Genetic Variability and Population Structure of Pakistani Potato Genotypes Using Retrotransposon-Based Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Genomic DNA Extraction
2.2. Amplification Profile of Retrotransposon-Based iPBS Primers
2.3. Band Counting and Statistical Measurement
3. Results
3.1. Molecular Assessment of iPBS Markers
3.2. Heterozygosity and Molecular Variance (AMOVA) of 80 Potato Genotypes
3.3. Principal Coordinate analysis (PCoA) and Hierarchical Clustering of 80 Potato Genotypes
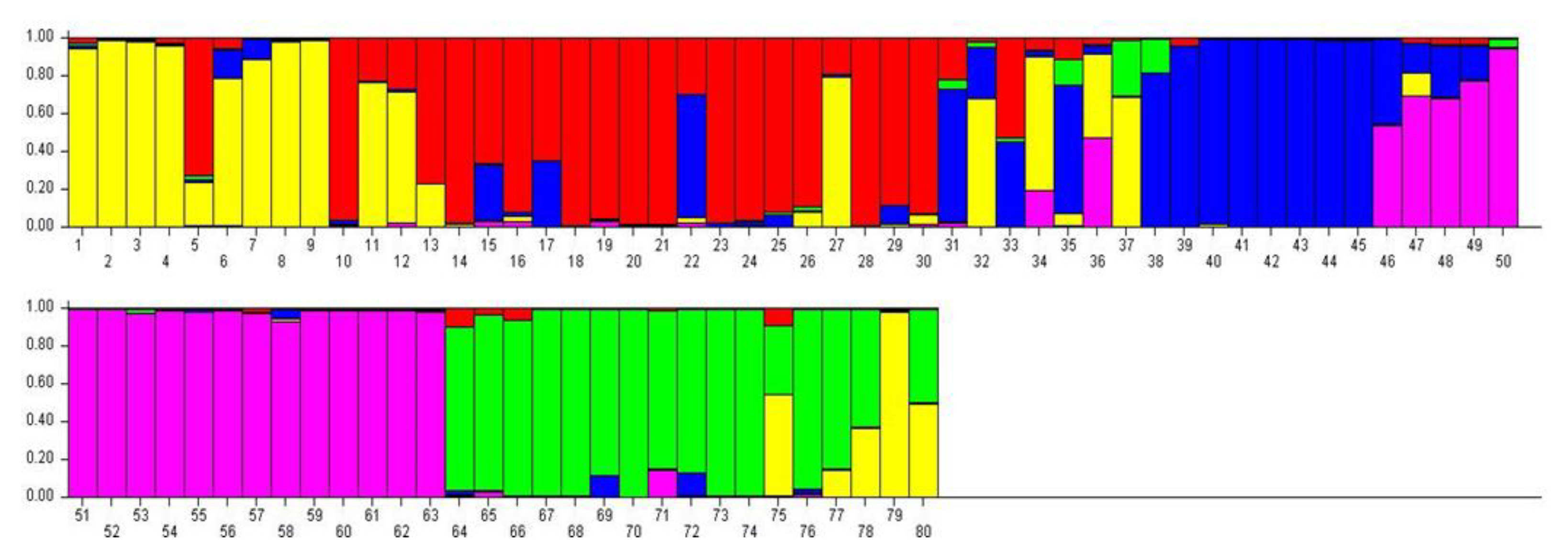
3.4. Genetic Structure of 80 Potato Genotypes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- De Haan, S.; Rodriguez, F. Potato origin and production. In Advances in Potato Chemistry and Technology, 2nd ed.; Singh, J., Kaur, L., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 1–32. [Google Scholar]
- FAO. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 18 June 2022).
- Dongyu, Q. Role and Potential of Potato in Global Food Security; FAO: Rome, Italy, 2022; pp. 23–30. Available online: https://www.fao.org/3/cc0330en/cc0330en.pdf (accessed on 2 July 2022).
- FAOSTAT. Food and Agricultural Organization Statistical Database, Crop Production. 2020. Available online: http://faostat3.fao.org/download/Q/QC/E (accessed on 22 March 2022).
