ScRpb4, Encoding an RNA Polymerase Subunit from Sugarcane, Is Ubiquitously Expressed and Resilient to Changes in Response to Stress Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval and Phylogenetic Reconstruction
2.2. Structural Analysis of the Sugarcane RPB4 Protein (ScRpb4)
2.3. Cloning of the Sugarcane RNA Polymerase II
2.4. Induction and Expression Conditions of ScRpb4
2.5. ScRpb4 Gene Expression in Vegetative and Reproductive Tissues under Stress Conditions
2.6. In Situ Hybridization
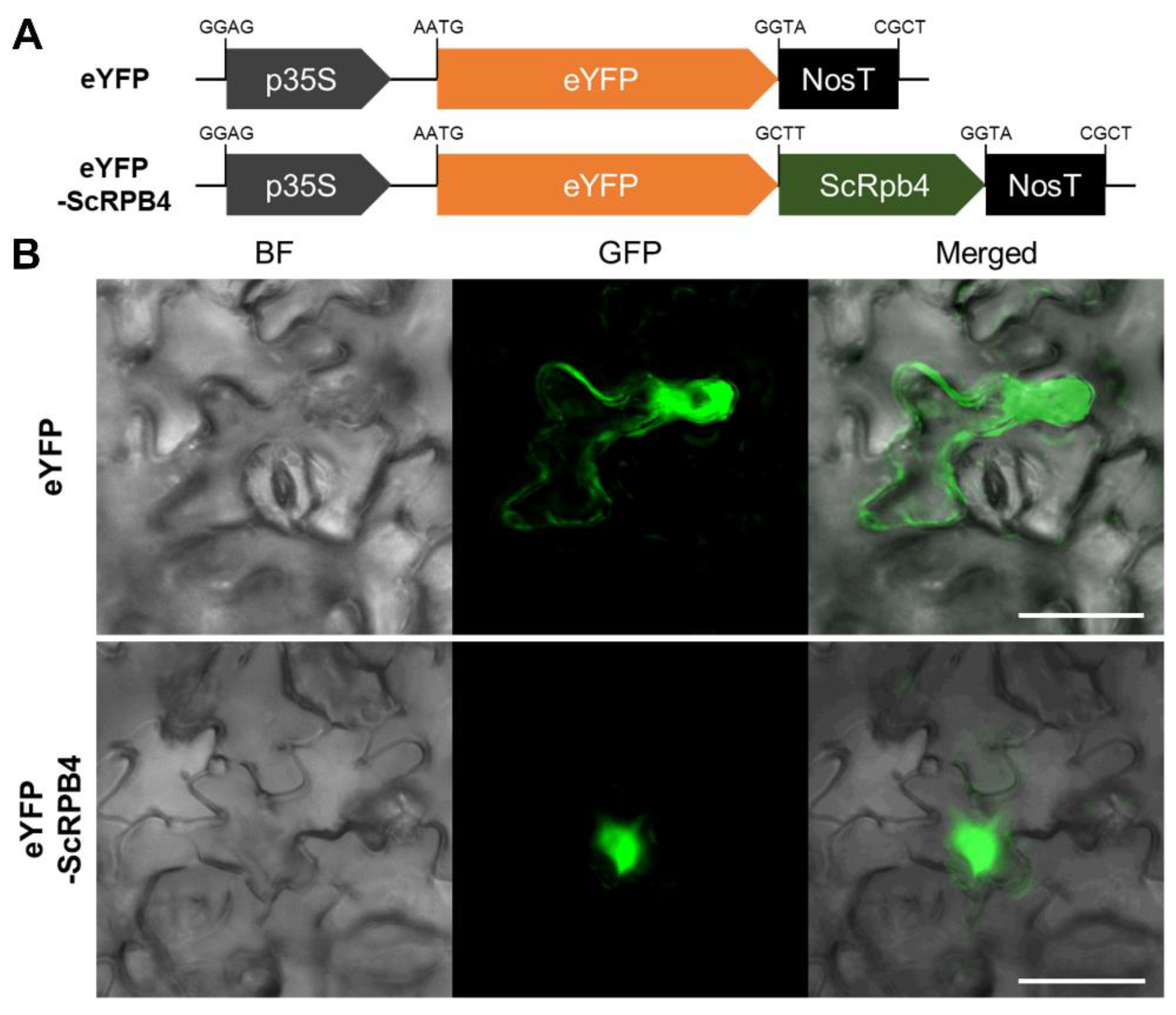
2.7. ScRpb4 Subcellular Localization
3. Results
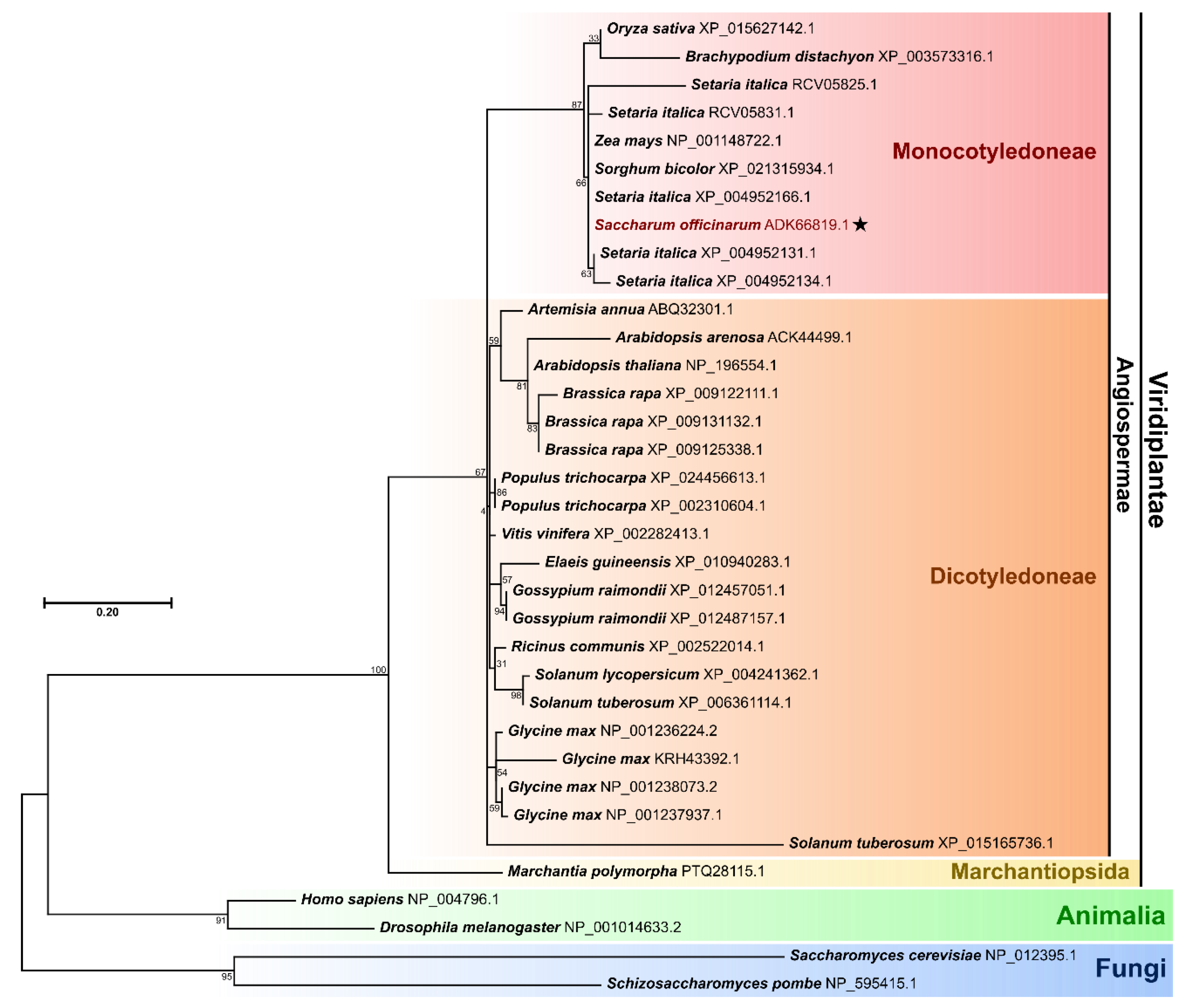
3.1. Sugarcane Rpb4 Is Conserved across Kingdoms
3.2. Nuclear ScRpb4 Localization
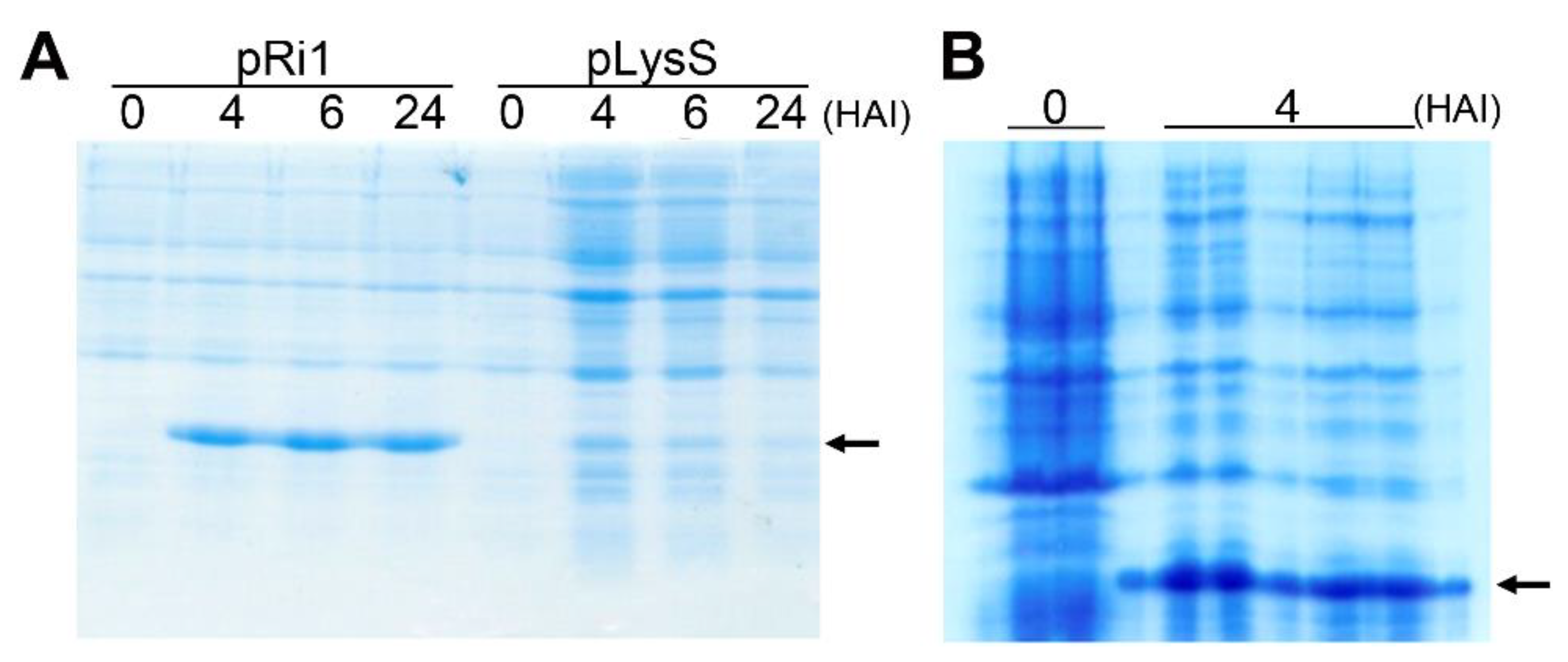
3.3. In Vitro Expression of the ScRpb4 Protein
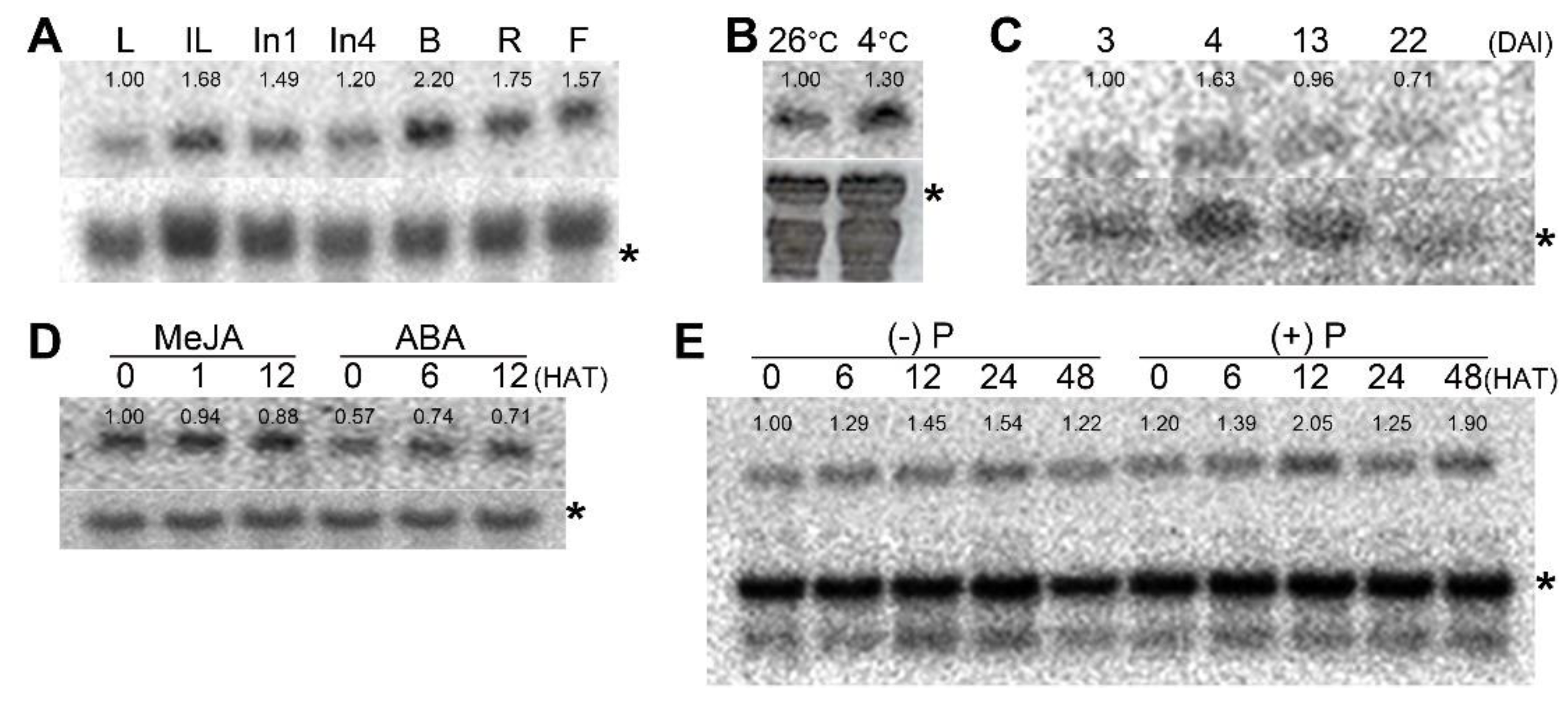
3.4. ScRpb4 Is Expressed in Vegetative and Reproductive Tissues
3.5. ScRpb4 Expression Is Maintained under Stress Conditions in Different Plant Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaufmann, K.; Pajoro, A.; Angenent, G.C. Regulation of Transcription in Plants: Mechanisms Controlling Developmental Switches. Nat. Rev. Genet. 2010, 11, 830–842. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Networks in Response to Abiotic Stresses in Arabidopsis and Grasses. Plant. Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eick, D.; Geyer, M. The RNA Polymerase II Carboxy-Terminal Domain (CTD) Code. Chem. Rev. 2013, 113, 8456–8490. [Google Scholar] [CrossRef]
- Antosz, W.; Deforges, J.; Begcy, K.; Bruckmann, A.; Poirier, Y.; Dresselhaus, T.; Grasser, K.D. Critical Role of Transcript Cleavage in Arabidopsis RNA Polymerase II Transcriptional Elongation. Plant. Cell 2020, 32, 1449–1463. [Google Scholar] [CrossRef] [Green Version]
