Prospects of Developing Novel Genetic Resources by Chemical and Physical Mutagenesis to Enlarge the Genetic Window in Bread Wheat Varieties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Material
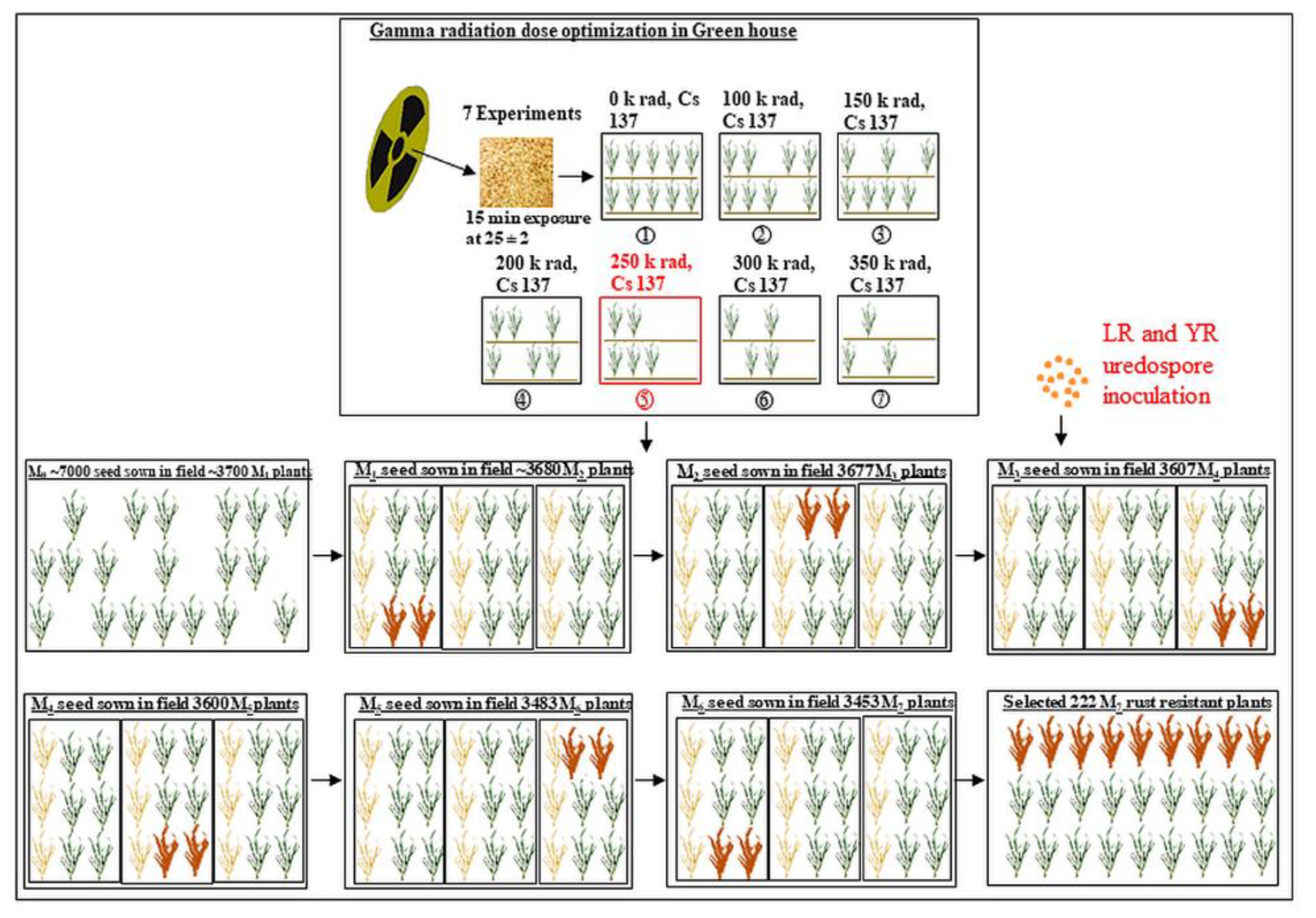
2.2. Development of Mutant Populations
2.3. Rust Inoculation
2.4. Collection of Data from Mutant Generations
2.5. Selection Strategy
2.6. Mutagen Effectiveness
2.7. ANOVA, Heritability, and Correlation Analyses
3. Results
3.1. Prospects of NN-Gandum-1 Derived Population
3.2. Prospects of Punjab-11-Derived Population
3.3. Correlations among Generations
3.4. Comparison of the Selected Mutant Lines with the Wild-Type Parents
3.4.1. Days to Heading
3.4.2. Spike Length
3.4.3. Plant Height
3.4.4. Tillers per Plant
3.5. Correlation among Traits of the Selected Mutant Lines
3.6. Mutagen Effectiveness
3.7. Heritability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arumuganathan, K.; Earle, E. Nuclear DNA content of some important plant species. Plant. Mol. Biol Rep. 1991, 9, 208–218. [Google Scholar] [CrossRef]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.; Steuernagel, B.; Mayer, K.F.; Olsen, O.-A. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 6194. [Google Scholar] [CrossRef] [PubMed]
- Conway, G. The Doubly Green Revolution: Food for all in the Twenty-First Century; Cornell University Press: Ithaca, NY, USA, 1998. [Google Scholar]
- Mukhtar, S.; Rahman, M.; Zafar, Y. Assessment of genetic diversity among wheat cultivars using random amplified polymorphic DNA (RAPD) analysis. Euphytica 2002, 128, 417–425. [Google Scholar] [CrossRef]
- Rahman, M.; Malik, T.; Chowdhary, M.; Iqbal, M.; Zafar, Y. Application of random amplified polymorphic DNA (RAPD) technique for the identification of markers linked to salinity tolerance in wheat (Triticum aestivum L.). Pak. J. Bot. 2004, 36, 595–602. [Google Scholar]
- Malik, R.; Tiwari, R.; Arora, A.; Kumar, P.; Sheoran, S.; Sharma, P.; Singh, R.; Tiwari, V.; Sharma, I. Genotypic characterization of elite Indian wheat genotypes using molecular markers and their pedigree analysis. Aust. J. Crop. Sci. 2013, 7, 561. [Google Scholar]
- Huerta-Espino, J.; Singh, R.; German, S.; McCallum, B.; Park, R.; Chen, W.Q.; Bhardwaj, S.; Goyeau, H. Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 2011, 179, 143–160. [Google Scholar] [CrossRef]
- Beddow, J.M.; Pardey, P.G.; Chai, Y.; Hurley, T.M.; Kriticos, D.J.; Braun, H.-J.; Park, R.F.; Cuddy, W.S.; Yonow, T. Research investment implications of shifts in the global geography of wheat stripe rust. Nat. Plants 2015, 1, 15132. [Google Scholar] [CrossRef]
- Vergara-Diaz, O.; Kefauver, S.C.; Elazab, A.; Nieto-Taladriz, M.T.; Araus, J.L. Grain yield losses in yellow-rusted durum wheat estimated using digital and conventional parameters under field conditions. Crop. J. 2015, 3, 200–210. [Google Scholar] [CrossRef] [Green Version]
