Identification and Validation of QTLs for Yield and Yield Components under Long-Term Salt Stress Using IR64 CSSLs in the Genetic Background of Koshihikari and Their Backcross Progenies
Abstract
:1. Introduction
2. Materials and Methods
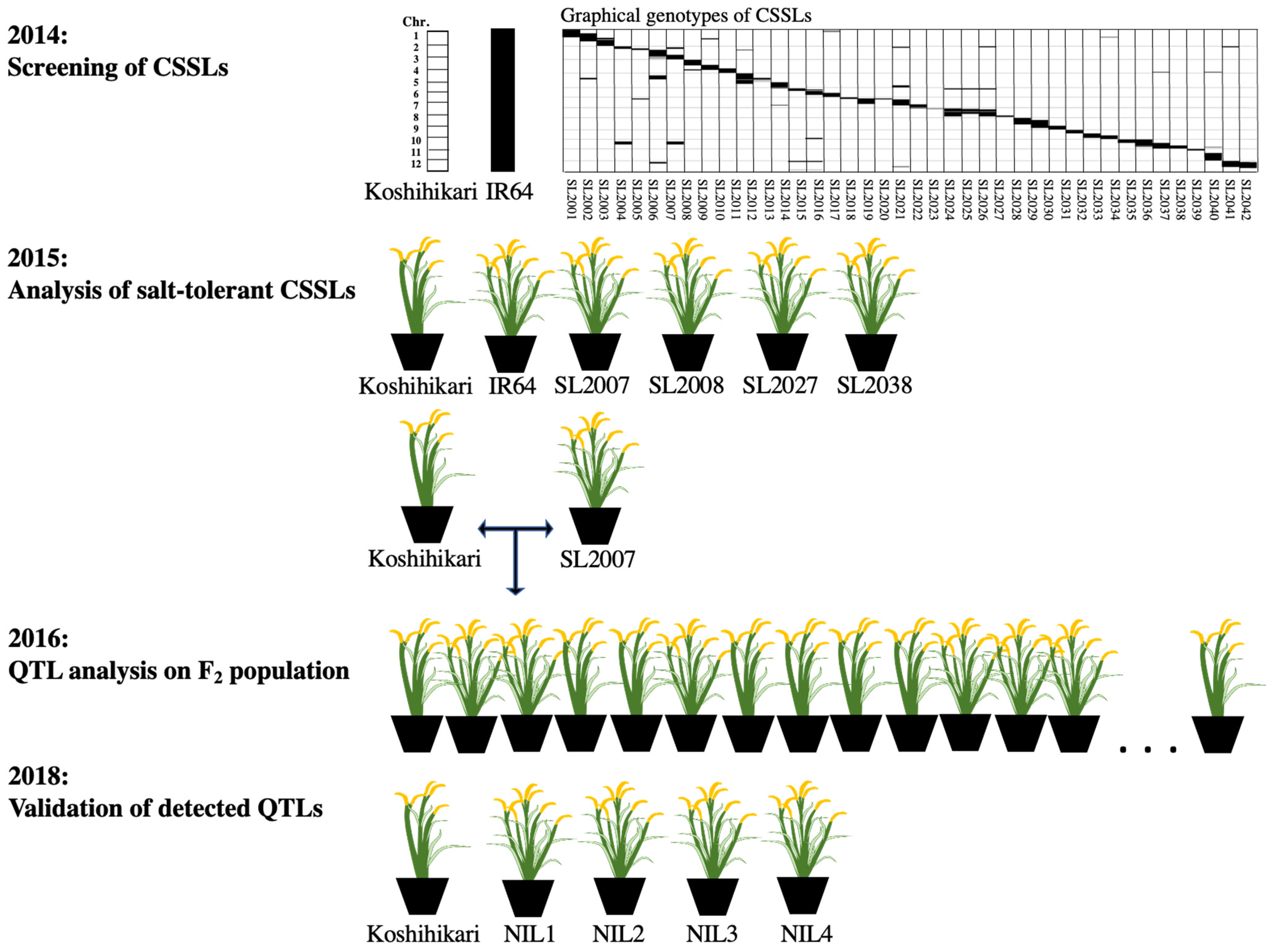
2.1. Plant Materials
2.2. Growing Conditions and Salt Treatment
2.3. Phenotypic Evaluation
2.4. Ion Measurement of Selected Chromosome Segment Substitution Lines (CSSLs)
2.5. DNA Extraction and Polymerase Chain Reaction (PCR) Analysis
2.6. Quantitative Trait Locus (QTL) Mapping
2.7. Statistical Analysis
3. Results
3.1. Salt Tolerance Evaluation of CSSLs
3.2. Physio-Morphological Analysis of Salt-Tolerant CSSLs
3.2.1. Yield and Yield Components
3.2.2. Ion Contents
3.2.3. Correlation between Traits
3.3. Detection of QTLs for Salt Tolerance
3.4. Validation and Fine Mapping of Detected QTLs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- GRiSP. Rice Almanac, 4th ed.; International Rice Research Institute: Los Baños, Philippines, 2013. [Google Scholar]
- United Nations, Department of Economic and Social Affairs, Population Division. World Population Prospects: The 2012 Revision Highlights and Advance Tables; Working Paper No. ESA/P/WP.228; Department of Economic & Social Affairs: New York, NY, USA, 2013. [Google Scholar]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pessarakli, M. Handbook of Plant and Crop Stress, 2nd ed.; Marcel Dekker Inc.: New York, NY, USA, 1999. [Google Scholar]
- Rehman, S.; Harris, P.J.C.; Ashraf, A. Stress environments and their impact on crop production. In Abiotic Stress: Plant Resistance Through Breeding and Molecular Approaches; Food Products Press: New York, NY, USA, 2005. [Google Scholar]
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Singh, A.; Mithra, S.V.A.; Krishnamurthy, S.L.; Parida, S.K.; Jain, S.; Tiwari, K.K.; Kumar, P.; Rao, A.R.; Sharma, S.K.; et al. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 2015, 22, 133–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, I.N.B.L.; Kim, B.K.; Yoon, I.S.; Kim, K.H.; Kwon, T.R. Salt tolerance in rice: Focus on mechanisms and approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Heenan, D.P.; Lewin, L.G.; McCaffery, D.W. Salinity tolerance in rice varieties at different growth stages. Aust. J. Exp. Agric. 1988, 28, 343–349. [Google Scholar] [CrossRef]
- Makihara, D.; Tsuda, M.; Morita, M.; Hirai, Y.; Kuroda, T. Effect of salinity on the growth and development on rice (Oryza sativa L.) varieties. Jpn. J. Trop. Agric. 1999, 43, 285–294. [Google Scholar]
- Singh, R.K.; Flowers, T.J. Physiology and Molecular Biology of the Effects of Salinity on Rice, 3rd ed.; Taylor & Francis: Boca Raton, FL, USA, 2010. [Google Scholar]
- Zeng, L.; Shannon, M.C.; Grieve, C.M. Evaluation of salt tolerance in rice genotypes by multiple agronomic characters. Euphytica 2002, 127, 235–245. [Google Scholar] [CrossRef]
