Abstract
Seeds are the most fundamental and significant production tool in agriculture. They play a critical role in boosting the output and revenue of agriculture. To achieve rapid identification and protection of maize seeds, 3938 images of 11 different types of maize seeds were collected for the experiment, along with a combination of germ and non-germ surface datasets. The training set, validation set, and test set were randomly divided by a ratio of 7:2:1. The experiment introduced the CBAM (Convolutional Block Attention Module) attention mechanism into MobileNetV2, improving the CBAM by replacing the cascade connection with a parallel connection, thus building an advanced mixed attention module, I_CBAM, and establishing a new model, I_CBAM_MobileNetV2. The proposed I_CBAM_MobileNetV2 achieved an accuracy of 98.21%, which was 4.88% higher than that of MobileNetV2. Compared to Xception, MobileNetV3, DenseNet121, E-AlexNet, and ResNet50, the accuracy was increased by 9.24%, 6.42%, 3.85%, 3.59%, and 2.57%, respectively. Gradient-Weighted Class Activation Mapping (Grad-CAM) network visualization demonstrates that I_CBAM_MobileNetV2 focuses more on distinguishing features in maize seed images, thereby boosting the accuracy of the model. Furthermore, the model is only 25.1 MB, making it suitable for portable deployment on mobile terminals. This study provides effective strategies and experimental methods for identifying maize seed varieties using deep learning technology. This research provides technical assistance for the non-destructive detection and automatic identification of maize seed varieties.
1. Introduction
Maize (Zea mays L.) is an annual herb in the Poaceae family, and is one of the world’s most important food crops, grown in the United States, China, Brazil, and other countries. Maize is also an important source of feed for animal husbandry and farming, as well as one of the most important raw materials in various industries [1,2]. Variety purity is an important criterion for seed quality testing. Because of the morphological characteristics and visual similarity of maize seeds, even experts struggle to identify them with the naked eye, necessitating a significant amount of labor and time for identification.
In recent years, computer-vision technology has been widely adopted in agriculture. Yang Hang [3] applied the Wilk-lambda stepwise discriminant method for band selection, and established a discriminant model. The average recognition accuracy of five types of maize seed was found to be 91.6%, except GAOYOU115, which was found to be 87%. Cheng Hong [4] used the Support Vector Machine (SVM) algorithm to train the maize seed images, and the recognition accuracy reached 92.3%. Yang Shuqin [5] used the BP artificial neural network method to perform the training recognition, and the results showed that the overall identification was 93% for four varieties of maize. Moges T.G. [6] proposed a hybrid Convolutional Neural Network (CNN) and HOG features trained with an SVM classifier, and the recognition rate was observed to be 99%.
Deep learning is a crucial branch of machine learning. Compared with conventional machine learning, deep learning reduces the incompleteness of the manually extracted features, and can automatically extract several complex features from the input data, with stronger objectivity [7,8,9]. The VGG16 network was used by Tu K. [10] to identify the authenticity of the maize variety “JINGKE 968”, and the results highlighted that the best detection accuracy exceeded 99%. Zhou Q. [11] proposed a method for CNN to reshape the pixel spectral images. The results demonstrated that for six varieties of common maize seeds, the test accuracy of germ and non-germ surfaces was 93.33% and 95.56%, respectively. For six varieties of sweet maize seeds, the test accuracy of germ and non-germ surfaces was 97.78% and 98.15%, respectively. Kurtulmus F. [12] used multiple deep-learning methods to identify four varieties of sunflower seeds, and GoogleNet succeeded in achieving the highest classification accuracy (95%).
With the development of transfer learning [13] and CNN, unlike networks with huge parameters such as VGG16(Simonyan and Zisserman 2015), more and more deep lightweight neural networks such as DenseNet [14], NASNet [15], MobileNetV2 [16], SqueezeNet [17], and Xception [18] are being examined and focused on for ease of deployment on mobile terminals.
To classify crop diseases, Moyazzoma R. [19] used the MobileNetV2 network and obtained a validated accuracy of 90.38%. He further applied this method in agriculture to help farmers classify diseases from the harvest. Khan E. [20] proposed a deep learning-based technique to classify six different citrus diseases that severely impact the yield and quality of citrus fruits, using the whale optimization algorithm for optimization. The SqueezeNet model outperformed MobileNetV2, achieving an accuracy of 96%. Feng Xiao [21] used MobileNetV2 to construct maize seed images for variety identification. The results showed that the double-sided recognition accuracy of maize-kernel double-sided characteristics modeling was 99.83%, which was better than single-sided characteristics modeling and recognition. Elfatimi E. [22] classified the bean leaf dataset using the MobileNetV2 model, and achieved an average classification accuracy of over 97% on the training dataset, and over 92% on the test data. Jaithavil D. [23] used multiple CNNs to classify rice seed varieties, and the experimental results illustrated that the overall accuracy of VGG16, InceptionV3, and MobileNetV2 was 80.00%, 83.33%, and 83.33%, respectively. Zhang Z [24] proposed a rice disease identification system using lightweight MobileNetV2. He further compared it with the original model, and found that the memory usage was reduced by 74%, the number of floating-point operations per second was reduced by 49%, the number of parameters was reduced by 50%, and the accuracy of rice disease recognition was improved from 0.16% to 90.84%. Hamid Y. [25] used MobileNetV2 to classify 14 different categories of seeds, and the results demonstrated an accuracy of 98% and 95% for the training and test sets, respectively.
In recent years, attention mechanisms such as SENet [26], CBAM [27], ECA [28] have attracted extensive research, adding CNN to enhance learning ability. Wang S.H. [29] proposed a novel VGG-Style Base Network (VSBN) as a backbone network, introduced a Convolutional Block Attention Module (CBAM), and achieved sensitivity/precision/F1 all above 95%. Jia L [30] proposed an improved MobileNetV3 model that can perform near real-time detection on mobile terminals with an accuracy of 97%. Shahi T.B. [31] combined CBAM with MobileNetV2, and the results proved that it could outperform other methods, with fewer trainable parameters and higher classification accuracy. Zhu X. [32] established EfficientNet-B4-CBAM to identify Camellia oleifera cultivars. The overall accuracy of the EfficientNet-B4-CBAM model on the test dataset reached 97.02%, with a kappa coefficient of 0.96, which was significantly higher than that of the other methods used in the comparison experiments. Wang Meihua [33] proposed a new parallel mixed attention module, I_CBAM, which exhibited a better identification effect on the fine-grained classification of pests and diseases.
Yonis Gulzar [34] proposed using migration learning and CNN networks to classify 14 seeds with 99% accuracy, but the common seeds used in the paper have very different properties and are not very effective in identifying maize seeds with similar properties. Yahya [35] proposed using migration learning with VGG19 to identify haploid and polyploid maize seeds, with all metrics greater than 93%; however, the VGG19 model has a large number of parameters, a long training time, and the resulting model is unsuitable for mobile deployment. Aqib Ali [36] first used a machine learning algorithm to obtain nine optimized features from the collected maize images, and then used a random forest to construct the model; Zhou [11] used hyperspectral maize images combined with CNN to identify different varieties of maize seeds; and Cao Weishi [37] proposed a combination of DWT and BP neural networks to identify the purity of maize seeds with good results, but our proposed method eliminates the complex hyperspectral image acquisition steps and significantly reduces research costs. Kha-lied [38] proposed a model based on the MobileNetV2 architecture that could identify eight date species with 99% accuracy, demonstrating the superiority of CNNin image recognition. The distinction between the dates specified in this study was more obvious, and good recognition results were obtained by fine-tuning the MobileNetV2 network with a fully connected layer, but the improved method will still have a large model.
In this study, a new mixed attention module, I_CBAM, was constructed with MobileNetV2 as the benchmark model, and a more effective CNN I_CBAM_MobileNetV2 was proposed to achieve adaptive refinement of feature channels and spaces. This helps to solve the problem of mutual interference between two types of attention when CBAM is cascade-connected. In this experiment, based on the self-created dataset containing 3938 images of 11 maize seed varieties, several comparative experiments were conducted between I_CBAM_MobileNetV2, MobileNetV2, MobileNetV3, DenseNet121, Xception, ResNet50, and E-AlexNet. The results highlighted that the proposed I_CBAM_MobileNetV2 significantly outperforms other methods, and provides technical support for the automatic identification and non-destructive detection of maize seed varieties.