- Acevedo, M.; Zurn, J.D.; Molero, G.; Singh, P.; He, X.; Aoun, M.; Juliana, P.; Bockleman, H.; Bonman, M.; El-Sohl, M. The role of wheat in global food security. In Agricultural Development and Sustainable Intensification; Routledge: London, UK, 2018; pp. 81–110. [Google Scholar]
- Miedaner, T.; Schmid, J.E.; Flath, K.; Koch, S.; Jacobi, A.; Ebmeyer, E.; Taylor, M. A multiple disease test for field-based phenotyping of resistances to Fusarium head blight, yellow rust and stem rust in wheat. Eur. J. Plant. Pathol. 2018, 151, 451–461. [Google Scholar] [CrossRef]
- Barua, P.; You, M.; Bayliss, K.; Lanoiselet, V.; Barbetti, M. Extended survival of Puccinia graminis f. sp. tritici urediniospores: Implications for biosecurity and on-farm management. Plant. Pathol. 2018, 67, 799–809. [Google Scholar] [CrossRef]
- Hovmøller, M.S.; Walter, S.; Bayles, R.A.; Hubbard, A.; Flath, K.; Sommerfeldt, N.; Leconte, M.; Czembor, P.; Rodriguez-Algaba, J.; Thach, T. Replacement of the European wheat yellow rust population by new races from the centre of diversity in the near-Himalayan region. Plant. Pathol. 2016, 65, 402–411. [Google Scholar] [CrossRef] [Green Version]
- Riaz, A.; Athiyannan, N.; Periyannan, S.K.; Afanasenko, O.; Mitrofanova, O.P.; Platz, G.J.; Aitken, E.A.; Snowdon, R.J.; Lagudah, E.S.; Hickey, L.T. Unlocking new alleles for leaf rust resistance in the Vavilov wheat collection. Theor. Appl. Genet. 2018, 131, 127–144. [Google Scholar] [CrossRef] [PubMed]
- Wallace, J. Increasing agricultural water use efficiency to meet future food production. Agric. Ecosyst. Environ. 2000, 82, 105–119. [Google Scholar] [CrossRef]
- Skovmand, B.; Reynolds, M.; Delacy, I. Searching genetic resources for physiological traits with potential for increasing yield. In Application of Physiology in Wheat Breeding; CIMMYT: Mexico City, Mexico, 2001; pp. 17–28. [Google Scholar]
- Eynard, A.; Lal, R.; Wiebe, K. Crop response in salt-affected soils. J. Sustain. Agric. 2005, 27, 5–50. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant. Biol 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- Krasileva, K.V.; Vasquez-Gross, H.A.; Howell, T.; Bailey, P.; Paraiso, F.; Clissold, L.; Simmonds, J.; Ramirez-Gonzalez, R.H.; Wang, X.; Borrill, P. Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. USA 2017, 114, E913–E921. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Pan, Q.; He, F.; Akhunova, A.; Chao, S.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 2018, 1, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Muqaddasi, Q.H.; Brassac, J.; Koppolu, R.; Plieske, J.; Ganal, M.W.; Röder, M.S. TaAPO-A1, an ortholog of rice ABERRANT PANICLE ORGANIZATION 1, is associated with total spikelet number per spike in elite European hexaploid winter wheat (Triticum aestivum L.) varieties. Sci. Rep. 2019, 9, 13853. [Google Scholar] [CrossRef] [Green Version]