- Watanabe, K. Potato genetics, genomics, and applications. Breed. Sci. 2015, 65, 53–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.J.; Sebastin, R.; Cho, G.T.; Yoon, M.; Lee, G.A.; Hyun, D.Y. Genetic Diversity and Population Structure of Potato Germplasm in RDA-Genebank: Utilization for Breeding and Conservation. Plants 2021, 10, 752. [Google Scholar] [CrossRef] [PubMed]
- Schulman, A.H. Molecular markers to assess genetic diversity. Euphytica 2007, 158, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Nadeem, M.A.; Muhammad, A.N.; Muhammad, Q.S.; Yıldız, D.; Gonul, C.; Mehtap, Y.; Rüştü, H.; Fiaz, A.; Ahmad, A.; Nitin, L.; et al. DNA molecular markers in plant breeding: Status and recent advancements in genomic selection and genome editing. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef] [Green Version]
- Kalendar, R.; Antonius, K.; Smýkal, P.; Schulman, A.H. iPBS: A universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 2010, 121, 1419–1430. [Google Scholar] [CrossRef]
- Yildiz, M.; Kocak, M.; Nadeem, M.A.; Cavagnaro, P.; Barboza, K.; Baloch, F.S.; Argün, D.; Keleş, D. Genetic diversity analysis in the Turkish pepper germplasm using iPBS retrotransposon-based markers. Turk. J. Agric. For. 2020, 44, 1–14. [Google Scholar] [CrossRef]
- Mehmood, A.; Jaskani, M.J.; Ahmad, S.; Ahmad, R. Evaluation of genetic diversity in open pollinated guava by iPBS primers. Pak. J. Agric. Sci. 2013, 50, 591–597. [Google Scholar]
- Baloch, F.S.; Ahmad, A.; Luis, E.; Sáenzde, M.; Rüştü, H.; Vahdettin, Ç.; Tolga, K.; Mehtap, Y.; Hakan, Ö. DNA based iPBS-retrotransposon markers for investigating the population structure of pea (Pisum sativum) germplasm from Turkey. Biochem. Syst. Ecol. 2015, 61, 244–252. [Google Scholar] [CrossRef]
- Yıldız, M.; Koçak, M.; Baloch, F.S. Genetic bottlenecks in Turkish okra germplasm and utility of iPBS retrotransposon markers for genetic diversity assessment. Genet. Mol. Res. 2015, 14, 10588–10602. [Google Scholar] [CrossRef]
- Al-Najm, A.; Luo, S.; Ahmad, N.M.; Trethowan, R. Molecular variability and genetic relationships of date palm (Phoenix dactylifera L.) cultivars based on inter-primer binding site (iPBS) markers. Aust. J. Crop Sci. 2016, 10, 732–740. [Google Scholar] [CrossRef]