- Barba-Aliaga, M.; Alepuz, P.; Pérez-Ortín, J.E. Eukaryotic RNA Polymerases: The Many Ways to Transcribe a Gene. Front. Mol. Biosci. 2021, 8, 663209. [Google Scholar] [CrossRef] [PubMed]
- Allepuz-Fuster, P.; Martínez-Fernández, V.; Garrido-Godino, A.I.; Alonso-Aguado, S.; Hanes, S.D.; Navarro, F.; Calvo, O. Rpb4/7 Facilitates RNA Polymerase II CTD Dephosphorylation. Nucleic Acids Res. 2014, 42, 13674–13688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKune, K.; Richards, K.L.; Edwards, A.M.; Young, R.A.; Woychik, N.A. RPB7, One of Two Dissociable Subunits of Yeast RNA Polymerase II, Is Essential for Cell Viability. Yeast 1993, 9, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Woychik, N.A.; Young, R.A. RNA Polymerase II Subunit RPB4 Is Essential for High- and Low-Temperature Yeast Cell Growth. Mol. Cell. Biol. 1989, 9, 2854–2859. [Google Scholar] [PubMed] [Green Version]
- Choder, M.; Young, R.A. A Portion of RNA Polymerase II Molecules Has a Component Essential for Stress Responses and Stress Survival. Mol. Cell. Biol. 1993, 13, 6984–6991. [Google Scholar] [PubMed] [Green Version]
- Choder, M. Rpb4 and Rpb7: Subunits of RNA Polymerase II and Beyond. Trends Biochem. Sci. 2004, 29, 674–681. [Google Scholar] [CrossRef]
- Sampath, V.; Rekha, N.; Srinivasan, N.; Sadhale, P. The Conserved and Non-Conserved Regions of Rpb4 Are Involved in Multiple Phenotypes in Saccharomyces Cerevisiae. J. Biol. Chem. 2003, 278, 51566–51576. [Google Scholar] [CrossRef] [Green Version]
- Meka, H.; Werner, F.; Cordell, S.C.; Onesti, S.; Brick, P. Crystal Structure and RNA Binding of the Rpb4/Rpb7 Subunits of Human RNA Polymerase II. Nucleic Acids Res. 2005, 33, 6435–6444. [Google Scholar] [CrossRef]