- Bhat, R.S.; Upadhyaya, N.M.; Chaudhury, A.; Raghavan, C.; Qiu, F.; Wang, H.; Wu, J.; McNally, K.; Leung, H.; Till, B. Chemical-and irradiation-induced mutants and TILLING. In Rice Functional Genomics; Springer: Berlin/Heidelberg, Germany, 2007; pp. 148–180. [Google Scholar]
- Ponce-Molina, L.J.; Huerta-Espino, J.; Singh, R.P.; Basnet, B.R.; Lagudah, E.; Aguilar-Rincón, V.H.; Alvarado, G.; Lobato-Ortiz, R.; García-Zavala, J.; Lan, C. Characterization of adult plant resistance to leaf rust and stripe rust in Indian wheat cultivar ‘New Pusa 876’. Crop. Sci. 2018, 58, 630–638. [Google Scholar] [CrossRef]
- Brini, F.; Masmoudi, K. Biotechnology for drought and salinity tolerance of crops. In Physiological Mechanisms and Adaptation Strategies in Plants Under Changing Environment; Springer: Berlin/Heidelberg, Germany, 2014; pp. 97–113. [Google Scholar]
- Dumanovć, J.; Denić, M.; Ehrenberg, L.; Bergstrand, K. Radiation-induced heritable variation of quantiative characters in wheat. Hereditas 1969, 62, 221–238. [Google Scholar] [CrossRef]
- Naito, K.; Kusaba, M.; Shikazono, N.; Takano, T.; Tanaka, A.; Tanisaka, T.; Nishimura, M. Transmissible and nontransmissible mutations induced by irradiating Arabidopsis thaliana pollen with γ-rays and carbon ions. Genetics 2005, 169, 881–889. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.; Missirian, V.; Ngo, K.J.; Tran, R.K.; Chan, S.R.; Sundaresan, V.; Comai, L. Production of a high-efficiency TILLING population through polyploidization. Plant. Physiol. 2013, 161, 1604–1614. [Google Scholar] [CrossRef] [Green Version]
- Peterson, R.F.; Campbell, A.; Hannah, A. A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 1948, 26, 496–500. [Google Scholar] [CrossRef]
- Tollenaar, D. Untersuchungen ueber Mutation bei Tabak. Genetica 1934, 16, 111–152. [Google Scholar] [CrossRef]
- Chawade, A.; Sikora, P.; Bräutigam, M.; Larsson, M.; Vivekanand, V.; Nakash, M.A.; Chen, T.; Olsson, O. Development and characterization of an oat TILLING-population and identification of mutations in lignin and β-glucan biosynthesis genes. BMC Plant. Biol. 2010, 10, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Huang, L.; Min, D.; Phillips, A.; Wang, S.; Madgwick, P.J.; Parry, M.A.; Hu, Y.-G. Development and characterization of a new TILLING population of common bread wheat (Triticum aestivum L.). PLoS ONE 2012, 7, e41570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawat, N.; Sehgal, S.K.; Joshi, A.; Rothe, N.; Wilson, D.L.; McGraw, N.; Vadlani, P.V.; Li, W.; Gill, B.S. A diploid wheat TILLING resource for wheat functional genomics. BMC Plant. Biol. 2012, 12, 205. [Google Scholar] [CrossRef] [Green Version]
