.; Mills, D.A.; Lamuela-Raventos, R.M.; Waterhouse, A.L. Phenolic metabolites and substantial microbiome changes in pig feces by ingesting grape seed proanthocyanidins. Food Funct. 2014, 5, 2298–2308. [Google Scholar] [CrossRef]
- Herrero-Encinas, J.; Blanch, M.; Pastor, J.J.; Mereu, A.; Ipharraguerre, I.R.; Menoyo, D. Effects of a bioactive olive pomace extract from Olea europaea on growth performance, gut function, and intestinal microbiota in broiler chickens. Poult. Sci. 2020, 99, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Hassan, Y.I.; Das, Q.; Lepp, D.; Hernandez, M.; Godfrey, D.V.; Orban, S.; Ross, K.; Delaquis, P.; Diarra, M.S. Dietary organic cranberry pomace influences multiple blood biochemical parameters and cecal microbiota in pasture-raised broiler chickens. J. Funct. Foods 2020, 72, 104053. [Google Scholar] [CrossRef]
- Ghasemi, K.; Ghasemi, Y.; Ebrahimzadeh, M.A. Antioxidant activity, phenol and flavonoid contents of 13 citrus species peels and tissues. Pak. J. Pharm. Sci. 2009, 22, 277–281. [Google Scholar] [PubMed]
- Mahato, N.; Sharma, K.; Sinha, M.; Cho, M.H. Citrus waste derived nutra-/pharmaceuticals for health benefits: Current trends and future perspectives. J. Funct. Foods 2018, 40, 307–316. [Google Scholar] [CrossRef]
- Yang, X.-Y.; Xie, J.-X.; Wang, F.-F.; Zhong, J.; Liu, Y.-Z.; Li, G.-H.; Peng, S.-A. Comparison of ascorbate metabolism in fruits of two citrus species with obvious difference in ascorbate content in pulp. J. Plant Physiol. 2011, 168, 2196–2205. [Google Scholar] [CrossRef] [PubMed]
- Bampidis, V.A.; Robinson, P.H. Citrus by-products as ruminant feeds: A review. Anim. Feed. Sci. Technol. 2006, 128, 175–217. [Google Scholar] [CrossRef]
- Maggiolino, A.; Lorenzo, J.M.; Quiñones, J.; Latorre, M.A.; Blando, F.; Centoducati, G.; Dahl, G.E.; De Palo, P. Effects of dietary supplementation with Pinus taeda hydrolyzed lignin on in vivo performances, in vitro nutrient apparent digestibility, and gas emission in beef steers. Anim. Feed. Sci. Technol. 2019, 255, 114217. [Google Scholar] [CrossRef]
- Maggiolino, A.; Bragaglio, A.; Salzano, A.; Rufrano, D.; Claps, S.; Sepe, L.; Damiano, S.; Ciarcia, R.; Dinardo, F.R.; Hopkins, D.L.; et al. Dietary supplementation of suckling lambs with anthocyanins: Effects on growth, carcass, oxidative and meat quality traits. Anim. Feed. Sci. Technol. 2021, 276, 114925. [Google Scholar] [CrossRef]
- Herbig, A.; Maixner, F.; Bos, K.I.; Zink, A.; Krause, J.; Huson, D.H. MALT: Fast alignment and analysis of metagenomic DNA sequence data applied to the Tyrolean iceman. bioRxiv 2016, 050559. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.; Tappu, R. MEGAN Community Edition-Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Beiko, R.G. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 2010, 26, 715–721. [Google Scholar] [CrossRef] [Green Version]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Mengoni, A.; Bazzicalupo, M. The statistical treatment of data and the analysis of Molecular Variance (AMOVA) in molecular microbial ecology. Ann. Microbiol. 2002, 52, 95–101. [Google Scholar]
- Tanca, A.; Fraumene, C.; Manghina, V.; Palomba, A.; Abbondio, M.; Deligios, M.; Pagnozzi, D.; Addis, M.F.; Uzzau, S. Diversity and functions of the sheep faecal microbiota: A multi-omic characterization. Microb. Biotechnol. 2017, 10, 541–554. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. Taxonomic Identification of Commensal Bacteria Associated with the Mucosa and Digesta throughout the Gastrointestinal Tracts of Preweaned Calves. J. Appl. Environ. Microbiol. 2014, 80, 2021–2028. [Google Scholar] [CrossRef] [Green Version]
- Mancabelli, L.; Ferrario, C.; Milani, C.; Mangifesta, M.; Turroni, F.; Duranti, S.; Lugli, G.A.; Viappiani, A.; Ossiprandi, M.C.; van Sinderen, D.; et al. Insights into the biodiversity of the gut microbiota of broiler chickens. Environ. Microbiol. 2016, 18, 4727–4738. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Lu, Q.; Forster, R.J.; Zhou, C.; Wang, M.; Kang, J.; Tan, Z. Age and feeding system (supplemental feeding versus grazing) modulates colonic bacterial succession and host mucosal immune maturation in goats1. J. Anim. Sci. 2016, 94, 2506–2518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Mehmood, K.; Zhang, H.; Jiang, X.; Shahzad, M.; Dong, X.; Li, J. Characterization of fungus microbial diversity in healthy and diarrheal yaks in Gannan region of Tibet Autonomous Prefecture. Acta Trop. 2018, 182, 14–26. [Google Scholar] [CrossRef]