2. Materials and Methods
2.1. Maize Seed Image Collection
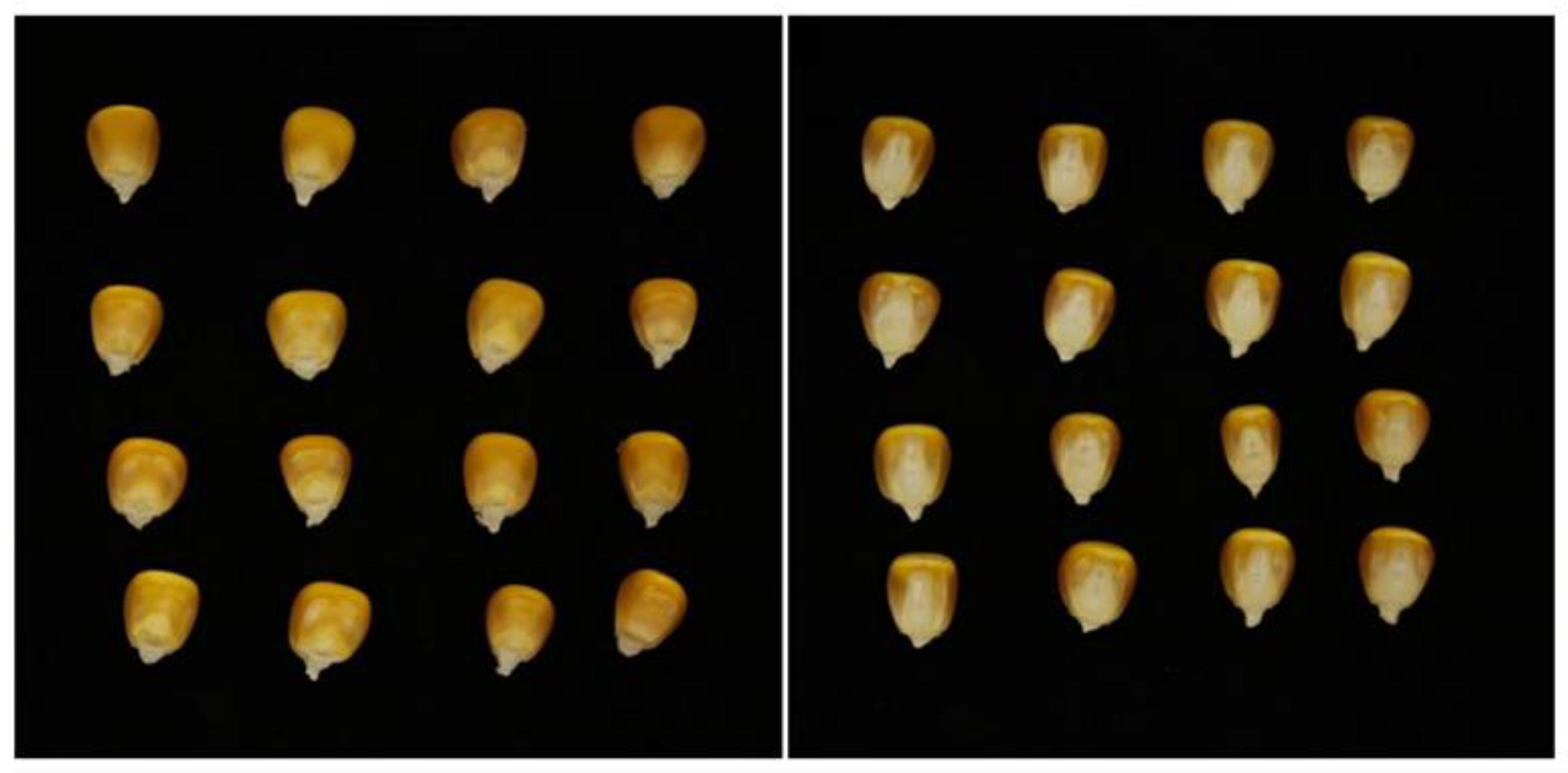
A total of 11 maize varieties were selected for this experiment, including AOYU 116, ZHENGDAN 958, XJH, JINGNIAN 1, KENUO 58, TIEYAN, DENGHAI 605, LIYUAN, JINYU 118, YUNYU, and BT506 (provided by Shandong Academy of Agricultural Sciences). A photographing console was built under natural lighting conditions in the laboratory, and a Canon EOS 80D digital camera at a distance of 50 cm was used to capture images of multi-grain maize seeds vertically, using black flannel as the background; the maize seeds were placed as represented in Figure 1.
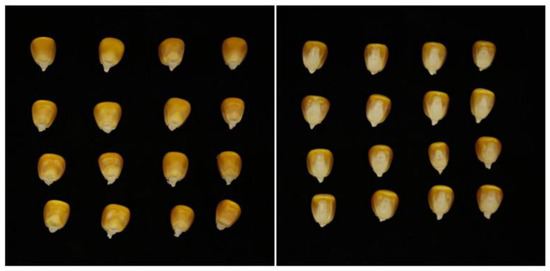
Figure 1.
Multi-grain maize seed images.
2.2. Image Processing
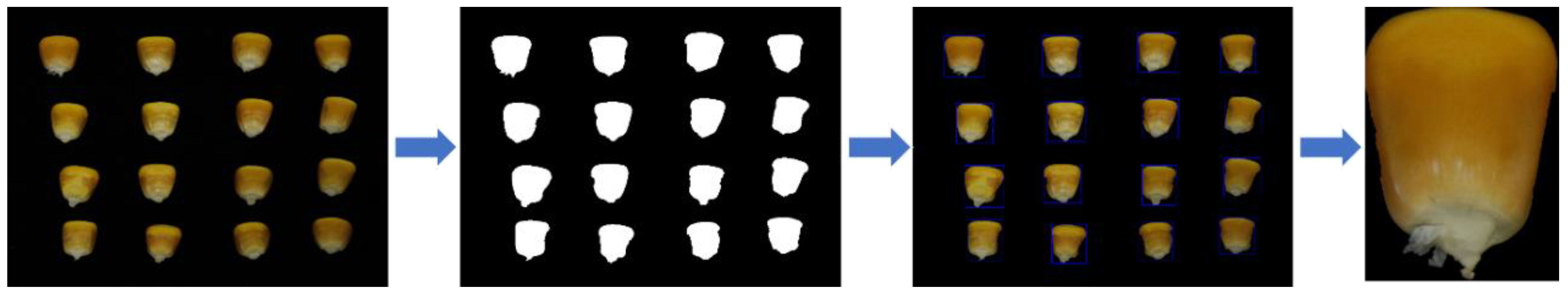
A single seed image for the identification of maize seeds was used in this experiment, so the single maize seed image needed to be segmented out, as demonstrated in Figure 2.
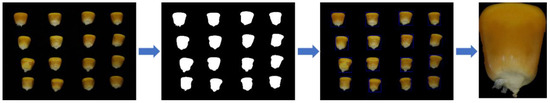
Figure 2.
Maize seed image segmentation.
To outline the maize seeds, the OpenCV contour algorithm was used. The image segmentation method is as follows:
- The multi-grain image is subjected to binarization, bilateral filtering, erosion expansion, and edge particle removal operations.
- The maize seeds are outlined using an OpenCV contour algorithm.
- For the original multi-grain seed image, repeat the preceding operations. The single seed image is segmented further to yield an 11-class maize seed image dataset.
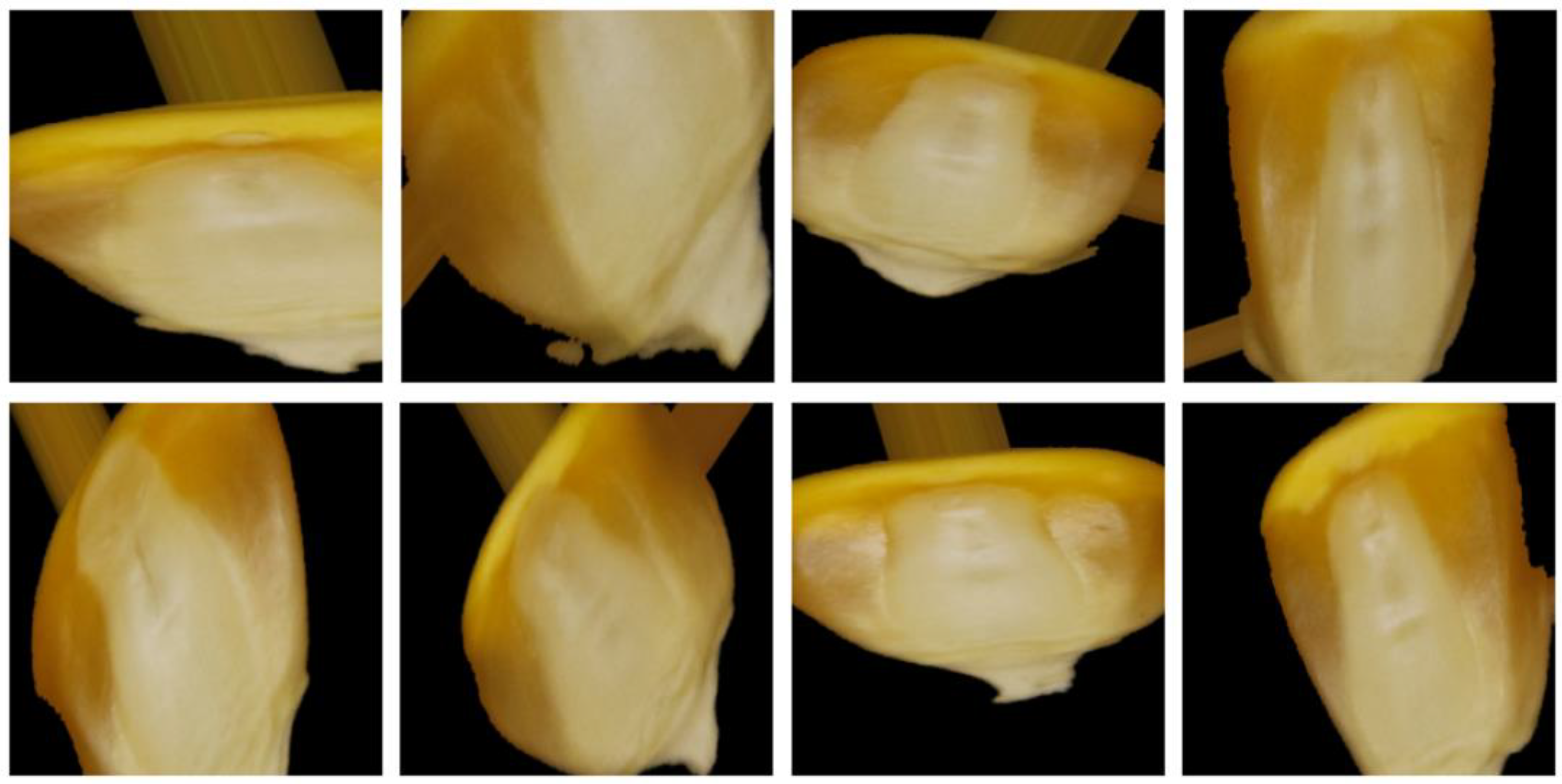
A double-sided mixed dataset of maize germ surface and non-germ surface was established based on previous research [21]. The original dataset was divided into training set, validation set, and test set by a ratio of 7:2:1. The dataset of maize seeds (double-sided mixture) is mentioned in Table 1. To address the problem of insufficient generalization ability due to small samples, the training set was augmented by rotation, height shift, width shift, shear, zoom, and horizontal flip. Figure 3 depicts a sample of some of the enhanced images.