- Joshi, A. Map-Based Cloning of the Hessian Fly Resistance Gene H13 in Wheat. 2018. Available online: https://www.proquest.com/openview/683f08ff0d8dd44fb6ebdf4a5ed95c0a/1?pq-origsite=gscholar&cbl=18750 (accessed on 1 July 2021).
- Chen, Y.L.; Liang, H.L.; Ma, X.L.; Lou, S.L.; Xie, Y.Y.; Liu, Z.L.; Chen, L.T.; Liu, Y.G. An Efficient Rice Mutagenesis System Based on Suspension-Cultured Cells. J. Integr. Plant. Biol. 2013, 55, 122–130. [Google Scholar] [CrossRef]
- Dorosti, H.; Zandi, P.; Basu, S.K.; Chalaras, S.K. Ethyl methane sulfonate and its effect on mutagenesis in rice crop improvement. In Proceedings of the VII International Scientific Agriculture Symposium, "Agrosym 2016", Jahorina, Bosnia and Herzegovina, 6–9 October 2016; pp. 1215–1220. [Google Scholar]
- Bora, A.; Choudhury, P.R.; Pande, V.; Mandal, A.B. Assessment of genetic purity in rice (Oryza sativa L.) hybrids using microsatellite markers. 3 Biotech 2016, 6, 50. [Google Scholar] [CrossRef] [Green Version]
- Burkart-Waco, D.; Tsai, H.; Ngo, K.; Henry, I.M.; Comai, L.; Tai, T.H. Next-generation sequencing for targeted discovery of rare mutations in rice. In Biotechnologies for Plant Mutation Breeding; Springer: Cham, Switzerland, 2017; pp. 323–340. [Google Scholar]
- Jiao, Y.; Burke, J.; Chopra, R.; Burow, G.; Chen, J.; Wang, B.; Hayes, C.; Emendack, Y.; Ware, D.; Xin, Z. A sorghum mutant resource as an efficient platform for gene discovery in grasses. Plant. Cell 2016, 28, 1551–1562. [Google Scholar] [CrossRef] [Green Version]
- Lababidi, S.; Mejlhede, N.; Rasmussen, S.K.; Backes, G.; Al-Said, W.; Baum, M.; Jahoor, A. Identification of barley mutants in the cultivar ‘Lux’at the Dhn loci through TILLING. Plant. Breed. 2009, 128, 332–336. [Google Scholar] [CrossRef]
- Caldwell, D.G.; McCallum, N.; Shaw, P.; Muehlbauer, G.J.; Marshall, D.F.; Waugh, R. A structured mutant population for forward and reverse genetics in Barley (Hordeum vulgare L.). Plant. J. 2004, 40, 143–150. [Google Scholar] [CrossRef]
- Slade, A.J.; Knauf, V.C. TILLING moves beyond functional genomics into crop improvement. Transgenic Res. 2005, 14, 109–115. [Google Scholar] [CrossRef]
- Rakszegi, M.; Kisgyörgy, B.; Tearall, K.; Shewry, P.; Láng, L.; Phillips, A.; Bedő, Z. Diversity of agronomic and morphological traits in a mutant population of bread wheat studied in the Healthgrain program. Euphytica 2010, 174, 409–421. [Google Scholar] [CrossRef]