- Larkin, R.M.; Guilfoyle, T.J. Two Small Subunits in Arabidopsis RNA Polymerase II Are Related to Yeast RPB4 and RPB7 and Interact with One Another. J. Biol. Chem. 1998, 273, 5631–5637. [Google Scholar] [CrossRef] [Green Version]
- Ream, T.S.; Haag, J.R.; Wierzbicki, A.T.; Nicora, C.D.; Norbeck, A.D.; Zhu, J.-K.; Hagen, G.; Guilfoyle, T.J.; Paša-Tolić, L.; Pikaard, C.S. Subunit Compositions of the RNA-Silencing Enzymes Pol IV and Pol V Reveal Their Origins as Specialized Forms of RNA Polymerase II. Mol. Cell 2009, 33, 192–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.-J.; Hsu, Y.-F.; Pontes, O.; Zhu, J.; Lu, J.; Bressan, R.A.; Pikaard, C.; Wang, C.-S.; Zhu, J.-K. NRPD4, a Protein Related to the RPB4 Subunit of RNA Polymerase II, Is a Component of RNA Polymerases IV and V and Is Required for RNA-Directed DNA Methylation. Genes Dev. 2009, 23, 318–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formann, S.; Hahn, A.; Janke, L.; Stinner, W.; Sträuber, H.; Logroño, W.; Nikolausz, M. Beyond Sugar and Ethanol Production: Value Generation Opportunities Through Sugarcane Residues. Front. Energy Res. 2020, 8, 579577. [Google Scholar] [CrossRef]
- Thirugnanasambandam, P.P.; Hoang, N.V.; Henry, R.J. The Challenge of Analyzing the Sugarcane Genome. Front. Plant. Sci. 2018, 9, 616. [Google Scholar] [CrossRef] [Green Version]
- Papini-Terzi, F.S. Transcription Profiling of Signal Transduction-Related Genes in Sugarcane Tissues. DNA Res. 2005, 12, 27–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, T.H.; Gentile, A.; Vilela, R.D.; Costa, G.G.L.; Dias, L.I.; Endres, L.; Menossi, M. MicroRNAs Associated with Drought Response in the Bioenergy Crop Sugarcane (Saccharum Spp.). PLoS ONE 2012, 7, e46703. [Google Scholar] [CrossRef] [Green Version]