Table 1.
Dataset of maize seeds (double-sided mixture).
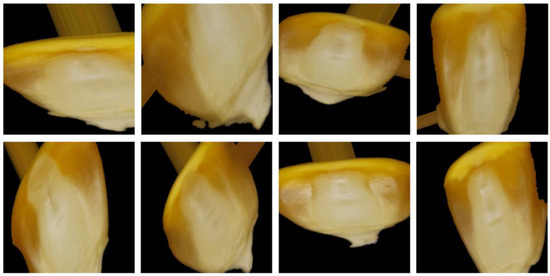
Figure 3.
Enhanced dataset images.
2.3. Improved Attention Mechanism CBAM
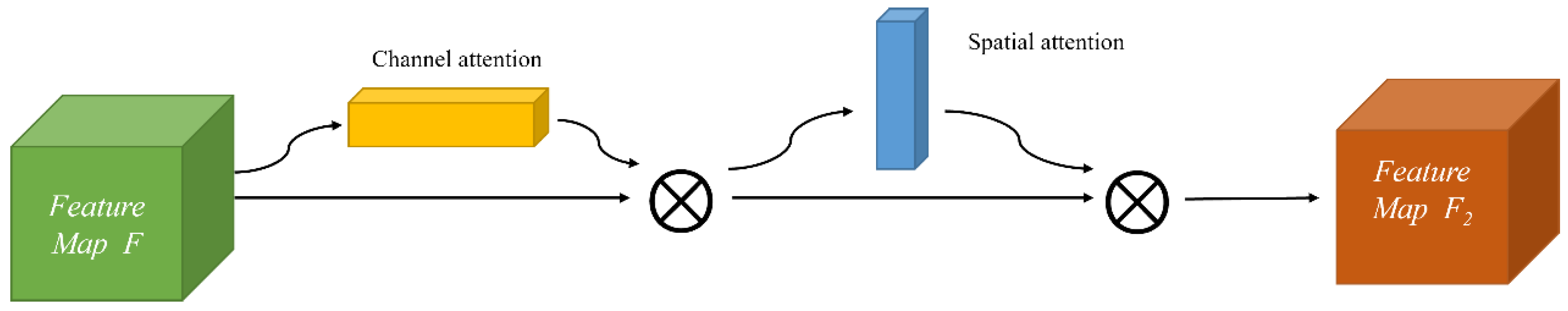
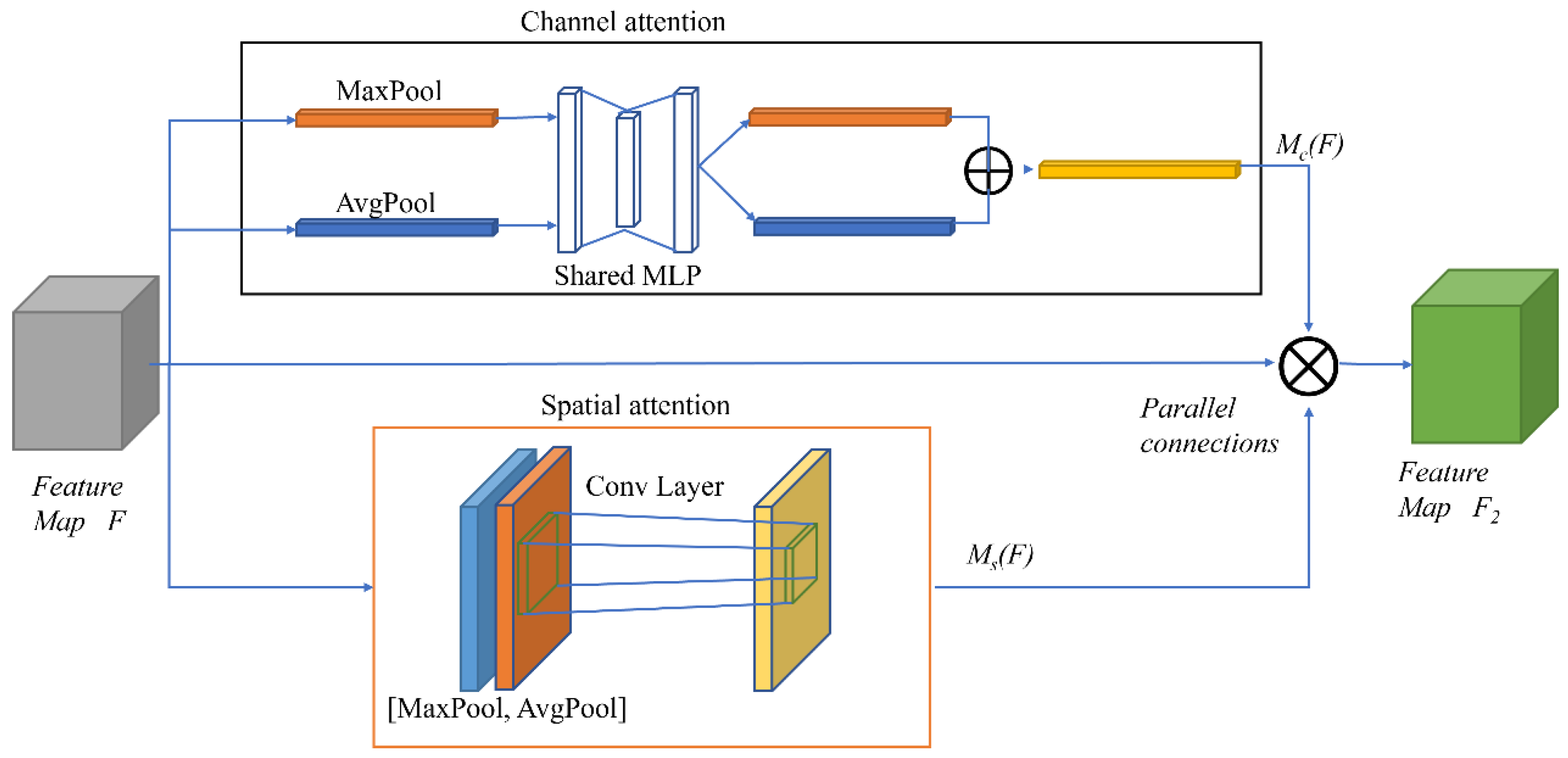
CABM is a lightweight attention mechanism module that can be easily embedded into the existing popular convolutional neural network structures without any extra computation. To achieve the rescaling operation of the original features, it uses two pooling methods: maximum pooling, and average pooling. It generates weights from the information of two dimensions (channel and space). The detailed structure of the CBAM module is represented in Figure 4.
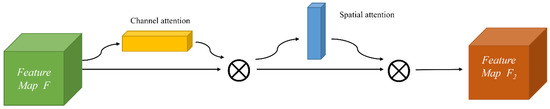
Figure 4.
Structure of CBAM.
The CBAM module first inputs the feature map F, which is weighted by the channel attention as F1; then, the output feature map F2 is weighted by the spatial attention, which is a “cascade connection” structure. The process formula is as below:
where: Mc(F) is the output power of F passing through the channel attention;
Ms(F1) denotes the output power of F1 passing through the spatial attention;
⊗ denotes the feature map weighted multiplicative operator.
Whether channel attention is enabled before spatial attention (channel → spatial, i.e., CBAM), or spatial attention is enabled before the channel attention (spatial → channel, i.e., reverse CBAM, or R_CBAM), the weights ranked later would be generated from the feature maps ranked earlier. To some extent, the input of the attentional features ranked at the back is influenced by the front-attentional mechanism, and this creates interference and makes the model unstable.
In this experiment, the original “cascade connection” was replaced with a “parallel connection” to improve the serial attention module of CBAM. Thus, both attention modules learn the original input feature map directly, without paying attention to the spatial and channel attention order, thereby leading to an improved CBAM (I_CBAM). The detailed structure of I_CBAM is represented in Figure 5.
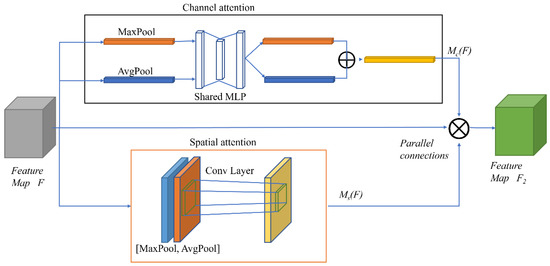
Figure 5.
The overall structure of I_CBAM.
The I_CBAM process formula is:
where Ms(F) indicates the output power of F passing through the spatial attention.
Channel attention is computed as follows:
Spatial attention is calculated as follows:
where:
The σ is the Sigmoid Function;
W0, and W1 indicate MLP weights;
and are the average-pooled features;
, and denote the max-pooled features;
Item f 7×7 is a convolution operation with a filter size of 7 × 7.
2.4. I_CBAM_MobileNetV2 Architecture
The Google team proposed the MobileNetV2 network in 2018 [16]. The accuracy of this network is higher than that of the MobileNetV1 network, and the model is smaller. MobileNetV2 incorporates inverted residuals and linear bottlenecks, significantly improving the model’s accuracy and efficiency.