- Guo, J.; Shi, W.; Zhang, Z.; Cheng, J.; Sun, D.; Yu, J.; Li, X.; Guo, P.; Hao, C. Association of yield-related traits in founder genotypes and derivatives of common wheat (Triticum aestivum L.). BMC Plant. Biol. 2018, 18, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talamè, V.; Bovina, R.; Sanguineti, M.C.; Tuberosa, R.; Lundqvist, U.; Salvi, S. TILLMore, a resource for the discovery of chemically induced mutants in barley. Plant. Biotechnol. J. 2008, 6, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Eiguchi, M.; Kumamaru, T.; Satoh, H.; Matsusaka, H.; Moriguchi, K.; Nagato, Y.; Kurata, N. MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genom. 2008, 279, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Sabetta, W.; Alba, V.; Blanco, A.; Montemurro, C. sunTILL: A TILLING resource for gene function analysis in sunflower. Plant. Methods 2011, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Hussain, M.; Iqbal, M.A.; Till, B.J.; Rahman, M.-U. Identification of induced mutations in hexaploid wheat genome using exome capture assay. PLoS ONE 2018, 13, e0201918. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Chai, L.; Chen, Z.; Xu, L.; Zhai, H.; Zhao, A.; Peng, H.; Yao, Y.; You, M.; Sun, Q. Identification and characterization of a high kernel weight mutant induced by gamma radiation in wheat (Triticum aestivum L.). BMC Genet. 2015, 16, 127. [Google Scholar] [CrossRef] [Green Version]
- Mohsin, T.; Khan, N.; Naqvi, F.N. Heritability, phenotypic correlation and path coefficient studies for some agronomic characters in synthetic elite lines of wheat. J. Food Agric. Environ. 2009, 7, 278–282. [Google Scholar]
- Patel, J.D.; Wright, R.J.; Auld, D.; Chandnani, R.; Goff, V.H.; Ingles, J.; Pierce, G.J.; Torres, M.J.; Paterson, A.H. Alleles conferring improved fiber quality from EMS mutagenesis of elite cotton genotypes. Theor. Appl. Genet. 2014, 127, 821–830. [Google Scholar] [CrossRef]
- Gurung, S.; Mamidi, S.; Bonman, J.M.; Xiong, M.; Brown-Guedira, G.; Adhikari, T.B. Genome-wide association study reveals novel quantitative trait loci associated with resistance to multiple leaf spot diseases of spring wheat. PLoS ONE 2014, 9, e108179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccaferri, M.; Zhang, J.; Bulli, P.; Abate, Z.; Chao, S.; Cantu, D.; Bossolini, E.; Chen, X.; Pumphrey, M.; Dubcovsky, J. A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3-Genes Genom. Genet. 2015, 5, 449–465. [Google Scholar]
- Hassani, I.; Marker, S.; Lal, G. Inter-relationship studies among grain yield and its component characters in wheat (Triticum aestivum L.). J. Pharmacog. Phytochem. 2017, 6, 186–191. [Google Scholar]
- Koo, B.-H.; Yoo, S.-C.; Park, J.-W.; Kwon, C.-T.; Lee, B.-D.; An, G.; Zhang, Z.; Li, J.; Li, Z.; Paek, N.-C. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant. 2013, 6, 1877–1888. [Google Scholar] [CrossRef] [Green Version]