- Manechini, J.R.V.; Santos, P.H.d.S.; Romanel, E.; Brito, M.d.S.; Scarpari, M.S.; Jackson, S.; Pinto, L.R.; Vicentini, R. Transcriptomic Analysis of Changes in Gene Expression During Flowering Induction in Sugarcane Under Controlled Photoperiodic Conditions. Front. Plant. Sci. 2021, 12, 635784. [Google Scholar] [CrossRef]
- Folsom, J.J.; Begcy, K.; Hao, X.; Wang, D.; Walia, H. Rice Fertilization-Independent Endosperm1 Regulates Seed Size under Heat Stress by Controlling Early Endosperm Development. Plant. Physiol. 2014, 165, 238–248. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Begcy, K.; Liu, K.; Folsom, J.J.; Wang, Z.; Zhang, C.; Walia, H. Heat Stress Yields a Unique MADS Box Transcription Factor in Determining Seed Size and Thermal Sensitivity. Plant. Physiol. 2016, 171, 606–622. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic Stress Responses in Plants. Nat. Rev. Genet. 2021. Available online: https://www.nature.com/articles/s41576-021-00413-0 (accessed on 6 January 2022).
- Chung, W.-H.; Craighead, J.L.; Chang, W.-H.; Ezeokonkwo, C.; Bareket-Samish, A.; Kornberg, R.D.; Asturias, F.J. RNA Polymerase II/TFIIF Structure and Conserved Organization of the Initiation Complex. Mol. Cell 2003, 12, 1003–1013. [Google Scholar] [CrossRef]
- He, Y.; Yan, C.; Fang, J.; Inouye, C.; Tjian, R.; Ivanov, I.; Nogales, E. Near-Atomic Resolution Visualization of Human Transcription Promoter Opening. Nature 2016, 533, 359–365. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.; Samraj, S.; Jiménez, J.; Gómez, C.; Liu, T.; Begcy, K. Genome-Wide Identification of Heat Shock Factors and Heat Shock Proteins in Response to UV and High Intensity Light Stress in Lettuce. BMC Plant. Biol. 2021, 21, 185. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for MacOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Begcy, K.; Mariano, E.D.; Lembke, C.G.; Zingaretti, S.M.; Souza, G.M.; Araújo, P.; Menossi, M. Overexpression of an Evolutionarily Conserved Drought-Responsive Sugarcane Gene Enhances Salinity and Drought Resilience. Ann. Bot. 2019, 124, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED Protein Analysis Workbench: 20 Years On. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef] [Green Version]
- Needleman, S.B.; Wunsch, C.D. A General Method Applicable to the Search for Similarities in the Amino Acid Sequence of Two Proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Vettore, A.L.; Silva, F.R.d.; Kemper, E.L.; Arruda, P. The Libraries That Made SUCEST. Genet. Mol. Biol. 2001, 24, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Werle, R.; Begcy, K.; Yerka, M.K.; Mower, J.P.; Dweikat, I.; Jhala, A.J.; Lindquist, J.L. Independent Evolution of Acetolactate Synthase–Inhibiting Herbicide Resistance in Weedy Sorghum Populations across Common Geographic Regions. Weed Sci. 2017, 65, 164–176. [Google Scholar] [CrossRef] [Green Version]
- Begcy, K.; Weigert, A.; Egesa, A.; Dresselhaus, T. Compared to Australian Cultivars, European Summer Wheat (Triticum Aestivum) Overreacts When Moderate Heat Stress Is Applied at the Pollen Development Stage. Agronomy 2018, 8, 99. [Google Scholar] [CrossRef] [Green Version]
- Behrendorff, J.B.Y.H.; Sandoval-Ibañez, O.A.; Sharma, A.; Pribil, M. Membrane-Bound Protein Scaffolding in Diverse Hosts Using Thylakoid Protein CURT1A. ACS Synth. Biol. 2019, 8, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S. A Modular Cloning System for Standardized Assembly of Multigene Constructs. PLoS ONE 2011, 6, e16765. [Google Scholar] [CrossRef]