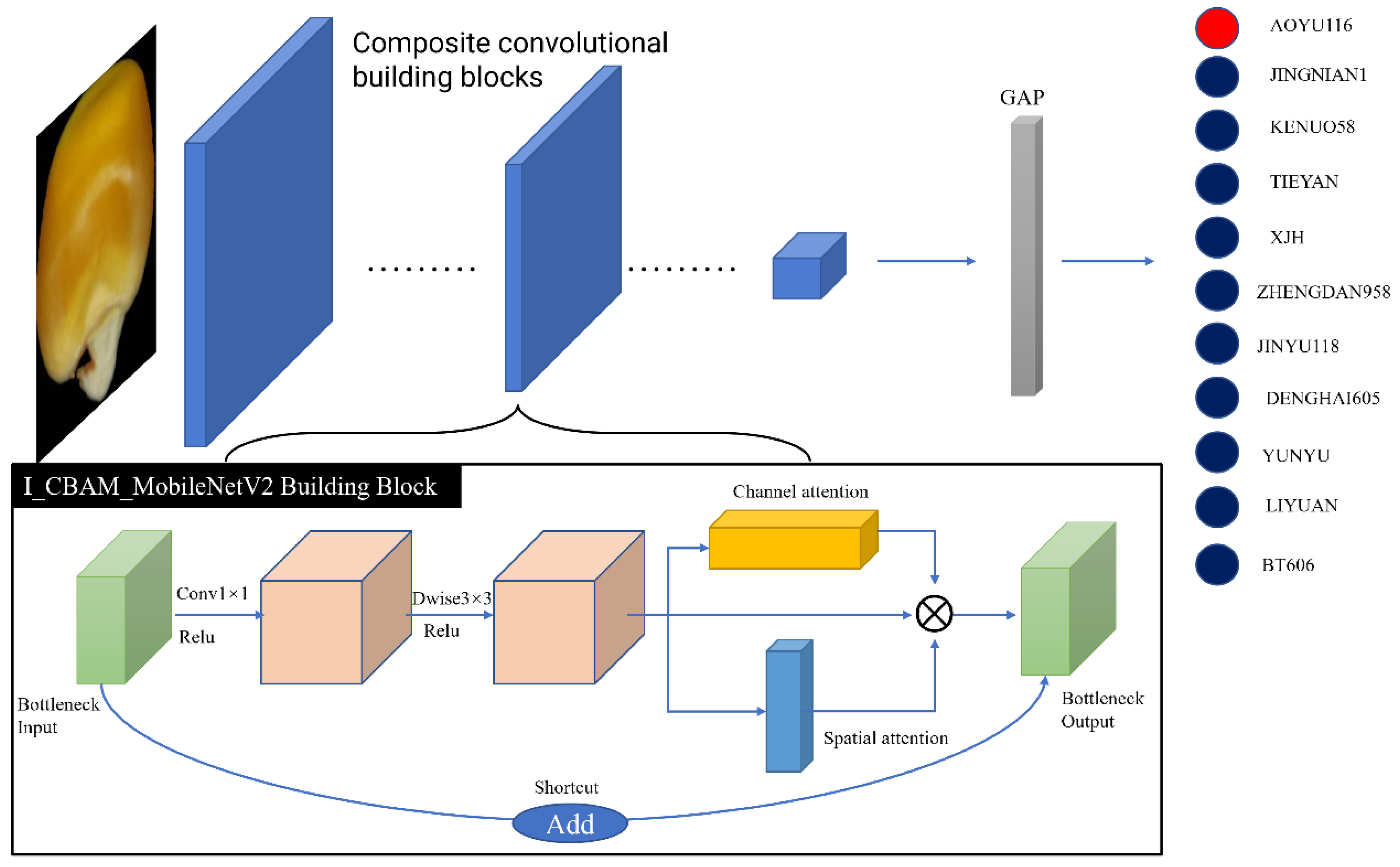
Images of maize seeds cannot be helpful in clearly distinguishing between different types, and the grain color is very similar. The MobileNetV2 network is strengthened in terms of representation power by adding the I_CBAM module, which enables the adaptive feature refinement of the pictures while emphasizing the local and global information about the images. To accurately identify various maize seed kinds, a new deep-learning model called I_CBAM_MobileNetV2 was created by fusing MobileNetV2 and the improved CBAM module (I_CBAM). The I_CBAM_MobileNetV2 model’s network topology is depicted in Figure 6.
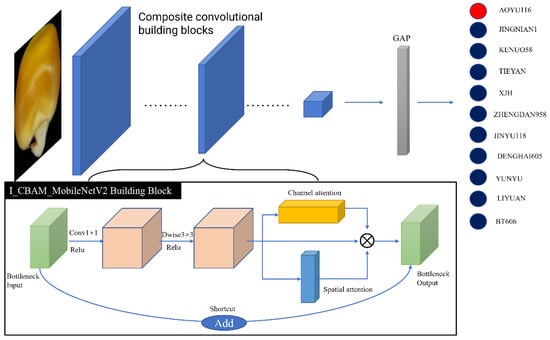
Figure 6.
I_CBAM_MobileNetV2 overall architecture.
In contrast to the inclusion of the attention mechanism before transfer learning, the attention module is added at the end of the entire convolutional neural network. In this experiment, the Inverted_res_block was enhanced with the improved CBAM to allow each module to extract more precise features; this contributed to the model’s precision for fine-grained recognition.
2.5. Model Training Environment Configuration
This experiment is based on the TensorFlow platform, using the Keras deep learning framework. Jupyter was used to build the model. The specific parameter configuration is listed in Table 2.

Table 2.
Detailed hyperparameters of the experiment.
2.6. Model Performance Evaluation Metrics
In this experiment, accuracy, precision, recall, F1-score, kappa coefficient, and confusion matrix are used as model performance evaluation metrics.
where:
TP is the number of samples that are positive but predicted to be positive;
FP is the number of samples that are negative but predicted to be positive;
FN denotes the number of samples that are positive but predicted to be negative;
TN is the number of samples that are negative but predicted to be negative;
N indicates the number of observation points of the confusion matrix;
Item n is the number of varieties;
Item aii represents elements of the i-th row and the column of the confusion matrix;
Item ai+ represents the sum elements of the i-th row of the confusion matrix;
Item a+i represents the sum elements of the i-th column of the confusion matrix.
3. Results
3.1. Accuracy Comparison of Each Attention Module
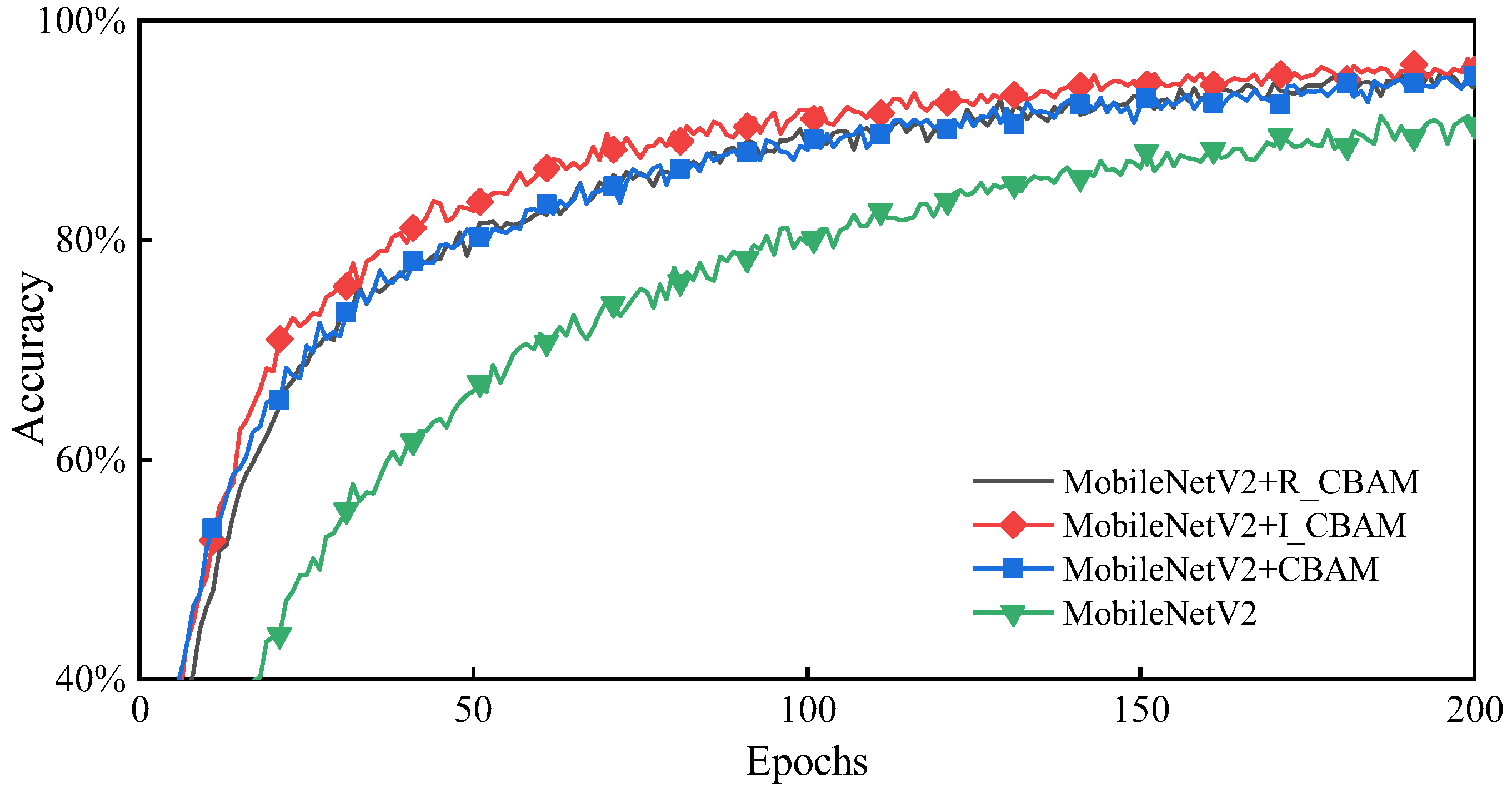
As highlighted in Figure 7 adding different attention mechanisms to MobileNetV2 significantly enhances the training effect compared to the network without attention mechanisms, wherein the MobileNetV2 + I_CBAM method achieves the optimal training effect with faster convergence and better accuracy. Therefore, it is practical to add the I_CBAM module to the MobileNetV2 network.