- Li, C.; Wang, W.; Liu, T.; Zhang, Q.; Wang, G.; Li, F.; Li, F.; Yue, X.; Li, T. Effect of Early Weaning on the Intestinal Microbiota and Expression of Genes Related to Barrier Function in Lambs. Front. Microbiol. 2018, 9, 1431. [Google Scholar] [CrossRef] [Green Version]
- Bi, Y.; Yang, C.; Diao, Q.; Tu, Y. Effects of dietary supplementation with two alternatives to antibiotics on intestinal microbiota of preweaned calves challenged with Escherichia coli K99. Sci. Rep. 2017, 7, 5439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, J.J.; Gomez, A.; White, B.A.; Mangian, H.J.; Loften, J.R.; Drackley, J.K. Changes in the intestinal bacterial community, short-chain fatty acid profile, and intestinal development of preweaned Holstein calves. 1. Effects of prebiotic supplementation depend on site and age. J. Dairy Sci. 2016, 99, 9682–9702. [Google Scholar] [CrossRef] [PubMed]
- Thoetkiattikul, H.; Mhuantong, W.; Laothanachareon, T.; Tangphatsornruang, S.; Pattarajinda, V.; Eurwilaichitr, L.; Champreda, V. Comparative Analysis of Microbial Profiles in Cow Rumen Fed with Different Dietary Fiber by Tagged 16S rRNA Gene Pyrosequencing. Curr. Microbiol. 2013, 67, 130–137. [Google Scholar] [CrossRef]
- Garneau, J.E.; Tremblay, D.M.; Moineau, S. Characterization of 1706, a virulent phage from Lactococcus lactis with similarities to prophages from other Firmicutes. Virology 2008, 373, 298–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spence, C.; Wells, W.G.; Smith, C.J. Characterization of the Primary Starch Utilization Operon in the Obligate Anaerobe Bacteroides fragilis: Regulation by Carbon Source and Oxygen. J. Bacteriol. 2006, 188, 4663–4672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuriel-Ohayon, M.; Neuman, H.; Koren, O. Microbial Changes during Pregnancy, Birth, and Infancy. Front. Microbiol. 2016, 7, 1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Xiao, Y.; Gui, G.; Li, J.; Wang, J.; Li, D. Microbial community and short-chain fatty acid profile in gastrointestinal tract of goose. Poult. Sci. 2018, 97, 1420–1428. [Google Scholar] [CrossRef]
- Min, B.R.; Solaiman, S.; Shange, R.; Eun, J.-S. Gastrointestinal Bacterial and Methanogenic Archaea Diversity Dynamics Associated with Condensed Tannin-Containing Pine Bark Diet in Goats Using 16S rDNA Amplicon Pyrosequencing. Int. J. Microbiol. 2014, 2014, 141909. [Google Scholar] [CrossRef]
- Kafantaris, I.; Kotsampasi, B.; Christodoulou, V.; Kokka, E.; Kouka, P.; Terzopoulou, Z.; Gerasopoulos, K.; Stagos, D.; Mitsagga, C.; Giavasis, I.; et al. Grape pomace improves antioxidant capacity and faecal microflora of lambs. J. Anim. Physiol. Anim. Nutr. 2017, 101, e108–e121. [Google Scholar] [CrossRef]
- Blake, D.P.; Hillman, K.; Fenlon, D.R.; Low, J.C. Transfer of antibiotic resistance between commensal and pathogenic members of the Enterobacteriaceae under ileal conditions. J. Appl. Microbiol. 2003, 95, 428–436. [Google Scholar] [CrossRef]
- Huang, Z.; Kraus, V.B. Does lipopolysaccharide-mediated inflammation have a role in OA? Nat. Rev. Rheumatol. 2016, 12, 123–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quijada, N.M.; Bodas, R.; Lorenzo, J.M.; Schmitz-Esser, S.; Rodríguez-Lázaro, D.; Hernández, M. Dietary Supplementation with Sugar Beet Fructooligosaccharides and Garlic Residues Promotes Growth of Beneficial Bacteria and Increases Weight Gain in Neonatal Lambs. Biomolecules 2020, 10, 1179. [Google Scholar] [CrossRef] [PubMed]
- Douillard, F.P.; de Vos, W.M. Functional genomics of lactic acid bacteria: From food to health. Microb. Cell Factories 2014, 13, S8. [Google Scholar] [CrossRef] [Green Version]