- Marillonnet, S.; Grützner, R. Synthetic DNA Assembly Using Golden Gate Cloning and the Hierarchical Modular Cloning Pipeline. Curr. Protoc. Mol. Biol. 2020, 130, e115. [Google Scholar] [CrossRef]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional Roles in Stress Tolerance. Front. Plant. Sci. 2016, 7, 813. [Google Scholar] [CrossRef] [Green Version]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front. Plant. Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Begcy, K.; Mariano, E.D.; Gentile, A.; Lembke, C.G.; Zingaretti, S.M.; Souza, G.M.; Menossi, M. A Novel Stress-Induced Sugarcane Gene Confers Tolerance to Drought, Salt and Oxidative Stress in Transgenic Tobacco Plants. PLoS ONE 2012, 7, e44697. [Google Scholar] [CrossRef] [Green Version]
- Garcia Tavares, R.; Lakshmanan, P.; Peiter, E.; O’Connell, A.; Caldana, C.; Vicentini, R.; Soares, J.S.; Menossi, M. ScGAI Is a Key Regulator of Culm Development in Sugarcane. J. Exp. Bot. 2018, 69, 3823–3837. [Google Scholar] [CrossRef] [Green Version]
- Thibaud-Nissen, F.; Ouyang, S.; Buell, C.R. Identification and Characterization of Pseudogenes in the Rice Gene Complement. BMC Genom. 2009, 10, 317. [Google Scholar] [CrossRef] [Green Version]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [Green Version]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol. Plant. 2018, 11, 414–428. [Google Scholar] [CrossRef] [Green Version]
- Bushnell, D.A.; Kornberg, R.D. Complete, 12-Subunit RNA Polymerase II at 4.1-A Resolution: Implications for the Initiation of Transcription. Proc. Natl. Acad. Sci. USA 2003, 100, 6969–6973. [Google Scholar] [CrossRef] [Green Version]
- Kamenski, T.; Heilmeier, S.; Meinhart, A.; Cramer, P. Structure and Mechanism of RNA Polymerase II CTD Phosphatases. Mol. Cell 2004, 15, 399–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, M.; Suzuki, H.; Ishihama, A. Formation of a Carboxy-Terminal Domain Phosphatase (Fcp1)/TFIIF/RNA Polymerase II (Pol II) Complex in Schizosaccharomyces Pombe Involves Direct Interaction between Fcp1 and the Rpb4 Subunit of Pol II. Mol. Cell Biol. 2002, 22, 1577–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suh, M.