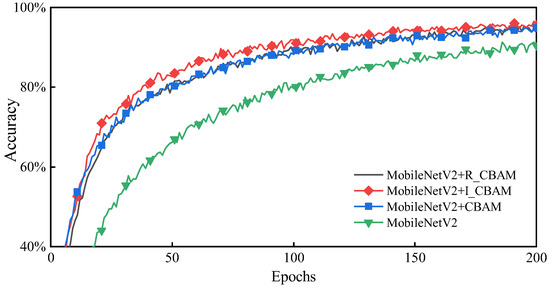
Figure 7.
Accuracy comparison after embedding the attention module.
3.2. Results of Reduction Ratios for I_CBAM_MobileNetV2
The experiments mentioned in this paper found that most of the parameters in the attention module I_CBAM are from channel attention. To further reduce the number of parameters and the model size after adding the attention module (to make the model as light as possible), a tuning experiment was conducted on the reduction ratio of channel attention on I_CBAM_MobileNetV2. The experimental results are mentioned in Table 3.

Table 3.
Results of using reduction ratios for I_CBAM_MobileNetV2.
It can be seen from Table 3 that when the reduction ratio is 16, the accuracy of the model reaches 98.21%, and the model size is somewhat reduced, which makes a decent balance between model size and accuracy. Therefore, using 16 as the model reduction ratio in I_CBAM_MobileNetV2 seems to be the best choice.
3.3. Analysis of Variety Identification Results
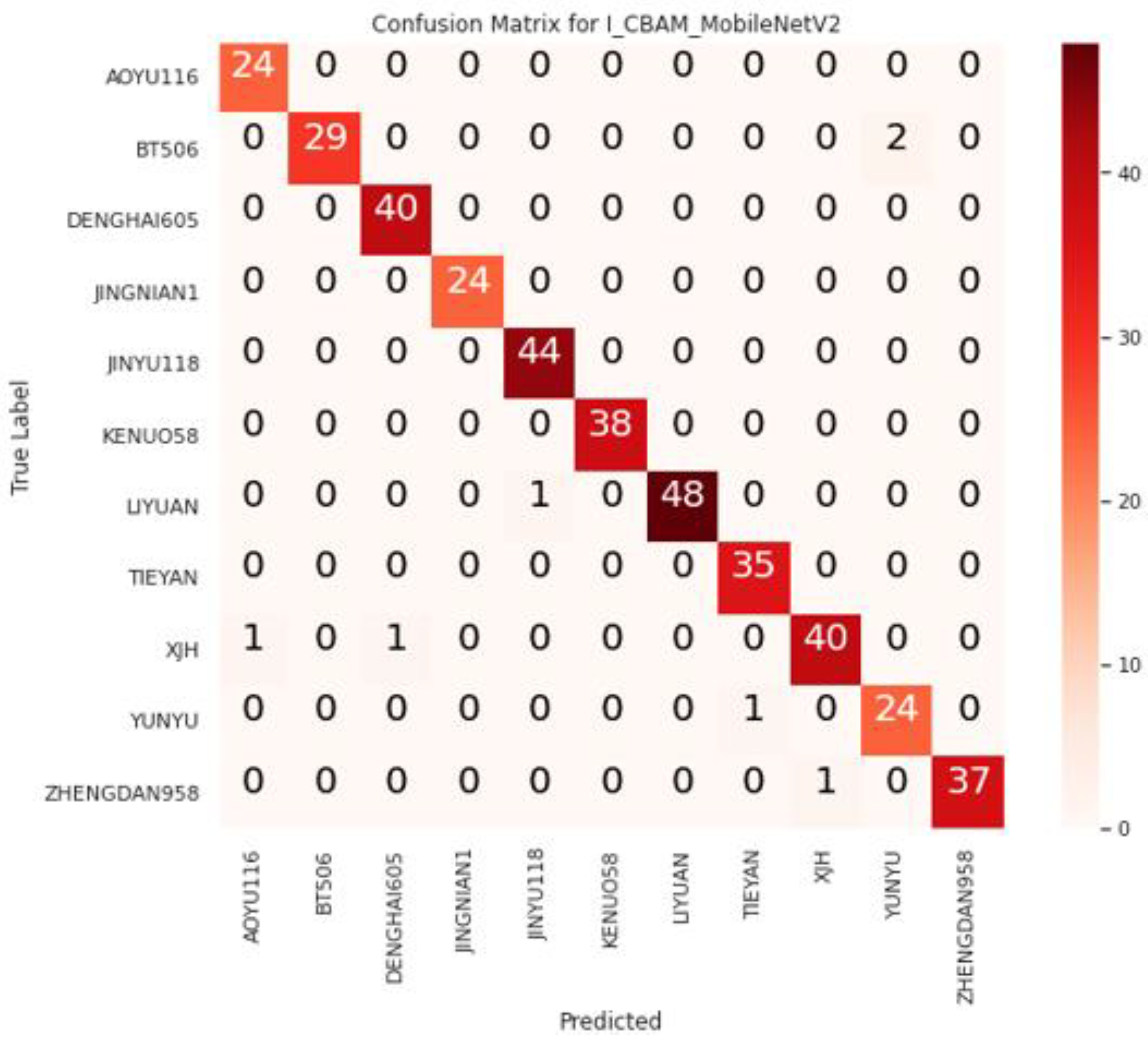
The I_CBAM_MobileNetV2 model was evaluated using the test dataset, and Figure 8 illustrates the confusion matrix of I_CBAM_MobileNetV2 for identifying maize seed varieties.
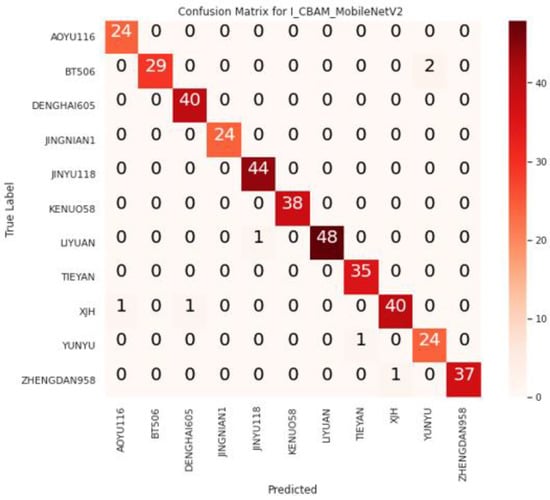
Figure 8.
I_CBAM_MobileNetV2 confusion matrix.
From the confusion matrix, it can be seen that two false identifications were generated in the BT506 variety identification, which were identified as YUNYU; two false identifications were generated in the XJH varieties, which were identified as AOYU116 and DENGHAI605; and one false identification was generated in LIYUAN, YUNYU, and ZHENGDAN958, respectively. This is because of the similarity in color among the varieties, and the minimal and subtle differences between the varieties.
For example, the colors of ZHENGDAN958 and XJH are nearly identical, with only a slight difference in shape and outline. Furthermore, the difference between LIYUAN and JINYU118 is in the surface pattern of the seeds, despite the fact that their contours are very similar. Table 4 shows the quantitative evaluation results of I_CBAM_MobileNetV2 on the test dataset.

Table 4.
I_CBAM_MobileNetV2 quantitative evaluation results.
As illustrated in Table 4, the proposed I_CBAM_MobileNetV2 significantly affects the maize seed variety identification in the test dataset. The identification effect of individual varieties highlights that the model has an accuracy higher than 99.23%, a precision higher than 92.31%, a recall higher than 93.55%, and an F1-score higher than 94.12% for 11 maize seed varieties. Considering KENUO58 as an example, the accuracy, precision, recall, and F1 score reached 100%. The above results depict that I_CBAM_MobileNetV2 can accurately identify each maize seed variety in the test set.