- Hori, K.; Nonoue, Y.; Ono, N.; Shibaya, T.; Ebana, K.; Matsubara, K.; Ogiso-Tanaka, E.; Tanabata, T.; Sugimoto, K.; Taguchi-Shiobara, F. Genetic architecture of variation in heading date among Asian rice accessions. BMC Plant. Biol 2015, 15, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasmine, F.; Ullah, M.A.; Ahmad, F.; Rahman, M.A.; Harun, A.R. Effects of chronic gamma irradiation on three rice varieties. J. Sains Nukl. Malays. 2019, 31, 1–10. [Google Scholar]
- Kakar, K.; Kakar, Z.; Shawani, M. Varietal dynamics of yield stability in wheat. Online J. Biol. Sci. 2003, 3, 137–140. [Google Scholar]
- Adnan, M.; Muhammad, F.; Hussain, Q.; Hussain, I.; Ali, F. Heritability estimates and correlation analysis in bread wheat (Triticum aestivum L.) under normal and late plantings. Pure Appl. Biol. 2017, 6, 1151–1160. [Google Scholar] [CrossRef]
- Iqbal, A.; Khalil, I.H.; Shah, S.; Kakar, M.S. Estimation of heritability, genetic advance and correlation for morphological traits in spring wheat. Sarhad J. Agric. 2017, 33, 674–679. [Google Scholar]
- Szurman-Zubrzycka, M.E.; Zbieszczyk, J.; Marzec, M.; Jelonek, J.; Chmielewska, B.; Kurowska, M.M.; Krok, M.; Daszkowska-Golec, A.; Guzy-Wrobelska, J.; Gruszka, D. HorTILLUS—A rich and renewable source of induced mutations for forward/reverse genetics and pre-breeding programs in barley (Hordeum vulgare L.). Front. Plant. Sci. 2018, 9, 216. [Google Scholar] [CrossRef]
- Singh, N.; Balyan, H. Induced mutations in bread wheat (Triticum aestivum L.) CV.‘Kharchia 65’for reduced plant height and improve grain quality traits. Adv. Biol. Res. 2009, 3, 215–221. [Google Scholar]
- Sikder, S.; Ravat, V.; Basfore, S.; Hazra, P. Isolation of induced mutants using gamma ray and ethyl methane sulphonate in Tomato (Solanum lycopersicum L.). Electron. J. Plant. Breed. 2015, 6, 464–471. [Google Scholar]
- Banjare, C. Induction of Mutation through Physical and Chemical Mutagens and Characterization of Muatant Lines in Garlic (Allium sativum L.). Ph.D. Thesis, Indira Gandhi Krishi Vishwavidhyalaya, Raipur, India, 2017. [Google Scholar]
- Devi, A.S.; Mullainathan, L. Physical and chemical mutagenesis for improvement of chilli (Capsicum annuum L.). World Appl. Sci. J. 2011, 15, 108–113. [Google Scholar]
- Kumar, P.; Singh, G.; Kumar, S.; Kumar, A.; Ojha, A. Genetic analysis of grain yield and its contributing traits for their implications in improvement of bread wheat cultivars. J. Appl. Nat. Sci. 2016, 8, 350–357. [Google Scholar] [CrossRef]
- Dhakshanamoorthy, D.; Selvaraj, R.; Chidambaram, A. Physical and chemical mutagenesis in Jatropha curcas L. to induce variability in seed germination, growth and yield traits. Rom. J. Biol. Plant. Biol. 2010, 55, 113–125. [Google Scholar]
- Begum, T.; Dasgupta, T. A comparison of the effects of physical and chemical mutagens in sesame (Sesamum indicum L.). Genet. Mol. Biol. 2010, 33, 761–766. [Google Scholar] [CrossRef]