- Yaldiz, G.; Camlica, M.; Nadeem, M.A.; Nawaz, M.A.; Baloch, F.S. Genetic diversity assessment in Nicotiana tabacum L. with iPBS-retrotransposons. Turk. J. Agric. For. 2018, 42, 154–164. [Google Scholar] [CrossRef]
- Nemli, S.; Kianoosh, T.; Tanyolac, M.B. Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) accessions through retrotransposon-based interprimer binding sites (iPBSs) markers. Turk. J. Agric. For. 2015, 39, 940–948. [Google Scholar] [CrossRef]
- Demirel, U.; Tındaş, İ.; Yavuz, C.; Baloch, F.S.; Çalışkan, M.E. Assessing genetic diversity of potato genotypes using inter-PBS retrotransposon marker system. Plant Genet. Resour. 2018, 16, 137–145. [Google Scholar] [CrossRef]
- Khan, M.F.; Tabassum, N.; Latif, A.; Khaliq, A.; Malik, M. Morphological characterization of potato (Solanum tuberosum L.) germplasm under rainfed environment. Afr. J. Biotechnol. 2013, 21, 3214–3223. [Google Scholar]
- Abbas, S.J.; Rasool, G.; Shah, S.R.U.; Iqbal, A. Analysis of genetic diversity in Pakistani potato cultivars by using randomly amplified polymorphic DNA (RAPD) primers. Am.-Eurasian J. Sustain. Agric. 2008, 2, 50–54. [Google Scholar]
- Murray, M.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.; Smouse, P.E. GenAlEx tutorials-part 2: Genetic distance and analysis of molecular variance (AMOVA). Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Andeden, E.E.; Baloch, F.S.; Derya, M.; Kilian, B.; Özkan, H. iPBS-Retrotransposons-based genetic diversity and relationship among wild annual Cicer species. J. Plant Biochem. Biotechnol. 2013, 22, 453–466. [Google Scholar] [CrossRef]
- Gailīte, A.; Ievinsh, G.; Ruņģis, D. Genetic diversity analysis of Latvian and Estonian Saussurea esthonica populations. Environ. Exp. Bot. 2011, 9, 115–119. [Google Scholar]
- Raddová, J.; Ptáčková, H.; Čechová, J.; Ondrášek, I. Genetic analysis of the genus Diospyros ssp. using RAPD and i-PBS methods. Acta Univ. Agric. Silvic. Mendelianae Brun. 2012, 60, 205–216. [Google Scholar] [CrossRef]
- Fang-Yong, C.; Ji-Hong, L. Germplasm genetic diversity of Myrica rubra in Zhejiang Province studied using inter-primer binding site and start codon-targeted polymorphism markers. Sci. Hortic. 2014, 170, 169–175. [Google Scholar] [CrossRef]
- Guo, D.L.; Guo, M.X.; Hou, X.G.; Zhang, G.H. Molecular diversity analysis of grape varieties based on iPBS markers. Biochem. Syst. Ecol. 2014, 52, 27–32. [Google Scholar] [CrossRef]