- Mohammadi, R.; Mendioro, M.S.; Diaz, G.Q.; Gregorio, Q.B.; Singh, R.K. Genetic analysis of salt tolerance at seedling and reproductive stages in rice (Oryza sativa). Plant Breed. 2014, 133, 548–559. [Google Scholar] [CrossRef]
- Khatun, S.; Rizzo, C.A.; Flowers, T.J. Genotypic variation in the effect of salinity on fertility in rice. Plant Soil. 1995, 173, 239–250. [Google Scholar] [CrossRef]
- Lutts, S.; Kinet, J.M.; Bouharmont, J. Changes in plant response to NaCI during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J. Exp. Bot. 1995, 46, 1843–1852. [Google Scholar] [CrossRef]
- Kumar, V.; Khare, T. Differential growth and yield responses of salt-tolerant and susceptible rice cultivars to individual (Na+ and Cl− ) and additive stress effects of NaCl. Acta Physiol. Plant. 2016, 38, 7. [Google Scholar] [CrossRef]
- Hussain, S.; Jun-hua, Z.; Chu, Z.; Lian-Feng, Z.; Xiao-Chuang, C.; Sheng-Miao, Y.; James, A.B.; Ji-Jie, H.; Qian-Yu, J. Effects of salt stress on rice growth, development characteristics, and the regulating ways: A review. J. Integr. Agric. 2017, 16, 2357–2374. [Google Scholar] [CrossRef] [Green Version]
- Amirjani, M.R. Effect of Salinity stress on growth, sugar content, pigments and enzyme activity of rice. Int. J. Bot. 2011, 7, 73–81. [Google Scholar] [CrossRef] [Green Version]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genomics. 2014, 2014, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flowers, T.J.; Yeo, A.R. Variability in the resistance of sodium chloride salinity within rice (Oryza sativa L.) varieties. New Phytol. 1981, 88, 363–373. [Google Scholar] [CrossRef]
- Ueda, A.; Yahagi, H.; Fujikawa, Y.; Nagaoka, T.; Esaka, M.; Calcaño, M.; González, M.M.; Martich, J.D.H.; Saneoka, H. Comparative physiological analysis of salinity tolerance in rice. Soil Sci. Plant Nutr. 2013, 59, 896–903. [Google Scholar] [CrossRef]
- Platten, J.D.; Egdane, J.A.; Ismail, A.M. Salinity tolerance, Na+ exclusion and allele mining of HKT1;5 in Oryza sativa and O. glaberrima: Many sources, many genes, one mechanism? BMC Plant Biol. 2013, 13, 32. [Google Scholar] [CrossRef] [Green Version]
- Mekawy, A.M.M.; Assaha, D.V.M.; Yahagi, H.; Tada, Y.; Ueda, A.; Saneoka, H. Growth, physiological adaptation, and gene expression analysis of two Egyptian rice cultivars under salt stress. Plant Physiol. Biochem. 2015, 87, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Gerona, M.E.B.; Deocampo, M.P.; Egdane, J.A.; Ismail, A.M.; Dionisio-Sese, M.L. Physiological Responses of Contrasting Rice Genotypes to Salt Stress at Reproductive Stage. Rice Sci. 2019, 26, 207–219. [Google Scholar] [CrossRef]
- Thuy, N.T.T.; Tokuyasu, M.; Mai, N.S.; Hirai, Y. Identification and characterization of chromosome regions associated with salinity tolerance in rice. J. Agric. Sci. 2018, 10, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.X.; Zhu, M.Z.; Yano, M.; Gao, J.P.; Liang, Z.W.; Su, W.A.; Hu, X.H.; Ren, Z.H.; Chao, D.Y. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 2004, 108, 253–260. [Google Scholar] [CrossRef]