-H.; Ye, P.; Zhang, M.; Hausmann, S.; Shuman, S.; Gnatt, A.L.; Fu, J. Fcp1 Directly Recognizes the C-Terminal Domain (CTD) and Interacts with a Site on RNA Polymerase II Distinct from the CTD. Proc. Natl. Acad. Sci. USA 2005, 102, 17314–17319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiina, T.; Tsunoyama, Y.; Nakahira, Y.; Khan, M.S. Plastid RNA Polymerases, Promoters, and Transcription Regulators in Higher Plants. In International Review of Cytology; Jeon, K.W., Ed.; Elsevier: Amsterdam, The Netherlands, 2005; Volume 244, pp. 1–68. [Google Scholar]
- Tudor, M.; Murray, P.J.; Onufryk, C.; Jaenisch, R.; Young, R.A. Ubiquitous Expression and Embryonic Requirement for RNA Polymerase II Coactivator Subunit Srb7 in Mice. Genes Dev. 1999, 13, 2365–2368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Patel, R.V.; Nahal, H.K.; Breit, R.; Provart, N.J. BAR Expressolog Identification: Expression Profile Similarity Ranking of Homologous Genes in Plant Species: Expression Profile Similarity Ranking of Homologous Genes. Plant. J. 2012, 71, 1038–1050. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-Box Proteins in Rice. Genome-Wide Analysis, Classification, Temporal and Spatial Gene Expression during Panicle and Seed Development, and Regulation by Light and Abiotic Stress. Plant. Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Ponnala, L.; Gandotra, N.; Wang, L.; Si, Y.; Tausta, S.L.; Kebrom, T.H.; Provart, N.; Patel, R.; Myers, C.R.; et al. The Developmental Dynamics of the Maize Leaf Transcriptome. Nat. Genet. 2010, 42, 1060–1067. [Google Scholar] [CrossRef]