- Bamberg, J.B.; del Rio, A. Conservation of Potato Genetic Resources. In Genetic Improvement of Solanaceous Crops, Volume I: Potato; Razdan, M.K., Mattoo, A.K., Eds.; Science Publishers, Inc.: Plymouth, UK, 2005; p. 476. [Google Scholar]
- D’hoop, B.B.; Paulo, M.J.; Mank, R.A.; van Eck, H.J.; van Eeuwijk, F.A. Association mapping of quality traits in potato (Solanum tuberosum L.). Euphytica 2008, 161, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Jansky, S. Breeding for Disease Resistance in Potato. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley & Sons, Inc.: Oxford, UK, 2010; Volume 19, pp. 69–155. [Google Scholar] [CrossRef]
- Meirmans, P.G.; Liu, S.; van Tienderen, P.H. The analysis of polyploid genetic data. J. Hered. 2018, 109, 283–296. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Mei, M.; Jin, F.; Xu, J.; Duan, S.; Bian, C.; Li, G.; Wang, X.; Jin, L. Phenotypic variability and genetic diversity analysis of cultivated potatoes in China. Front. Plant Sci. 2022, 13, 954162. [Google Scholar] [CrossRef]
- Bohn, M.; Utz, H.F.; Melchinger, A.E. Genetic similarities among winter wheat cultivars determined on the basis of RFLPs, AFLPs, and SSRs and their use for predicting progeny variance. Crop Sci. 1999, 39, 228–237. [Google Scholar] [CrossRef]
- Khodadadi, M.; Fotokian, M.H.; Miransari, M. Genetic diversity of wheat (Triticum aestivum L.) genotypes based on cluster and principal component analyses for breeding strategies. Aust. J. Crop Sci. 2011, 5, 17–24. [Google Scholar]
- Akkale, C.; Yildirim, Z.; Yildirim, M.B.; Kaya, C.; Öztürk, G.; Tanyolaç, B. Assessing genetic diversity of some potato (Solanum tuberosum L.) genotypes grown in Turkey using the AFLP marker technique. Turk. J. Field Crop. 2010, 15, 73–78. [Google Scholar]
- Kandemir, N.; Yılmaz, G.; Karan, Y.B.; Borazan, D. Development of a simple sequence repeat (SSR) marker set to fingerprint local and modern potato varieties grown in central Anatolian Plateau in Turkey. Afr. J. Biotechnol. 2010, 9, 5516–5522. [Google Scholar]
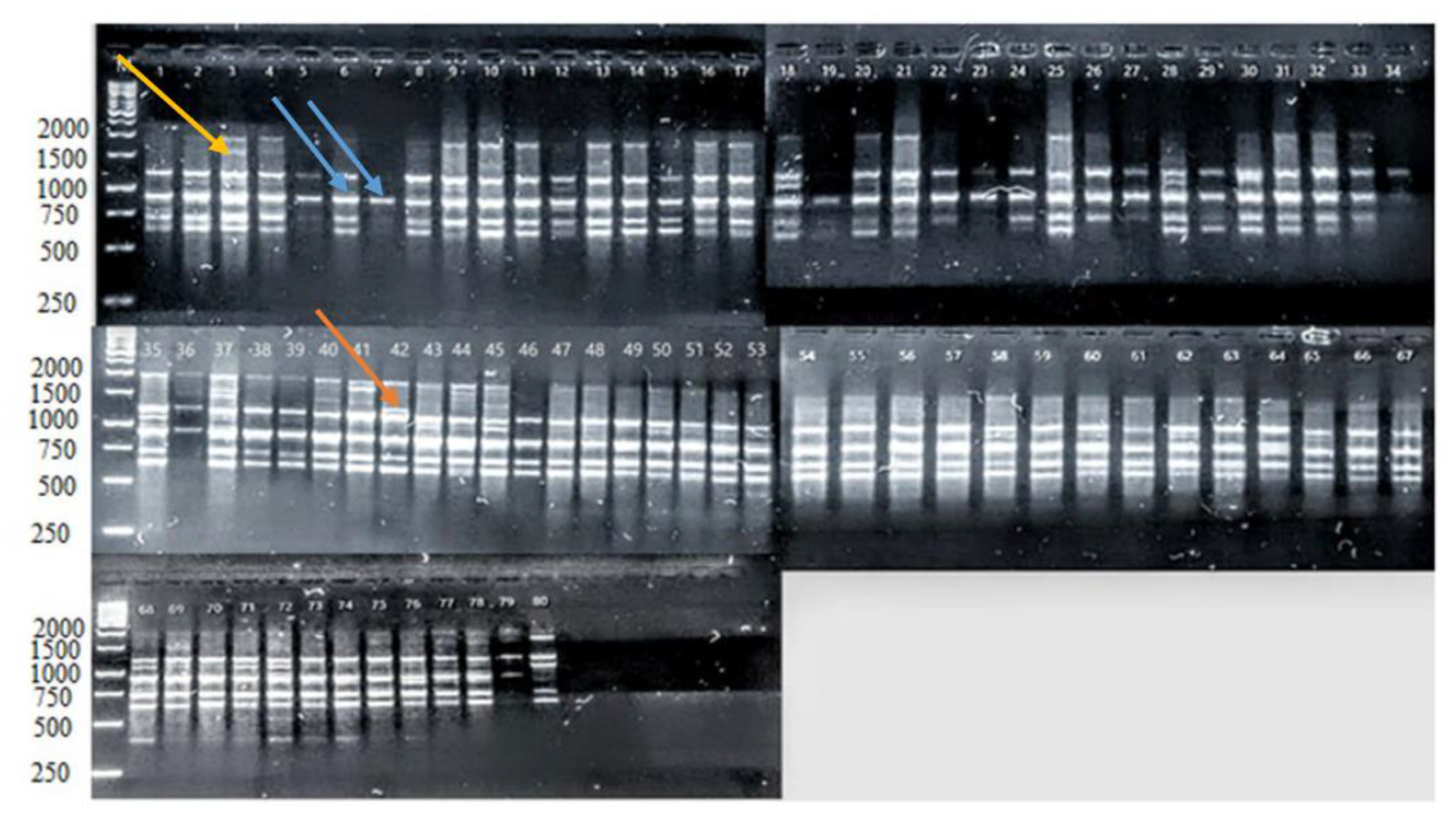

 ), local strains; LC (
), local strains; LC ( ), local cultivars; NARC (
), local cultivars; NARC ( ), and EC (
), and EC ( ), exotic cultivars.
), exotic cultivars.
 ), local strains; LC (
), local strains; LC ( ), local cultivars; NARC (
), local cultivars; NARC ( ), and EC (
), and EC ( ), exotic cultivars.