- Lee, S.Y.; Ahn, J.H.; Cha, Y.S.; Yun, D.W.; Lee, M.C.; Ko, J.C.; Lee, K.S.; Eun, M.Y. Mapping of quantitative trait loci for salt tolerance at the seedling stage in rice. Mol. Cells. 2006, 21, 192–196. [Google Scholar] [PubMed]
- Kim, D.M.; Ju, H.G.; Kwon, T.R.; Oh, C.S.; Ahn, S.N. Mapping QTLs for salt tolerance in an introgression line population between Japonica cultivars in rice. J. Crop Sci. Biotechnol. 2009, 12, 121–128. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, Y.; Meng, L.; Hu, X.; Cui, Y.; Sun, Y.; Zhu, L.; Ali, J.; Xu, J.; Li, Z. Identification of salt-tolerant QTLs with strong genetic background effect using two sets of reciprocal introgression lines in rice. Genome 2012, 55, 45–55. [Google Scholar] [CrossRef]
- Leon, T.B.D.; Linscombe, S.; Subudhi, P.K. Identification and validation of QTLs for seedling salinity tolerance in introgression lines of a salt tolerant rice landrace ‘Pokkali’. PLoS ONE 2017, 12, 1–30. [Google Scholar]
- Puram, V.R.R.; Ontoy, J.; Subudhi, P.K. Identification of QTLs for salt tolerance traits and prebreeding lines with enhanced salt tolerance in an introgression line population of rice. Plant Mol. Biol. Report. 2018, 36, 695–709. [Google Scholar] [CrossRef]
- Rahman, M.A.; Thomson, M.J.; Ocampo, M.D.; Egdane, J.A.; Salam, M.A.; Shah-E-Alam, M.; Ismail, A.M. Assessing trait contribution and mapping novel QTL for salinity tolerance using the Bangladeshi rice landrace Capsule. Rice 2019, 12, 12–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Yang, J.; Yu, D.; Xu, P. Identification and validation a major QTL from ‘Sea Rice 86’ seedlings conferred salt tolerance. Agronomy 2020, 10, 410. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Wang, X.; Zhao, Y.; Khan, N.U.; Zhao, Z.; Zhang, Y.; Wen, X.; Tang, F.; Wang, F.; Li, Z. Genetic basis and identification of candidate genes for salt tolerance in rice by GWAS. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nakhla, W.R.; Sun, W.; Fan, K.; Yang, K.; Zhang, C.; Yu, S. Identification of QTLs for salt tolerance at the germination and seedling stages in rice. Plants 2021, 10, 428. [Google Scholar] [CrossRef]
- Bonilla, P.; Dvorak, J.; Mackill, D.; Deal, K.; Gregorio, G. RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp. Agric. Sci. 2002, 65, 68–76. [Google Scholar]
- Sevanthi, A.M.; Prakash, C.; Shanmugavadivel, C. Recent progress in rice varietal development for abiotic stress tolerance. In Advances in Rice Research for Abiotic Stress Tolerance; Woodhead Publishing: Duxford, UK, 2019; pp. 47–68. [Google Scholar]
- Krishnamurthy, S.L.; Pundir, P.; Warraich, A.S.; Rathor, S.; Lokeshkumar, B.M.; Singh, N.K.; Sharma, P.C. Introgressed Saltol QTL lines improves the salinity tolerance in rice at seedling stage. Front. Plant Sci. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Deng, P.; Cao, C.; Zhang, W. Fine mapping of qSKC-1, a major quantitative trait locus for shoot K+ concentration, in rice seedlings grown under salt stress. Breed. Sci. 2017, 67, 286–295. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, R.; Mendioro, M.S.; Diaz, G.Q.; Gregorio, G.B.; Singh, R.K. Mapping quantitative trait loci associated with yield and yield components under reproductive stage salinity stress in rice (Oryza sativa L.). J. Genet. 2013, 92, 433–443. [Google Scholar] [CrossRef]