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. The Gut Microbiome and Its Potential Role in the Development and Function of Newborn Calf Gastrointestinal Tract. Front. Veter. Sci. 2015, 2, 36. [Google Scholar] [CrossRef] [Green Version]
- Al-Saiady, M.Y. Effect of Probiotic Bacteria on Immunoglobulin G Concentration and Other Blood Components of Newborn Calves. J. Anim. Vet. Adv. 2010, 9, 604–609. [Google Scholar] [CrossRef] [Green Version]
- Rada, V.; Vlková, E.; Nevoral, J.; Trojanová, I. Comparison of bacterial flora and enzymatic activity in faeces of infants and calves. FEMS Microbiol. Lett. 2006, 258, 25–28. [Google Scholar] [CrossRef] [Green Version]
- Vlková, E.; Trojanová, I.; Rada, V. Distribution of bifidobacteria in the gastrointestinal tract of calves. Folia Microbiol. 2006, 51, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Salzano, A.; Damiano, S.; D’Angelo, L.; Ballistreri, G.; Claps, S.; Rufrano, D.; Maggiolino, A.; Neglia, G.; De Palo, P.; Ciarcia, R. Productive Performance and Meat Characteristics of Kids Fed a Red Orange and Lemon Extract. Animals 2021, 11, 809. [Google Scholar] [CrossRef]
- La Ragione, R.M.; Narbad, A.; Gasson, M.J.; Woodward, M.J. In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett. Appl. Microbiol. 2004, 38, 197–205. [Google Scholar] [CrossRef]
- Bi, Y.; Cox, M.S.; Zhang, F.; Suen, G.; Zhang, N.; Tu, Y.; Diao, Q. Feeding modes shape the acquisition and structure of the initial gut microbiota in newborn lambs. Environ. Microbiol. 2019, 21, 2333–2346. [Google Scholar] [CrossRef] [Green Version]
- Hammerum, A.M.; Heuer, O.E. Human Health Hazards from Antimicrobial-Resistant Escherichia coli of Animal Origin. J. Clin. Infect. Dis. 2009, 48, 916–921. [Google Scholar] [CrossRef] [Green Version]
- Lacombe, A.; Wu, V.C.H.; Tyler, S.; Edwards, K. Antimicrobial action of the American cranberry constituents; phenolics, anthocyanins, and organic acids, against Escherichia coli O157:H7. Int. J. Food Microbiol. 2010, 139, 102–107. [Google Scholar] [CrossRef]
- Benyacoub, J.; Rochat, F.; Saudan, K.-Y.; Rochat, I.; Antille, N.; Cherbut, C.; von der Weid, T.; Schiffrin, E.J.; Blum, S. Feeding a Diet Containing a Fructooligosaccharide Mix Can Enhance Salmonella Vaccine Efficacy in Mice. J. Nutr. 2008, 138, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. J. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Compound | [M]+ (m/z) | MSn (m/z) | Anthocyanin | Relative Composition (%) (a) |
|---|---|---|---|---|
| 1 | 611 | 449/287 | cyanidin 3,5-diglucoside | 1.29 |
| 2 | 465 | 303 | delphinidin 3-glucoside | 2.67 |
| 3 | 611 | 287 | cyanidin 3-sophoroside | 0.41 |
| 4 | 449 | 287 | cyanidin 3-glucoside | 39.97 |
| 5 | 595 | 287 | cyanidin 3-rutinoside | 1.30 |
| 6 | 479 | 317 | petunidin 3-glucoside | 1.59 |
| 7 | 551 | 465/303 | delphinidin 3-(6”-malonyl)glucoside | 1.43 |
| 8 | 463 | 301 | peonidin 3-glucoside | 2.98 |
| 9 | 565 | 479/317 | petunidin 3-(6”-malonyl)glucoside | 1.45 |
| 10 | 535 | 449/287 | cyanidin 3-(6”-malonyl)glucoside | 21.76 |
| 11 | - | 271 | pelargonidin derivative | 1.44 |
| 12 | 549 | 463/301 | peonidin 3-(6”-malonyl)glucoside | 13.80 |
| 13 | - | 287 | cyanidin derivative | 2.39 |
| 14 | - | 301 | peonitin derivative | 1.82 |
| Total anthocyanins (g CGE/100 g) | 2.66 ± 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrara, M.; Sgarro, M.F.; Maggiolino, A.; Damiano, S.; Iannaccone, F.; Mulè, G.; De Palo, P. Effect of Red Orange and Lemon Extract-Enriched Diet in Suckling Lambs’ Fecal Microbiota. Agriculture 2021, 11, 572. https://doi.org/10.3390/agriculture11070572
Ferrara M, Sgarro MF, Maggiolino A, Damiano S, Iannaccone F, Mulè G, De Palo P. Effect of Red Orange and Lemon Extract-Enriched Diet in Suckling Lambs’ Fecal Microbiota. Agriculture. 2021; 11(7):572. https://doi.org/10.3390/agriculture11070572
Chicago/Turabian StyleFerrara, Massimo, Maria Federica Sgarro, Aristide Maggiolino, Sara Damiano, Francesco Iannaccone, Giuseppina Mulè, and Pasquale De Palo. 2021. "Effect of Red Orange and Lemon Extract-Enriched Diet in Suckling Lambs’ Fecal Microbiota" Agriculture 11, no. 7: 572. https://doi.org/10.3390/agriculture11070572