), exotic cultivars.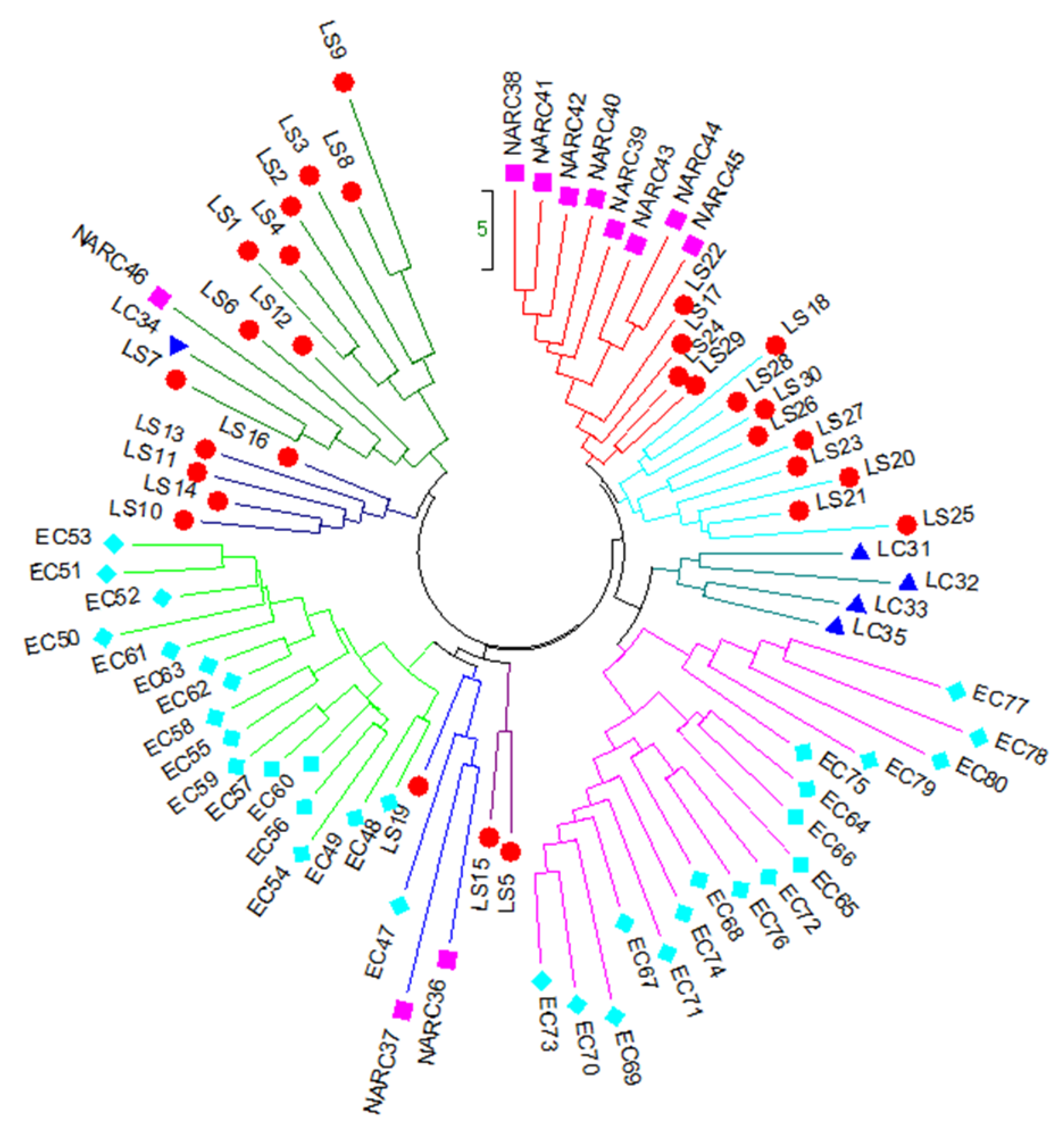

| Accession | Variety Name | Source | Cross | Morphological Characters (Color, Shape, Eyes) |
|---|---|---|---|---|
| LS 1 | FD44-26 | Local Strains | 385270-163 × Dura | Dark red, oblong, shallow |
| LS 2 | FD48-4 | Local Strains | 384640-3 × 385270-163 | Red, round |
| LS 3 | FD48-54 | Local Strains | 384640-3 × 385270-163 | White, round, shallow |
| LS 4 | FD61-3 | Local Strains | Diamant × FD12-24 | White, round, shallow |
| LS 5 | FD63-4 | Local Strains | 384636-1 × FD1-8 | White, round, deep |
| LS 6 | FD69-2 | Local Strains | FD4-2 × SH-5 | Dark red, round, deep |
| LS 7 | FD71-1 | Local Strains | FD8-3 × ultimas | Red, round/oval, medium |
| LS 8 | FD73-75 | Local Strains | FD35-36 × SH-5 | Red, round, shallow |
| LS 9 | FD73-77 | Local Strains | FD35-36 × SH-5 | Red, round, deep |
| LS 10 | FD74-19 | Local Strains | 9619 × FD49-28 | White, round, shallow |
| LS 11 | FD74-33 | Local Strains | 9619 × FD49-28 | White, oblong, shallow |
| LS 12 | FD74-40 | Local Strains | 9619 × FD49-28 | White, round, medium |
| LS 13 | FD74-47 | Local Strains | 9619 × FD49-28 | White, round, deep |
| LS 14 | FD74-50 | Local Strains | 9619 × FD49-28 | White, oval, shallow |
| LS 15 | FD74-51 | Local Strains | 9619 × FD49-28 | White, oblong, shallow |
| LS 16 | FD75-3 | Local Strains | FD49-28 × SH-5 | White, round, shallow |
| LS 17 | FD75-55 | Local Strains | FD49-28 × SH-5 | Red, round, deep |
| LS 18 | FD76-13 | Local Strains | FD3-15 × SH- 5 | Red, oblong, medium |
| LS 19 | FD76-27 | Local Strains | FD3-15 × SH- 5 | Red, oblong, shallow |
| LS 20 | FD76-35 | Local Strains | FD3-15 × SH- 5 | Dark red, oblong, shallow |
| LS 21 | FD76-48 | Local Strains | FD3-15 × SH- 5 | White, round, shallow |
| LS 22 | FD77-62 | Local Strains | FD3-9 × SH-5 | Dark red, oblong, deep |
| LS 23 | FD80-6 | Local Strains | FD3-15 × FD35-36 | Light red, round, deep |
| LS 24 | FD51-5 | Local Strains | Dura × SH-5 | White, round, shallow |
| LS 25 | SL 1-4 | Local strains | SH-5 × Red fantasy | Red, oblong, medium |
| LS 26 | SL 1-47 | Local strains | SH-5 × Red fantasy | Red, oblong, shallow |
| LS 27 | SL 4-26 | Local strains | FD48-4 × SH-5 | Dark red, oblong, deep |
| LS 28 | SL13-64 | Local strains | SH -5 × FD 48- 54 | Red, oblong, shallow |
| LS 29 | SL 13-78 | Local strains | FD51-5 × FD69-1 | White, oblong, shallow |
| LS 30 | SL14-15 | Local strains | Saghitta × SH-5 | Red, round, deep |
| LC 31 | FSD white | Local Cultivar | -------------------- | White, round, shallow |