- Bimpong, I.K.; Manneh, B.; Diop, B.; Ghislain, K.; Sow, A.; Amoah, N.K.A.; Gregorio, G.; Singh, R.K.; Ortiz, R.; Wopereis, M. New quantitative trait loci for enhancing adaptation to salinity in rice from Hasawi, a Saudi landrace into three African cultivars at the reproductive stage. Euphytica 2014, 200, 45–60. [Google Scholar] [CrossRef]
- Hossain, H.; Rahman, M.A.; Alam, M.S.; Singh, R.K. Mapping of quantitative trait loci associated with reproductive-stage salt tolerance in rice. J. Agron. Crop Sci. 2015, 201, 17–31. [Google Scholar] [CrossRef]
- Chattopadhyay, K.; Mohanty, S.K.; Vijayan, J.; Marndi, B.C.; Sarkar, A.; Molla, K.A.; Chakraborty, K.; Ray, S.; Sarkar, R.K. Genetic dissection of component traits for salinity tolerance at reproductive stage in rice. Plant Mol. Biol. Report. 2021, 39, 386–402. [Google Scholar] [CrossRef]
- Mondal, S.; Borromeo, T.H.; Diaz, M.G.Q.; Amas, J.; Rahman, M.A.; Thomson, M.J.; Gregorio, G.B. Dissecting qtls for reproductive stage salinity tolerance in rice from BRRI dhan 47. Plant Breed. Biotechnol. 2019, 7, 302–312. [Google Scholar] [CrossRef]
- Noor, A.U.Z.; Jewel, G.M.N.A.; Haque, T.; Elias, S.M.; Biswas, S.; Rahman, M.S.; Seraj, Z.I. Validation of QTLs in Bangladeshi rice landrace Horkuch responsible for salt tolerance in seedling stage and maturation. Acta Physiol. Plant. 2019, 41, 1–15. [Google Scholar] [CrossRef]
- Yeo, A.R.; Yeo, M.E.; Flowers, S.A.; Flowers, T.J. Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor. Appl. Genet. 1990, 79, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Prusty, M.R.; Kim, S.R.; Vinarao, R.; Entila, F.; Egdane, J.; Diaz, M.G.Q.; Jena, K.K. Newly identified wild rice accessions conferring high salt tolerance might use a tissue tolerance mechanism in leaf. Front. Plant Sci. 2018, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Mackill, D.J.; Khush, G.S. IR64: A high-quality and high-yielding mega variety. Rice 2018, 2018, 11–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagata, K.; Ando, T.; Nonoue, Y.; Mizubayashi, T.; Kitazawa, N.; Shomura, A.; Matsubara, K.; Ono, N.; Mizobuchi, R.; Shibaya, T.; et al. Advanced backcross QTL analysis reveals complicated genetic control of rice grain shape in a japonica × indica cross. Breed. Sci. 2015, 65, 308–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shavrukov, Y.; Gupta, N.K.; Miyazaki, J.; Baho, M.N.; Chalmers, K.J.; Tester, M.; Langridge, P.; Collins, N.C. HvNax3—A locus controlling shoot sodium exclusion derived from wild barley (Hordeum vulgare ssp. spontaneum). Funct. Integr. Genomics 2010, 10, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Chai, L.; Zhang, J.; Pan, X.; Zhang, F.; Zheng, T.; Zhao, X.; Wang, W.; Jauhar, A.; Xu, J.; Li, Z. Advanced backcross QTL analysis for the whole plant growth duration salt tolerance in rice (Oryza sativa L.). J. Integr. Agric. 2014, 13, 1609–1620. [Google Scholar] [CrossRef]