- Shah, T.M.; Mirza, J.I.; Haq, M.A.; Atta, B.M. Induced genetic variability in chickpea (Cicer arietinum L.). II. Comparative mutagenic effectiveness and efficiency of physical and chemical mutagens. Pak. J. Bot. 2008, 40, 605–613. [Google Scholar]
- Kaul, M.; Bhan, A. Mutagenic effectiveness and efficiency of EMS, DES and gamma-rays in rice. Theor. Appl. Genet. 1977, 50, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Solanki, I.; Sharma, B. Mutagenic effectiveness and efficiency of gamma rays, ethylene imine and N-nitroso-N-ethyl urea in macrosperma lentil (Lens culinaris Medik.). Indian J. Genet. Plant. Breed. 1994, 54, 72–76. [Google Scholar]
- Khan, M.H.; Tyagi, S.D. Studies on effectiveness and efficiency of gamma rays, EMS and their combination in soybean [Glycine max (L.) Merrill.]. J. Plant. Breed. Crop. Sci. 2010, 2, 055–058. [Google Scholar]
- Sikora, P.; Chawade, A.; Larsson, M.; Olsson, J.; Olsson, O. Mutagenesis as a tool in plant genetics, functional genomics, and breeding. Int. J. Plant. Genom. 2011, 2011, 314829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogbonnaya, F.C.; Rasheed, A.; Okechukwu, E.C.; Jighly, A.; Makdis, F.; Wuletaw, T.; Hagras, A.; Uguru, M.I.; Agbo, C.U. Genome-wide association study for agronomic and physiological traits in spring wheat evaluated in a range of heat prone environments. Theor. Appl. Genet. 2017, 130, 1819–1835. [Google Scholar] [CrossRef]
- Lopes, M.; Reynolds, M.; Manes, Y.; Singh, R.; Crossa, J.; Braun, H. Genetic yield gains and changes in associated traits of CIMMYT spring bread wheat in a “historic” set representing 30 years of breeding. Crop. Sci 2012, 52, 1123–1131. [Google Scholar] [CrossRef]



| Traits | Population | Mean (SD) of M5 | Mean (SD) of M6 | Mean (SD) of M7 | SD |
|---|---|---|---|---|---|
| NN-1 | |||||
| 10 WT lines | 3634 | 3533 | 3502 | ||
| DTH (days) | 89 | 92.57 (1.68) | 92.82 (1.62) | 92.4 (1.96) | −4.43 to +4.57 |
| SL (cm) | 9.25 | 9.58 (0.68) | 9.67 (0.66) | 9.36 (0.97) | −6.07 to +2.33 |
| PH (cm) | 89.26 | 89.32 (4.08) | 89.52 (5.06) | 86.27(5.58) | −16.04 to +10.96 |
| TPP | 5 | 5.06 (0.62) | 5.13 (0.57) | 5.07 (0.59) | −0.94 to +1.06 |
| Punjab-11 | |||||
| 10 WT lines | 3600 | 3483 | 3453 | ||
| DTH (days) | 92 | 93.03 (1.30) | 93.12 (1.31) | 93.19 (1.33) | −1.97 to +4.03 |
| SL (cm) | 8.86 | 9.29 (1.05) | 9.3 (1.05) | 9.35 (1.05) | −5.27 to +2.01 |
| PH (cm) | 87.65 | 88.03 (3.13) | 88.15 (3.15) | 88.21 (3.13) | −8.62 to +4.49 |
| TPP | 4 | 5.08 (0.63) | 5.1 (0.65) | 5.13 (0.62) | −0.92 to +1.08 |
| Genotypes | Generation | DTH (days) | SL (cm) | PH (cm) | TPP |
|---|---|---|---|---|---|
| NN-1 | M5 versus M6 | 0.56 * | 0.65 * | 0.55 * | 0.36 * |
| M5 versus M7 | 0.52 * | 0.68 * | 0.5 * | 0.48 * | |