| LC 32 | FSD red | Local Cultivar | -------------------- | Red, round, deep |
| LC 33 | Sadaf | Local Cultivar | FD3-15 × FD35-36 | White, round, shallow |
| LC 34 | Ruby | Local Cultivar | 384636-1 × FD1-8 | Dark red, round, shallow |
| LC 35 | PRI Red | Local Cultivar | FD44-24 × FD12-24 | Red, oblong, shallow |
| NARC 36 | Potato 3 | NARC Cultivar | ----------------------- | ------------------- |
| NARC 37 | Potato2 | NARC Cultivar | ----------------------- | -------------------- |
| NARC 38 | NARC | NARC Cultivar | ||
| NARC 39 | N-13 | NARC Cultivar | ---------------------- | Light yellow, oval, medium |
| NARC 40 | N-15 | NARC Cultivar | ----------------------- | Light red, oval, medium |
| NARC 41 | N-18 | NARC Cultivar | ----------------- | Light red, oval, medium |
| NARC 42 | N-34 | NARC Cultivar | ------------------- | White, oval, medium |
| NARC 43 | N-2005-1 | NARC Cultivar | ------------------ | Red, oval, medium |
| NARC 44 | N-2005-4 | NARC Cultivar | ||
| NARC 45 | N-393619-44 | NARC Cultivar | -------------------- | Light red, round, deep |
| NARC 46 | N-4 | NARC Cultivar | --------------------- | White, round, medium |
| EC 47 | Aurea | Exotic Cultivar | Lady Rosetta × (Satruna × Pentland dell) | White, round, medium |
| EC 48 | Dolly | Exotic Cultivar | Lady Rosetta × Britta | Light red, round, deep |
| EC 49 | Elbieda | Exotic Cultivar | -------------------- | White, oval, shallow |
| EC 50 | Elodie | Exotic Cultivar | 80F66.25 × 81F145.14 | White, oval, shallow |
| EC 51 | Eldorodo | Exotic Cultivar | -------------------- | Red, oval, medium |
| EC 52 | Estima | Exotic Cultivar | Nopol × G3014 | Light yellow, oval, medium |
| EC 53 | El-mundo | Exotic Cultivar | ------------------ | White, oval, medium |
| EC 54 | Erora | Exotic Cultivar | -------------------- | White, oval, medium |
| EC 55 | Paramount | Exotic Cultivar | Janat × Dutch seeding | Red, oval, medium |
| EC 56 | Hybrid 202-05-01 | Exotic Cultivar | ------------------- | Light red, oval, medium |
| EC 57 | Simply red | Exotic Cultivar | Asterix × HZ86 AM75 | Light red, oblong, medium |
| EC 58 | Santana | Exotic Cultivar | Spunta × VK69-491 | White, oblong, shallow |
| EC 59 | Romera | Exotic Cultivar | Belladonna × Laura | Light red, oval, medium |
| EC 60 | Monika | Exotic Cultivar | Krasa × Velox | White, oval, medium |
| EC 61 | Verdi | Exotic Cultivar | Tomnsa × Diana | White, oval, medium |
| EC 62 | Safrane | Exotic Cultivar | ----------------- | White, oval, medium |
| EC 63 | Terka | Exotic Cultivar | Fabula × Pamir | Light yellow, round, shallow |