- Downs, G.S.; Bi, Y.-M.; Colasanti, J.; Wu, W.; Chen, X.; Zhu, T.; Rothstein, S.J.; Lukens, L.N. A Developmental Transcriptional Network for Maize Defines Coexpression Modules. Plant. Physiol. 2013, 161, 1830–1843. [Google Scholar] [CrossRef] [Green Version]
- Schier, A.C.; Taatjes, D.J. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Genes Dev. 2020, 34, 465–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, D.; Pirkl, N.; Lehmann, E.; Cramer, P. Rpb4 Subunit Functions Mainly in MRNA Synthesis by RNA Polymerase II. J. Biol. Chem. 2014, 289, 17446–17452. [Google Scholar] [CrossRef] [Green Version]
- Zeller, G.; Henz, S.R.; Widmer, C.K.; Sachsenberg, T.; Rätsch, G.; Weigel, D.; Laubinger, S. Stress-Induced Changes in the Arabidopsis Thaliana Transcriptome Analyzed Using Whole-Genome Tiling Arrays. Plant. J. 2009, 58, 1068–1082. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.; Dias, F.O.; Gentile, A.; Menossi, M.; Begcy, K. ScRpb4, Encoding an RNA Polymerase Subunit from Sugarcane, Is Ubiquitously Expressed and Resilient to Changes in Response to Stress Conditions. Agriculture 2022, 12, 81. https://doi.org/10.3390/agriculture12010081
Kim T, Dias FO, Gentile A, Menossi M, Begcy K. ScRpb4, Encoding an RNA Polymerase Subunit from Sugarcane, Is Ubiquitously Expressed and Resilient to Changes in Response to Stress Conditions. Agriculture. 2022; 12(1):81. https://doi.org/10.3390/agriculture12010081
Chicago/Turabian StyleKim, Taehoon, Fábio Ometto Dias, Agustina Gentile, Marcelo Menossi, and Kevin Begcy. 2022. "ScRpb4, Encoding an RNA Polymerase Subunit from Sugarcane, Is Ubiquitously Expressed and Resilient to Changes in Response to Stress Conditions" Agriculture 12, no. 1: 81. https://doi.org/10.3390/agriculture12010081





