Variability in Estimating Crop Model Genotypic Parameters: The Impact of Different Sampling Methods and Sizes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Experiments
2.2. DSSAT-SUBSTOR Model Inputs
2.3. Sampling Methods and Sampling Design Framework
2.3.1. Introduction to Sampling Methods
2.3.2. Sampling Design Framework
2.4. Genotypic Parameter Calibration and Validation
3. Results
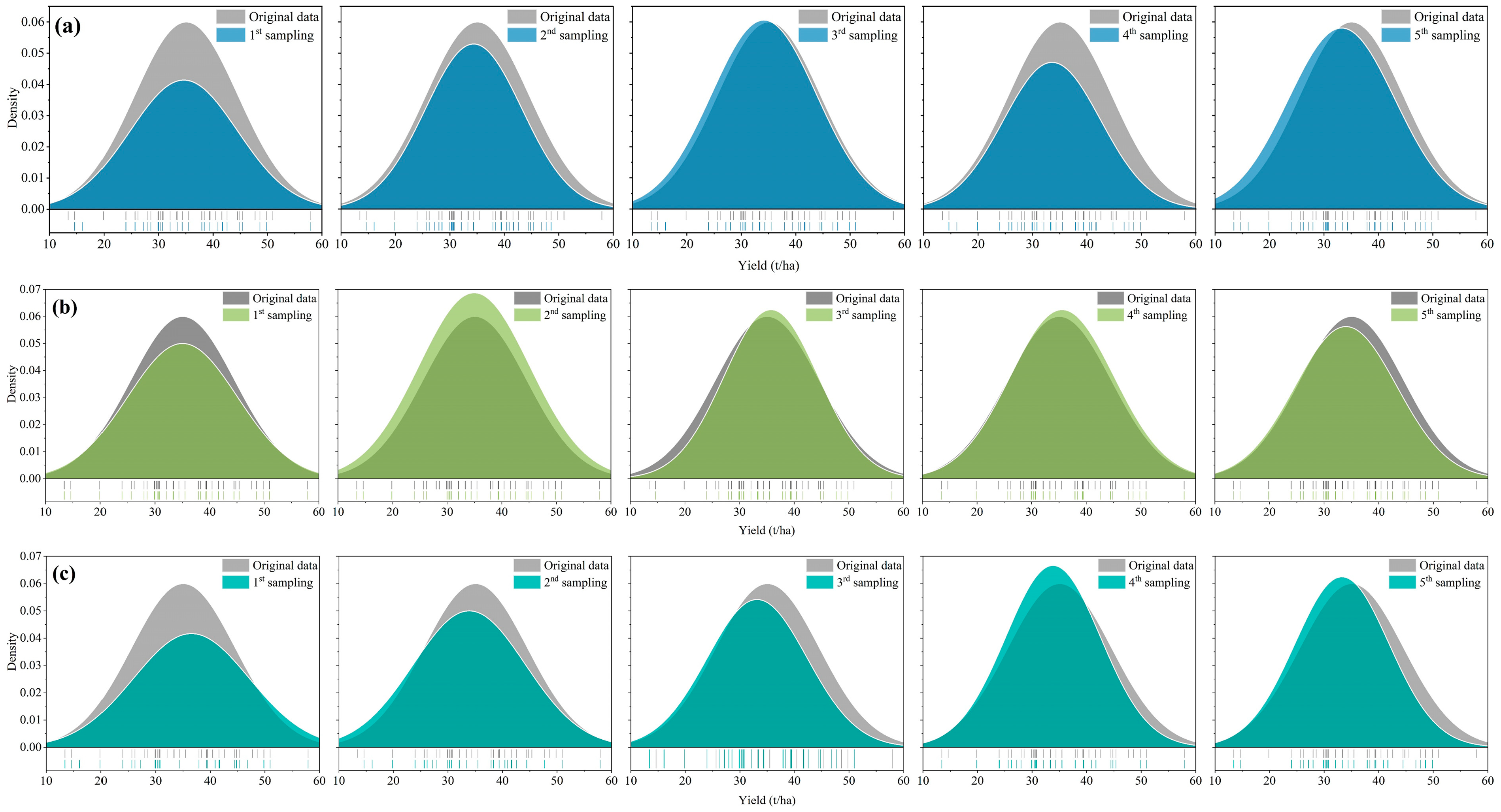
3.1. Statistical Distribution Consistency Analysis of Calibration Set and Original Sample Data
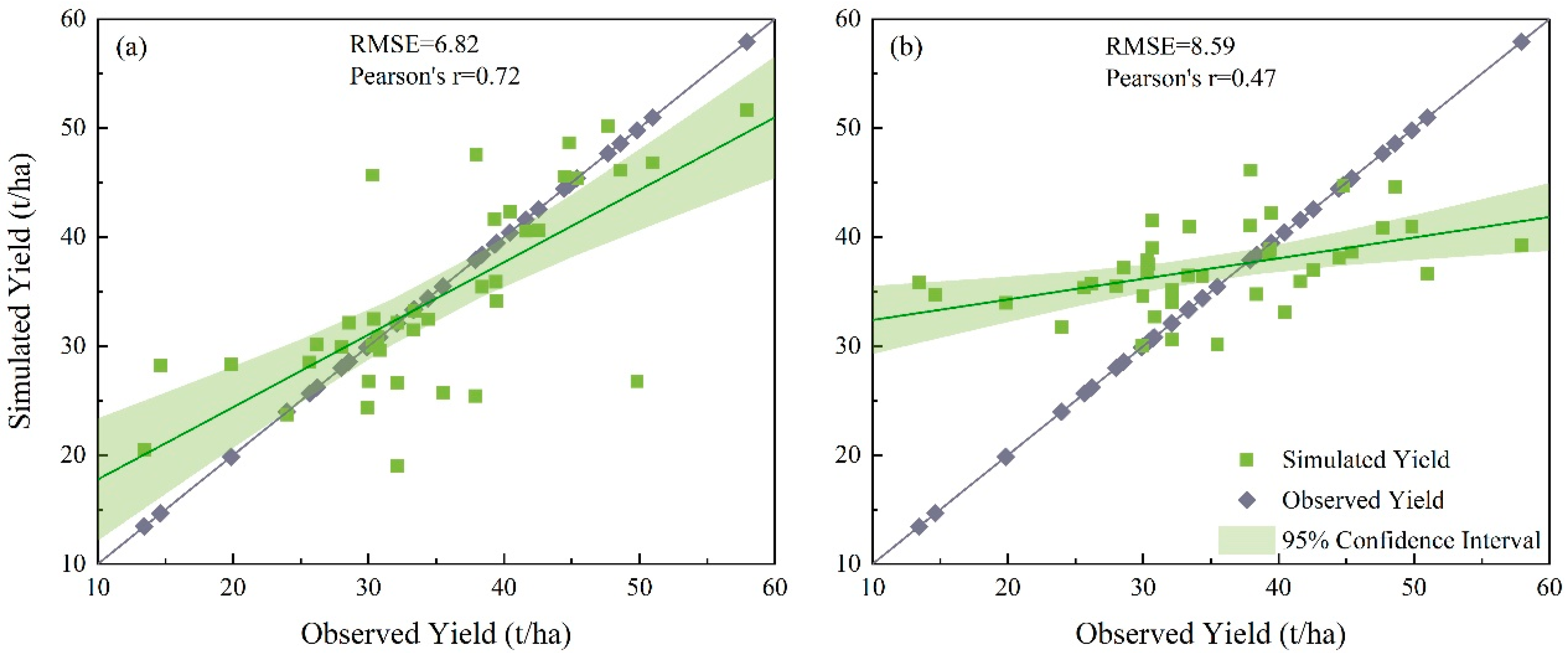
3.2. Validation of the Calibrated Yield Results Obtained with the Three Sampling Methods for Data from Each of the Observation Stations
3.3. CV Values of Calibrated Genotypic Parameters for the Three Sampling Methods
3.4. Comparison of Validation Results for Calibrated Genotypic Parameters Obtained with the Three Sampling Methods Using All 48 Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A


References