- Pundir, P.; Devi, A.; Krishnamurthy, S.L.; Sharma, P.C.; Vinaykumar, N.M. QTLs in salt rice variety CSR10 reveals salinity tolerance at reproductive stage. Acta Physiol. Plant. 2021, 43, 1–15. [Google Scholar] [CrossRef]
- Mai, N.S.; Hirai, Y. QTL mapping for salt tolerance at reproductive stage in rice: A Minireview. J. Environ. Sci. Sustain. Soc. 2021, 10, 31–34. [Google Scholar]
- Takai, T.; Nonoue, Y.; Yamamoto, S.; Yamanouchi, U.; Matsubara, K.; Liang, Z.W.; Lin, H.; Ono, N.; Uga, Y.; Yano, M. Development of Chromosome segment substitution lines derived from backcross between indica donor rice cultivar ‘Nona Bokar’ and japonica recipient cultivar ‘Koshihikari’. Jpn. Soc. Breed. 2007, 57, 257–261. [Google Scholar] [CrossRef] [Green Version]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horie, T.; Karahara, I.; Katsuhara, M. Salinity tolerance mechanisms in glycophytes: An overview with the central focus on rice plants. Rice 2012, 5, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; Shannon, M.C. Salinity effects on seedling growth and yield components of rice. Crop Sci. 2000, 40, 996–1003. [Google Scholar] [CrossRef] [Green Version]
- Fahad, S.; Adnan, M.; Noor, M.; Arif, M.; Alam, M.; Khan, I.A.; Ullah, H.; Wahid, F.; Mian, I.A.; Jamal, Y.; et al. Major constrains for global rice production. In Advances in Rice Research for Abiotic Stress Tolerance; Woodhead Publishing: Duxford, UK, 2019. [Google Scholar]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.F.; Chao, D.Y.; Shi, M.; Zhu, M.Z.; Gao, J.P.; Lin, H.X. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008, 228, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Fukumoto, T.; Matsumoto, T.; Gena, P.; Frascaria, D.; Kaneko, T.; Katsuhara, M.; Zhong, S.; Sun, X.; Zhu, Y.; et al. Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination. Plant Physiol. Biochem. 2013, 63, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Lee, S.C.; Kim, J.Y.; Kim, S.J.; Aye, S.S.; Kim, S.R. Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J. Plant Biol. 2014, 57, 383–393. [Google Scholar] [CrossRef]
- Mizuno, H.; Kawahara, Y.; Sakai, H.; Kanamori, H.; Wakimoto, H.; Yamagata, H.; Oono, Y.; Wu, J.; Ikawa, H.; Itoh, T.; et al. Massive parallel sequencing of mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.). BMC Genomics 2010, 11, 683. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Na+ Content (mmol/g) | Cl− Content (mmol/g) | |||||
|---|---|---|---|---|---|---|
| LB | LS | WP | LB | LS | WP | |
| Koshihikari | 1.19 ± 0.11 | 1.41 ± 0.17 | 1.26 ± 0.16 | 1.70 ± 0.12 | 1.43 ± 0.12 | 1.25 ± 0.16 |
| IR64 | 0.70 ± 0.14 * | 0.77 ± 0.15 *** | 0.70 ± 0.10 *** | 1.04 ± 0.17 *** | 1.01 ± 0.09 ** | 0.90 ± 0.06 * |
| SL2007 | 0.72 ± 0.07 * | 0.92 ± 0.09 ** | 0.77 ± 0.06 ** | 1.20 ± 0.10 ** | 1.04 ± 0.05 ** | 0.81 ± 0.04 ** |