4. Discussion
4.1. Comparison of Different Models’ Identification Results
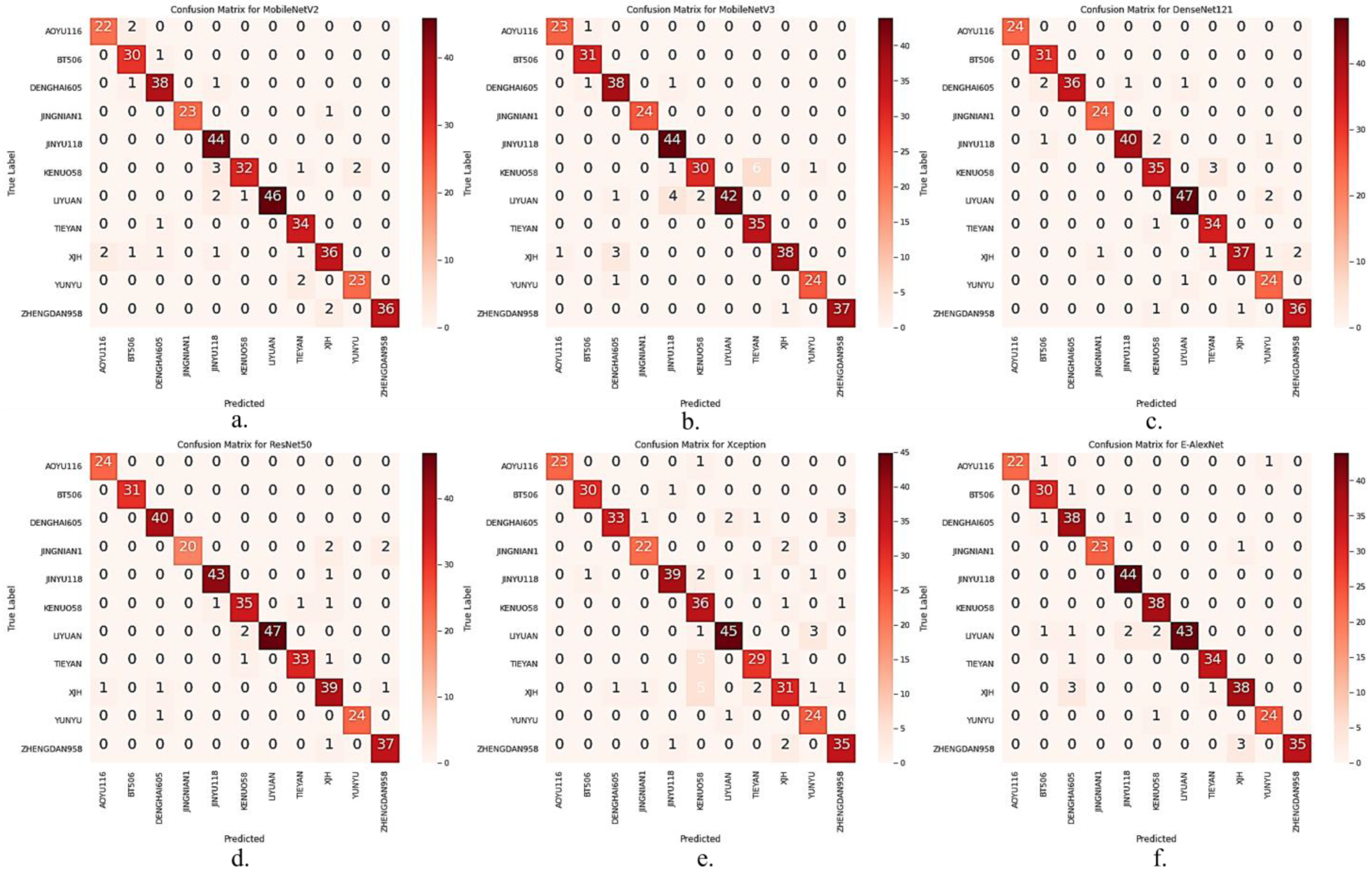
I_CBAM_MobileNetV2 was compared with MobileNetV2, MobileNetV3 [39], DenseNet121, Xception, ResNet50 [40], and E-AlexNet [41] in the test set, to demonstrate the seed variety identification performance of the I_CBAM_MobileNetV2 model more intuitively. The confusion matrices of the other six models are represented in Figure 9
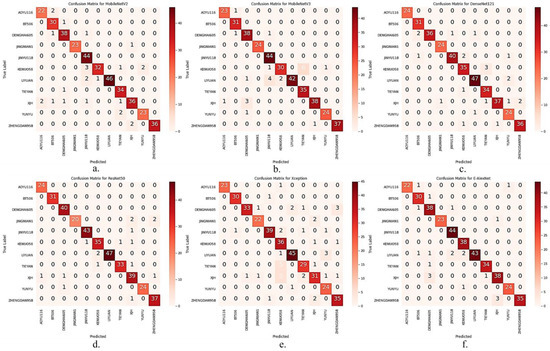
Figure 9.
Comparison of confusion matrices for the other six models. (a) MobileNetV2; (b) MobileNetV3; (c) DenseNet121; (d) ResNet50; (e) Xception; (f) E-AlexNet.
As represented in Figure 9 the number of incorrect identifications of I_CBAM_MobileNetV2 in the test set was much lower than that of the other models in the comparison experiments. The overall number of correct identifications increased by 19 compared with MobileNetV2, after adding the I_CBAM module to the MobileNetV2 model. The test results suggest that the I_CBAM module can effectively improve the maize seed variety identification ability of the MobileNetV2 model.
Overall accuracy (OA), kappa coefficient, and model size were introduced to the I_CBAM_MobileNetV2 model as evaluation metrics, to evaluate all methods of the comparison experiment quantitatively. The evaluation results are mentioned in Table 5.

Table 5.
Quantitative evaluation results of different models.
It can be seen from Table 5 that I_CBAM_MobileNetV2 achieved the highest overall accuracy and kappa coefficient, being 4.88% higher in accuracy and 0.0539 higher in kappa coefficient than MobileNetV2, and it outperformed the other four models.
Comparing I_CBAM_MobileNetV2 and MobileNetV3 (with the introduction of the SE module in MobileNetV2), the model size of I_CBAM_MobileNetV2 is smaller than that of MobileNetV3, suggesting that I_CBAM_MobileNetV2 does not significantly increase the model size, while significantly improving the model accuracy—making it more suitable for deployment in mobile terminals.
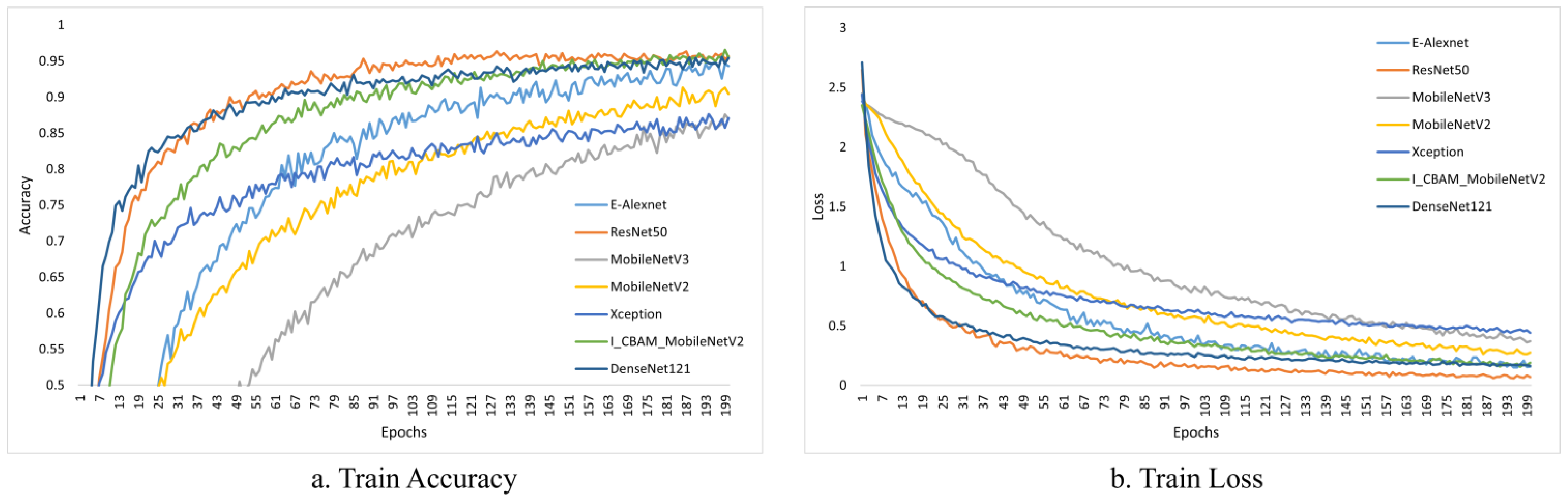
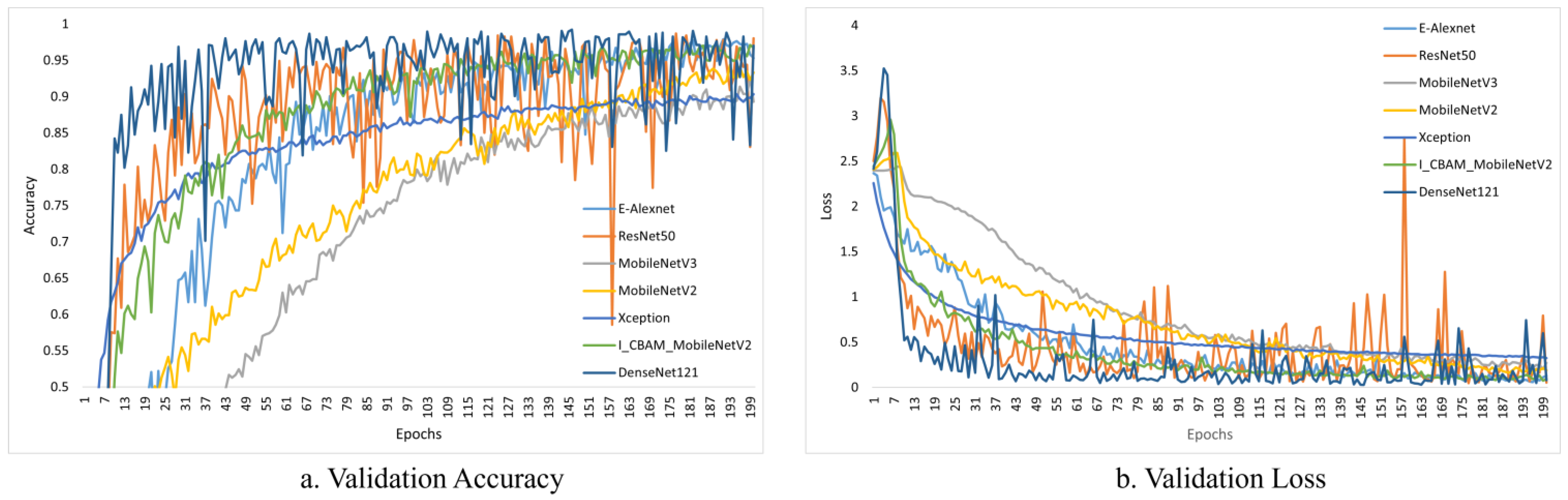
Figure 10 and Figure 11 depict the change curves for the various models during training and validation.