| M6 versus M7 | 0.59 * | 0.69 * | 0.61 * | 0.53 * | |
| Pb-11 | M5 versus M6 | 0.99 * | 0.92 * | 0.79 * | 0.96 * |
| M5 versus M7 | 0.99 * | 0.91 * | 0.79 * | 0.97 * | |
| M6 versus M7 | 0.99 * | 0.93 * | 0.79 * | 0.97 * |
| Population | Parameter | DTH (Days) | SL (cm) | PH (cm) | TPP |
|---|---|---|---|---|---|
| NN-1 | Wild type | 89 | 9.25 | 89.26 | 5 |
| Selected lines | 93.05 | 9.81 | 90.24 | 5.23 | |
| SD | 1.8 | 1.2 | 5.3 | 0.7 | |
| Z score range | −1.42 to +1.54 | −1.14 to +2.98 | −1.71 to +1.60 | −1.18 to +1.58 | |
| Lines with highest Z score | 9 | 12 | 12 | 6 | |
| Pb-11 | Wild type | 92 | 8.86 | 87.65 | 4 |
| Selected lines | 92.06 | 9.09 | 88.37 | 4.68 | |
| SD | 1.41 | 1.13 | 3 | 0.82 | |
| Z score range | −1.65 to +1.36 | −1.97 to +2.12 | −1.46 to +2.38 | −1.82 to +1.40 | |
| Lines with highest Z score | 14 | 81 | 137 | 52 |
| Traits | Variants (%) in M7 Generation | |||||
|---|---|---|---|---|---|---|
| NN-1 (n = 3502) | Pb-11 (n = 3453) | Gain over EMS/γ Rays | ||||
| Variants | % | Variants | % | NN-1 (EMS) | Pb-11 (γ Rays) | |
| DTH | 590 | 16.85 | 662 | 19.17 | 2.32 | |
| LR | 1172 | 33.47 | 794 | 22.99 | 10.47 | |
| YR | 587 | 16.76 | 858 | 24.85 | 8.09 | |
| SL | 813 | 23.22 | 1102 | 31.91 | 8.7 | |
| PH | 430 | 12.28 | 961 | 27.83 | 15.55 | |
| TPP | 1190 | 33.98 | 1047 | 30.32 | 3.66 | |
| Average | 22.76 | 26.18 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, M.; Gul, M.; Kamal, R.; Iqbal, M.A.; Zulfiqar, S.; Abbas, A.; Röder, M.S.; Muqaddasi, Q.H.; Mehboob-ur-Rahman. Prospects of Developing Novel Genetic Resources by Chemical and Physical Mutagenesis to Enlarge the Genetic Window in Bread Wheat Varieties. Agriculture 2021, 11, 621. https://doi.org/10.3390/agriculture11070621
Hussain M, Gul M, Kamal R, Iqbal MA, Zulfiqar S, Abbas A, Röder MS, Muqaddasi QH, Mehboob-ur-Rahman. Prospects of Developing Novel Genetic Resources by Chemical and Physical Mutagenesis to Enlarge the Genetic Window in Bread Wheat Varieties. Agriculture. 2021; 11(7):621. https://doi.org/10.3390/agriculture11070621
Chicago/Turabian StyleHussain, Momina, Maryyam Gul, Roop Kamal, Muhammad Atif Iqbal, Sana Zulfiqar, Ammad Abbas, Marion S. Röder, Quddoos H. Muqaddasi, and Mehboob-ur-Rahman. 2021. "Prospects of Developing Novel Genetic Resources by Chemical and Physical Mutagenesis to Enlarge the Genetic Window in Bread Wheat Varieties" Agriculture 11, no. 7: 621. https://doi.org/10.3390/agriculture11070621
APA StyleHussain, M., Gul, M., Kamal, R., Iqbal, M. A., Zulfiqar, S., Abbas, A., Röder, M. S., Muqaddasi, Q. H., & Mehboob-ur-Rahman. (2021). Prospects of Developing Novel Genetic Resources by Chemical and Physical Mutagenesis to Enlarge the Genetic Window in Bread Wheat Varieties. Agriculture, 11(7), 621. https://doi.org/10.3390/agriculture11070621