| SL2008 | 0.76 ± 0.05 * | 1.06 ± 0.04 * | 0.89 ± 0.07 * | 1.18 ± 0.01 ** | 1.19 ± 0.05 | 1.17 ± 0.08 |
| SL2027 | 0.81 ± 0.10 | 1.39 ± 0.10 | 1.15 ± 0.14 | 1.26 ± 0.12 * | 1.43 ± 0.07 | 1.44 ± 0.10 |
| SL2038 | 0.83 ± 0.10 | 0.97 ± 0.14 * | 0.85 ± 0.15 * | 1.34 ± 0.11 * | 1.14 ± 0.10 * | 0.97 ± 0.11 |
| DW | PW | PN | GY | SN | TGW | GF | ||
|---|---|---|---|---|---|---|---|---|
| Na+ content | LB | −0.510 * | −0.352 | −0.475 * | −0.358 | 0.215 | −0.257 | −0.634 ** |
| LS | −0.615 ** | −0.540 * | −0.640 ** | −0.535 * | 0.009 | −0.596 ** | −0.589 ** | |
| WP | −0.643 ** | −0.536 * | −0.671 ** | −0.535 * | 0.049 | −0.557 * | −0.602 ** | |
| Cl− content | LB | −0.439 | −0.275 | −0.397 | −0.263 | 0.298 | −0.196 | −0.543 * |
| LS | −0.621 ** | −0.548 * | −0.659 ** | −0.549 * | −0.029 | −0.558 * | −0.585 ** | |
| WP | −0.602 ** | −0.579 ** | −0.634 ** | −0.582 ** | −0.301 | −0.669 ** | −0.536 * |
| Trait | QTL | Marker Interval (Mb) | LOD | Additive | R2 | Parental Source a |
|---|---|---|---|---|---|---|
| Plant dry weight | qSTDW2 | RM13648-RM13755 (25.47–27.45) | 2.89 | −6.44 | 9.91 | I |
| Panicle number | qSTPN2 | RM13648-RM13893 (25.47–29.71) | 4.70 | −3.67 | 33.09 | I |
| Panicle weight | qSTPW2.1 | RM12674-RM12857 (5.69–8.63) | 2.71 | 1.07 | 10.39 | K |
| qSTPW2.2 | RM13755-RM13893 (27.45–29.71) | 2.83 | −1.06 | 24.51 | I | |
| Grain yield | qSTGY2.1 | RM12674-RM12857 (5.69–8.63) | 3.85 | 0.50 | 23.64 | K |
| qSTGY2.2 | RM13796-RM5607 (28.27–33.27) | 3.72 | −0.47 | 31.84 | I |
| DW | PN | PW | GY | GF | TGW | SN | |
|---|---|---|---|---|---|---|---|
| DW | 1 | ||||||
| PN | 0.675 * | 1 | |||||
| PW | 0.433 | 0.368 | 1 | ||||
| GY | 0.275 | 0.173 | 0.950 *** | 1 | |||
| GF | −0.061 | −0.289 | 0.698 *** | 0.859 *** | 1 | ||
| TGW | 0.072 | 0.163 | −0.312 | −0.323 | −0.438 | 1 | |
| SN | 0.250 | −0.204 | 0.582 ** | 0.548 * | 0.542 * | −0.431 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mai, N.S.; Hanh, D.D.; Nakashima, M.; Kumamoto, K.; Thuy, N.T.T.; Kobata, T.; Saitoh, K.; Hirai, Y. Identification and Validation of QTLs for Yield and Yield Components under Long-Term Salt Stress Using IR64 CSSLs in the Genetic Background of Koshihikari and Their Backcross Progenies. Agriculture 2021, 11, 777. https://doi.org/10.3390/agriculture11080777
Mai NS, Hanh DD, Nakashima M, Kumamoto K, Thuy NTT, Kobata T, Saitoh K, Hirai Y. Identification and Validation of QTLs for Yield and Yield Components under Long-Term Salt Stress Using IR64 CSSLs in the Genetic Background of Koshihikari and Their Backcross Progenies. Agriculture. 2021; 11(8):777. https://doi.org/10.3390/agriculture11080777
Chicago/Turabian StyleMai, Nguyen Sao, Dao Duy Hanh, Mai Nakashima, Kotaro Kumamoto, Nguyen Thi Thu Thuy, Tohru Kobata, Kuniyuki Saitoh, and Yoshihiko Hirai. 2021. "Identification and Validation of QTLs for Yield and Yield Components under Long-Term Salt Stress Using IR64 CSSLs in the Genetic Background of Koshihikari and Their Backcross Progenies" Agriculture 11, no. 8: 777. https://doi.org/10.3390/agriculture11080777