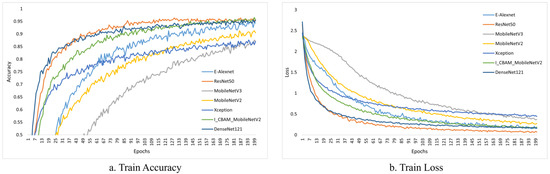
Figure 10.
Training accuracy and loss of different models.
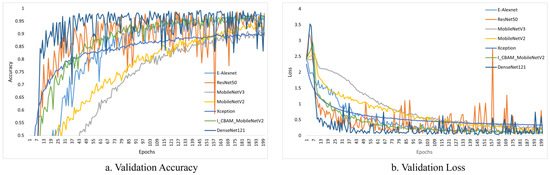
Figure 11.
Validation accuracy and loss of different models.
ResNet50 and DenseNet121 achieved stable values faster after initial training, as shown in Figure 10; while our models are slower to fit, they can also achieve stable accuracy after several iterations.
Although the various models perform well during training, the process can be deceptive, and may fail on the validation dataset. This is because the model only sees supervised data, and fails to learn well. To be considered the best fit, a model must perform well on the training dataset and the validation dataset. Figure 11 depicts the validation accuracy and loss of various models.
The curves of ResNet50 and DenseNet121 fluctuated significantly during the validation process, as shown in Figure 11, indicating that the models were not well learned and that the fit was poor. On the other hand, our model did not fluctuate significantly, and performed well in terms of fitting. In conclusion, our model outperformed the others.
According to a thorough comparison of various models on the test dataset, I_CBAM_ MobileNetV2 proposed in this paper can accurately identify the varieties of maize seed, offering an objective, accurate, efficient, and non-destructive model construction method for maize seed variety identification that is suitable for mobile deployment.
4.2. Visual Analysis of Maize Seed Variety Identification Results
Gradient-weighted Class Activation Mapping (Grad-CAM) [42,43] is a method for feature visualization in the form of a class activation heatmap. Grad-CAM can compute the classification gradient of the final convolutional feature map, where the position with the higher gradient is the most dependent feature position for classification, which is indicated as a strongly activated area on the feature map.
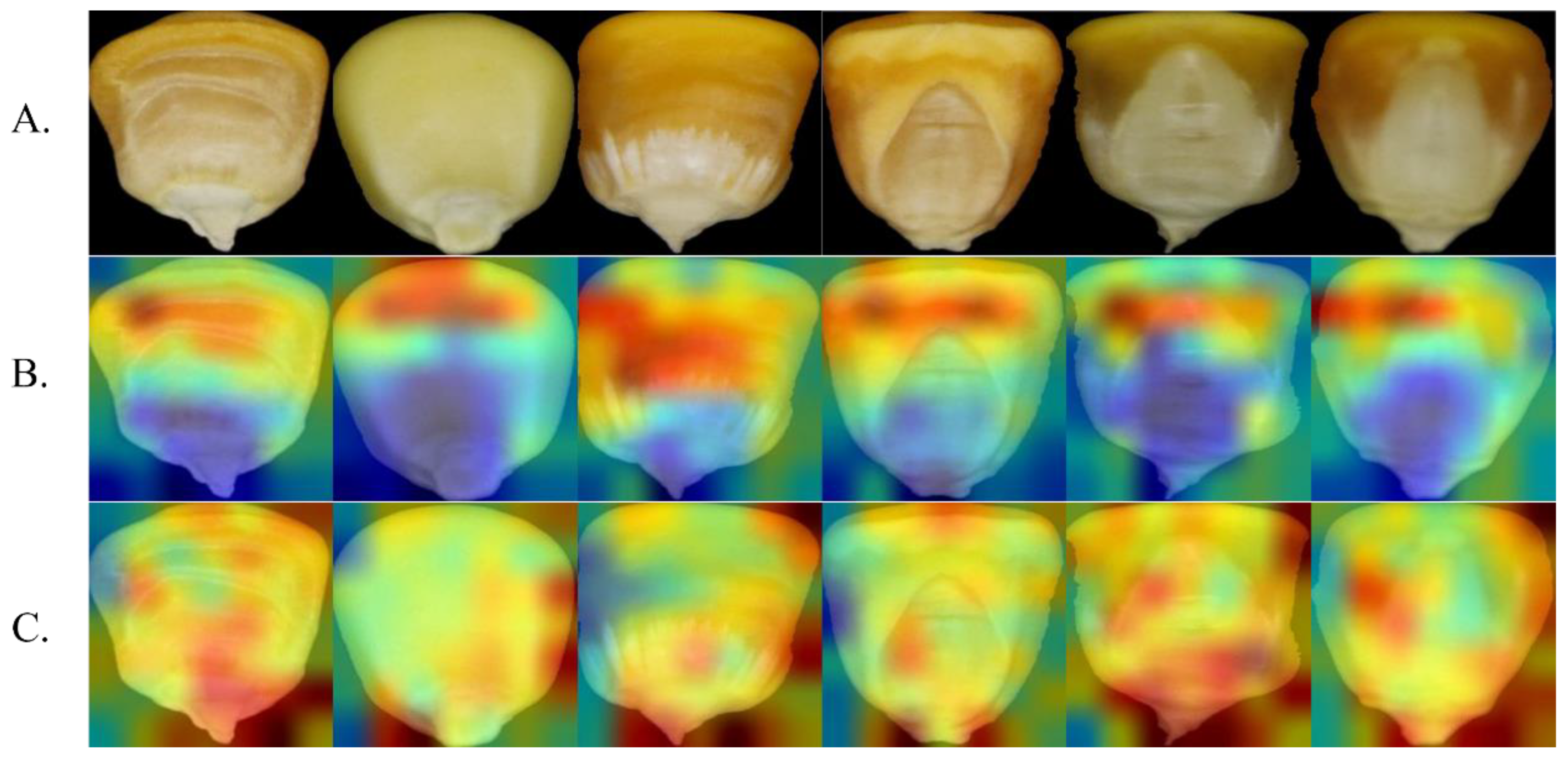
The red area in the figure depicts the strong activation area, and the blue area represents the weak activation area. The larger the gradient, the redder the color of the area and the higher the influence of the area on the classification result. Figure 12 highlights the attention heatmap of MobileNetV2 and I_CBAM_MobileNetV2.
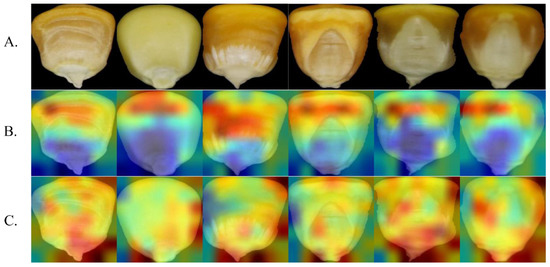
Figure 12.
Comparison of MobileNetV2 and I_CBAM_MobileNetV2 attentional thermograms. (A) Original image. (B) MobileNetV2 heatmap. (C) I_CBAM_MobileNetV2 heatmap.
MobileNetV2 focuses on the middle and upper areas of the seeds during identification, as shown in Figure 12, and ignores the tip area of the grains. I_CBAM_MobileNetV2, on the other hand, focuses on most of the grain image—particularly the special areas, such as the tip and edge—and thus can extract more critical information from the grain image.
Heatmap analysis experiments demonstrate the ability of I_CBAM_MobileNetV2 to identify maize seed varieties from a visual perspective. By comparing the heatmap represented in Figure 12, it can be inferred that after adding the I_CBAM module in MobileNetV2, the model’s attention focuses on more areas of the grain image. Considering this rule, it can be concluded that the I_CBAM module uses both the global and local information of the maize seed images to identify key information of the images, and improves the information representation of key regions in identifying maize seed varieties, thereby improving the model’s identification ability.
4.3. The Impact of Deleting a Certain Number of Layers
In this experiment, different model strategies were executed to reduce the model size further. Deleting the convolutional layer or the Inverted_res_block module in the model did not significantly impact the model’s accuracy. The results of the experiments are shown in Table 6.

Table 6.
Effect of different reduction strategies on model accuracy and model size.
In strategy A, the model’s penultimate Inverted_res_block module was removed, the number of convolutional kernels was increased to 160, and the number of parameters was significantly increased. The model size was reduced by about 20% by deleting this module, when compared to the original model. Despite the reduced accuracy, it still outperformed the unimproved MobileNetV2 by more than 95%.
In strategies B, C, and D, the modules with fewer convolution kernels were deleted, and the number of model parameters was increased compared with strategy A. The accuracy rate was improved compared with strategy A, indicating that after optimization and reduction, the model can achieve lightweight and accurate balance, providing a basis for further optimization of the model.
From Table 6, it can be inferred that the model optimization reduction strategy can reduce the training time of the model. It takes about 4 h to train the original model for 200 epochs, while after optimization, the training time is approximately 3.1 h, which is a significant reduction in training time, and there is no significant reduction in model accuracy. Therefore, in future research, the focus can be on reducing the unnecessary convolutional layers or convolutional modules, to enhance the efficiency of the model effectively.
5. Conclusions
In this experiment, 11 maize seed varieties were identified using deep-learning-based computer vision technology, and the maize seed variety identification model I_CBAM_MobileNetV2 was proposed. The overall accuracy and kappa coefficient of I_CBAM_MobileNetV2 were 98.21% and 0.9802, respectively.
Compared with the other CNN models in comparison experiments, I_CBAM_MobileNetV2 has several advantages in various evaluation metrics. Meanwhile, the I_CBAM module properly induced the network to focus on the target area. The results of Grad-CAM visual analysis experiments demonstrate that I_CBAM_MobileNetV2 can not only accurately locate the key information of the seeds and expand the activation area when identifying maize seed varieties, but also can fully express the information of the activation area.