- Farina, R.; Sándor, R.; Abdalla, M.; Álvaro-Fuentes, J.; Bechini, L.; Bolinder, M.A.; Brilli, L.; Chenu, C.; Clivot, H.; De Antoni Migliorati, M.; et al. Ensemble Modelling, Uncertainty and Robust Predictions of Organic Carbon in Long-term Bare-fallow Soils. Glob. Chang. Biol. 2021, 27, 904–928. [Google Scholar] [CrossRef] [PubMed]
- Sándor, R.; Ehrhardt, F.; Grace, P.; Recous, S.; Smith, P.; Snow, V.; Soussana, J.-F.; Basso, B.; Bhatia, A.; Brilli, L.; et al. Ensemble Modelling of Carbon Fluxes in Grasslands and Croplands. Field Crops Res. 2020, 252, 107791. [Google Scholar] [CrossRef]
- Seidel, S.J.; Palosuo, T.; Thorburn, P.; Wallach, D. Towards Improved Calibration of Crop Models—Where Are We Now and Where Should We Go? Eur. J. Agron. 2018, 94, 25–35. [Google Scholar] [CrossRef]
- Porwollik, V.; Müller, C.; Elliott, J.; Chryssanthacopoulos, J.; Iizumi, T.; Ray, D.K.; Ruane, A.C.; Arneth, A.; Balkovič, J.; Ciais, P.; et al. Spatial and Temporal Uncertainty of Crop Yield Aggregations. Eur. J. Agron. 2017, 88, 10–21. [Google Scholar] [CrossRef]
- Huang, J.; Gómez-Dans, J.L.; Huang, H.; Ma, H.; Wu, Q.; Lewis, P.E.; Liang, S.; Chen, Z.; Xue, J.-H.; Wu, Y.; et al. Assimilation of Remote Sensing into Crop Growth Models: Current Status and Perspectives. Agric. For. Meteorol. 2019, 276–277, 107609. [Google Scholar] [CrossRef]