| EC 64 | Red Valentine | Exotic Cultivar | Mondial × Amadeus | Red, oval, medium |
| EC 65 | Red Sonia | Exotic Cultivar | ------------------ | Red, oval, medium |
| EC 66 | Red Sun | Exotic Cultivar | Inova × Amadeus | Red, oblong, medium |
| EC 67 | Rositta | Exotic Cultivar | ------------------- | Light red, oval, medium |
| EC 68 | Sagitta | Exotic Cultivar | Gallia × RZ-86-2918 | Light yellow, oblong, medium |
| EC 69 | Suzen | Exotic Cultivar | ----------------- | White, oval, medium |
| EC 70 | Sassy | Exotic Cultivar | G82TTT37 × Propmesse | White, round, medium |
| EC 71 | Focus | Exotic Cultivar | Agria × Bru 82-78 | Light yellow, round, shallow |
| EC 72 | Florice | Exotic Cultivar | Fanette × Inra72.68.5 | Light yellow, oblong, medium |
| EC 73 | HZD-04-684 | Exotic Cultivar | ----------------- | Red, oval, shallow |
| EC 74 | Shepody | Exotic Cultivar | Bake king × F58050 | White, oval, medium |
| EC 75 | Orchestra | Exotic Cultivar | Maradonna × Cupido | Light yellow, oval, medium |
| EC 76 | Jitka | Exotic Cultivar | M22/12 × Bonita | Light yellow, round, medium |
| EC 77 | Red river | Exotic Cultivar | ------------------ | Red, oval, medium |
| EC 78 | Kuroda | Exotic Cultivar | AR76-199-3 × Konst80-1407 | Red, oval, medium |
| EC 79 | KWS-06-125 | Exotic Cultivar | ------------- | Dark red, oval, medium |
| EC 80 | Pirol | Exotic Cultivar | Agriax × 1.214.226-84 |
| Serial No | iPBS Primers | Base Pair Sequence (5′-3′) | Annealing Temp (°C) | GC Content (%) |
|---|---|---|---|---|
| 1 | 2257 | CTCTCAATGAAAGCACCA | 46 | 44 |
| 2 | 2229 | CGACCTGTTCTGATACCA | 46 | 50 |
| 3 | 2252 | TCATGGCTCATGATACCA | 43 | 44 |
| 4 | 2277 | GGCGATGATACCA | 46 | 54 |
| 5 | 2375 | TCGCATCAACCA | 30 | 50 |
| 6 | 2376 | TAGATGGCACCA | 46 | 50 |
| 7 | 2387 | GCGCAATACCCA | 46 | 58 |
| 8 | 2391 | ATCTGTCAGCCA | 46 | 50 |
| 9 | 2374 | CCCAGCAAACCA | 30 | 58 |
| 10 | 2377 | ACGAAGGGACCA | 46 | 67 |
| 11 | 2383 | GCATGGCCTCCA | 46 | 66 |
| 12 | 2232 | AGAGAGGCTCGGATACCA | 48 | 56 |
| 13 | 2239 | ACCTAGGCTCGGATGCCA | 50 | 61 |
| 14 | 2272 | GGCTCAGATGCCA | 46 | 62 |
| 15 | 2373 | GCTCATCATGCCA | 46 | 54 |
| 16 | 2390 | GCAACAACCCCA | 46 | 58 |
| iPBS Primers | AN | Size Range (bp) | PM | % PM | MM | PIC | Na | Ne | I | He | µHe | f |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2229 | 86 | 500–2000 | 85 | 99 | 1 | 0.37 | 1.83 | 1.58 | 0.48 | 0.33 | 0.34 | 0.45 |
| 2232 | 86 | 550–1350 | 86 | 100 | 0 | 0.33 | 1.61 | 1.40 | 0.36 | 0.24 | 0.25 | 0.46 |
| 2239 | 86 | 600–1500 | 80 | 93 | 6 | 0.24 | 1.45 | 1.37 | 0.33 | 0.22 | 0.23 | 0.35 |