This experiment provides an advanced technical scheme for maize seed variety identification, and lays the groundwork for future non-destructive varieties. In the future, the maize seed variety dataset can be expanded, and images of maize seeds with more complex backgrounds can be collected to provide enough data for the maize seed variety identification algorithm. I_CBAM_MobileNetV2’s speed can eventually be optimized, while attempting to remove some blocks from the model, to reduce the model size and deploy it on mobile devices.
Author Contributions
Conceptualization, R.M.; methodology, R.M. and J.W.; software: R.M., W.Z. and D.D.; validation, H.G. and D.M.; formal analysis, R.M.; investigation, J.W.; data curation, R.M. and W.Z.; writing—original draft preparation, R.M.; writing—review and editing, D.M. and Y.Y.; visualization, R.M.; supervision, D.M., F.H., L.L. and J.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No.42201458), the Shandong Provincial Natural Science Foundation (Grant No. ZR2022MC152), and the Shandong Province Colleges and Universities Young Innovative Talents Introduction and Cultivation Program (Grant No. 202202027).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
No applicable.
Acknowledgments
We thank the Shandong Academy of Agricultural Sciences for providing experimental support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hallauer, A.R.; Carena, M.J. Maize. In Cereals; Carena, M.J., Ed.; Plant breeding; Springer: New York, NY, USA, 2009; pp. 3–98. ISBN 978-0-387-72297-9. [Google Scholar]
- Dai, D.; Ma, Z.; Song, R. Maize Kernel Development. Mol. Breed. 2021, 41, 2. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, L.F.; Tong, Q.X. Discrimination of corn varieties using visible/near infrared imaging spectrometer system. Infrared Laser Eng. 2013, 42, 2437–2441. [Google Scholar]
- Cheng, H.; Shi, Z.X.; Yao, W.; Wang, L.; Pang, L.X. Corn Breed Recognition Based on Support Vector Machine. Trans. Chin. Soc. Agric. Mach. 2009, 40, 180–183. [Google Scholar]
- Yang, S.Q.; Ning, J.F.; He, D.J. Research on Recognition of Maize Varieties by BP Artificial Neural Network. J. Northwest Agric. For. Univ. (Nat. Sci.Ed.) 2004, 162–164. [Google Scholar] [CrossRef]
- Moges, T.G. Maize Seed Variety Identification Using Deep Learning Approach. Doctoral Thesis, Bahir Dar University, Bahir Dar, Ethiopia, 2021. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep Learning in Agriculture: A Survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine Learning and Deep Learning. Electron Mark. 2021, 31, 685–695. [Google Scholar] [CrossRef]
- Tu, K.; Wen, S.; Cheng, Y.; Zhang, T.; Pan, T.; Wang, J.; Wang, J.; Sun, Q. A Non-Destructive and Highly Efficient Model for Detecting the Genuineness of Maize Variety ’JINGKE 968′ Using Machine Vision Combined with Deep Learning. Comput. Electron. Agric. 2021, 182, 106002. [Google Scholar] [CrossRef]
- Zhou, Q.; Huang, W.; Tian, X.; Yang, Y.; Liang, D. Identification of the Variety of Maize Seeds Based on Hyperspectral Images Coupled with Convolutional Neural Networks and Subregional Voting. J. Sci. Food Agric. 2021, 101, 4532–4542. [Google Scholar] [CrossRef]
- Kurtulmuş, F. Identification of Sunflower Seeds with Deep Convolutional Neural Networks. Food Meas. 2021, 15, 1024–1033. [Google Scholar] [CrossRef]
- Torrey, L.; Shavlik, J. Transfer Learning. Available online: https://www.igi-global.com/chapter/transfer-learning/www.igi-global.com/chapter/transfer-learning/36988 (accessed on 29 May 2022).
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018, arXiv:1608.06993. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. arXiv 2018, arXiv:1707.07012. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2019, arXiv:1801.04381. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-Level Accuracy with 50x Fewer Parameters and <0.5 MB Model Size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. arXiv 2017, arXiv:1610.02357. [Google Scholar]
- Moyazzoma, R.; Hossain, M.A.A.; Anuz, M.H.; Sattar, A. Transfer Learning Approach for Plant Leaf Disease Detection Using CNN with Pre-Trained Feature Extraction Method Mobilnetv2. In Proceedings of the 2021 2nd International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST), Dhaka, Bangladesh, 5–7 January 2021; pp. 526–529. [Google Scholar]
- Khan, E.; Rehman, M.Z.U.; Ahmed, F.; Khan, M.A. Classification of Diseases in Citrus Fruits Using SqueezeNet. In Proceedings of the 2021 International Conference on Applied and Engineering Mathematics (ICAEM), London, UK, 7–9 July 2021; pp. 67–72. [Google Scholar]
- Feng, X.; Zhang, H.; Zhou, R.; Qiao, L.; Wei, D.; Li, D.D.; Zhang, Y.Y.; Zheng, G.Q. Variety Recognition Based on Deep Learning and Double-Sided Characteristics of Maize Kernel. J. Syst. Simul. 2021, 33, 2983–2991. [Google Scholar] [CrossRef]
- Elfatimi, E.; Eryigit, R.; Elfatimi, L. Beans Leaf Diseases Classification Using MobileNet Models. IEEE Access 2022, 10, 9471–9482. [Google Scholar] [CrossRef]
- Jaithavil, D.; Triamlumlerd, S.; Pracha, M. Paddy Seed Variety Classification Using Transfer Learning Based on Deep Learning. In Proceedings of the 2022 International Electrical Engineering Congress (iEECON), Khon Kaen, Thailand, 9–11 March 2022; pp. 1–4. [Google Scholar]
- Zhang, Z.; Gu, Y.; Hong, Q. Rice Disease Identification System Using Lightweight MobileNetV2. In Proceedings of the 8th International Conference on Intelligent Systems and Image Processing 2021, Chennai, India, 20–21 August 2021; The Institute of Industrial Applications Engineers: Kitakyushu, Japan, 2021; pp. 29–36. [Google Scholar]
- Hamid, Y.; Wani, S.; Soomro, A.B.; Alwan, A.A.; Gulzar, Y. Smart Seed Classification System Based on MobileNetV2 Architecture. In Proceedings of the 2022 2nd International Conference on Computing and Information Technology (ICCIT), Tabuk, Saudi Arabia, 25–27 January 2022; pp. 217–222. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2018, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–13 September 2018; pp. 3–19. [Google Scholar]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient Channel Attention for Deep Convolutional Neural Networks. arXiv 2020, arXiv:1910.03151. [Google Scholar]
- Wang, S.-H.; Fernandes, S.; Zhu, Z.; Zhang, Y.-D. AVNC: Attention-Based VGG-Style Network for COVID-19 Diagnosis by CBAM. IEEE Sens. J. 2021. [Google Scholar] [CrossRef]
- Jia, L.; Wang, Y.; Zang, Y.; Li, Q.; Leng, H.; Xiao, Z.; Long, W.; Jiang, L. MobileNetV3 With CBAM for Bamboo Stick Counting. IEEE Access 2022, 10, 53963–53971. [Google Scholar] [CrossRef]
- Shahi, T.B.; Sitaula, C.; Neupane, A.; Guo, W. Fruit Classification Using Attention-Based MobileNetV2 for Industrial Applications. PLoS ONE 2022, 17, e0264586. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yu, Y.; Zheng, Y.; Su, S.; Chen, F. Bilinear Attention Network for Image-Based Fine-Grained Recognition of Oil Tea (Camellia Oleifera Abel.) Cultivars. Agronomy 2022, 12, 1846. [Google Scholar] [CrossRef]
- Wang, M.H.; Wu, Z.X.; Zhou, Z.G. Fine-grained Identification Research of Crop Pests and Diseases Based on Improved CBAM via Attention. Trans. Chin. Soc. Agric. Mach. 2021, 52, 239–247. [Google Scholar]
- Gulzar, Y.; Hamid, Y.; Soomro, A.B.; Alwan, A.A.; Journaux, L. A Convolution Neural Network-Based Seed Classification System. Symmetry 2020, 12, 2018. [Google Scholar] [CrossRef]
- Altuntaş, Y.; Cömert, Z.; Kocamaz, A.F. Identification of Haploid and Diploid Maize Seeds Using Convolutional Neural Networks and a Transfer Learning Approach. Comput. Electron. Agric. 2019, 163, 104874. [Google Scholar] [CrossRef]
- Ali, A.; Qadri, S.; Mashwani, W.K.; Brahim Belhaouari, S.; Naeem, S.; Rafique, S.; Jamal, F.; Chesneau, C.; Anam, S. Machine Learning Approach for the Classification of Corn Seed Using Hybrid Features. Int. J. Food Prop. 2020, 23, 1110–1124. [Google Scholar] [CrossRef]
- Cao, W.; Zhang, C.; Wang, J.; Liu, S.; Xu, X. Purity Identification of Maize Seed Based on Discrete Wavelet Transform and BP Neural Network. Trans. Chin. Soc. Agric. Eng. 2012, 28, 253–258. [Google Scholar]
- Albarrak, K.; Gulzar, Y.; Hamid, Y.; Mehmood, A.; Soomro, A.B. A Deep Learning-Based Model for Date Fruit Classification. Sustainability 2022, 14, 6339. [Google Scholar] [CrossRef]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.-C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for MobileNetV3. In Proceedings of the IEEE/CVF International Conference on Computer Vision 2019, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ni, J.; Gao, J.; Li, J.; Yang, H.; Hao, Z.; Han, Z. E-AlexNet: Quality Evaluation of Strawberry Based on Machine Learning. Food Meas. 2021, 15, 4530–4541. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations From Deep Networks via Gradient-Based Localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Selvaraju, R.R.; Das, A.; Vedantam, R.; Cogswell, M.; Parikh, D.; Batra, D. Grad-CAM: Why Did You Say That? arXiv 2017, arXiv:1611.07450. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).