| 2252 | 86 | 650–2000 | 85 | 99 | 1 | 0.25 | 1.63 | 1.38 | 0.35 | 0.23 | 0.24 | 0.39 |
| 2272 | 84 | 500–1750 | 79 | 94 | 5 | 0.19 | 0.67 | 1.11 | 0.12 | 0.07 | 0.08 | 0.13 |
| 2277 | 80 | 450–1000 | 80 | 100 | 0 | 0.39 | 1.71 | 1.54 | 0.45 | 0.31 | 0.32 | 0.47 |
| 2374 | 82 | 400–1250 | 60 | 73 | 22 | 0.22 | 1.25 | 1.15 | 0.19 | 0.11 | 0.11 | 0.14 |
| 2375 | 80 | 350–3500 | 65 | 81 | 15 | 0.25 | 1.23 | 1.2 | 0.23 | 0.14 | 0.14 | 0.17 |
| 2377 | 83 | 300–3500 | 75 | 90 | 8 | 0.36 | 1.75 | 1.52 | 0.44 | 0.3 | 0.31 | 0.45 |
| 2390 | 85 | 280–1000 | 80 | 94 | 5 | 0.36 | 1.66 | 1.57 | 0,47 | 0.32 | 0.34 | 0.54 |
| 2391 | 75 | 250–1000 | 60 | 80 | 15 | 0.14 | 1.07 | 1.08 | 0.13 | 0.07 | 0.07 | 0.81 |
| Groups | N | Na | Ne | I | He | µHe |
|---|---|---|---|---|---|---|
| LS | 30 | 1.56 | 1.36 | 0.34 | 0.22 | 0.22 |
| LC | 5 | 1.11 | 1.35 | 0.29 | 0.2 | 0.22 |
| NARC | 11 | 1.36 | 1.30 | 0.29 | 0.19 | 0.19 |
| EC | 34 | 1.75 | 1.39 | 0.37 | 0.24 | 0.24 |
| Mean | 20 | 1.45 | 1.35 | 0.32 | 0.21 | 0.22 |
| SV | df | SS | MS | Est. Var. | % | PhiPT |
|---|---|---|---|---|---|---|
| Among Pops | 3 | 166.47 | 55.49 | 2.30 | 15% | 0.021 *** |
| Within Pops | 82 | 1056.16 | 12.88 | 12.88 | 85% | |
| Total | 85 | 1222.63 | 15.18 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehmood, A.; Dracatos, P.M.; Maqsood, L.; Yousafi, Q.; Hussain, A.; Jaskani, M.J.; Sajid, M.W.; Haider, M.S.; Hussain, M.M. Genetic Variability and Population Structure of Pakistani Potato Genotypes Using Retrotransposon-Based Markers. Agriculture 2023, 13, 185. https://doi.org/10.3390/agriculture13010185
Mehmood A, Dracatos PM, Maqsood L, Yousafi Q, Hussain A, Jaskani MJ, Sajid MW, Haider MS, Hussain MM. Genetic Variability and Population Structure of Pakistani Potato Genotypes Using Retrotransposon-Based Markers. Agriculture. 2023; 13(1):185. https://doi.org/10.3390/agriculture13010185
Chicago/Turabian StyleMehmood, Asim, Peter M. Dracatos, Linta Maqsood, Qudsia Yousafi, Abrar Hussain, Muhammad J. Jaskani, Muhammad W. Sajid, Muhammad S. Haider, and Muhammad M. Hussain. 2023. "Genetic Variability and Population Structure of Pakistani Potato Genotypes Using Retrotransposon-Based Markers" Agriculture 13, no. 1: 185. https://doi.org/10.3390/agriculture13010185
APA StyleMehmood, A., Dracatos, P. M., Maqsood, L., Yousafi, Q., Hussain, A., Jaskani, M. J., Sajid, M. W., Haider, M. S., & Hussain, M. M. (2023). Genetic Variability and Population Structure of Pakistani Potato Genotypes Using Retrotransposon-Based Markers. Agriculture, 13(1), 185. https://doi.org/10.3390/agriculture13010185