- Walker, W.E.; Harremoës, P.; Rotmans, J.; Sluijs, J.P.; van der Asselt, M.B.A.; van Janssen, P.; von Krauss, M.P.K. Defining Uncertainty: A Conceptual Basis for Uncertainty Management in Model-Based Decision Support. Integr. Assess. 2003, 4, 5–17. [Google Scholar] [CrossRef]
- Wallach, D.; Thorburn, P.; Asseng, S.; Challinor, A.J.; Ewert, F.; Jones, J.W.; Rotter, R.; Ruane, A. Estimating Model Prediction Error: Should You Treat Predictions as Fixed or Random? Environ. Model. Softw. 2016, 84, 529–539. [Google Scholar] [CrossRef]
- Tao, F.; Rötter, R.P.; Palosuo, T.; Gregorio Hernández Díaz-Ambrona, C.; Mínguez, M.I.; Semenov, M.A.; Kersebaum, K.C.; Nendel, C.; Specka, X.; Hoffmann, H.; et al. Contribution of Crop Model Structure, Parameters and Climate Projections to Uncertainty in Climate Change Impact Assessments. Glob. Chang. Biol. 2018, 24, 1291–1307. [Google Scholar] [CrossRef]
- Wallach, D.; Thorburn, P.J. Estimating Uncertainty in Crop Model Predictions: Current Situation and Future Prospects. Eur. J. Agron. 2017, 88, A1–A7. [Google Scholar] [CrossRef]
- Ramirez-Villegas, J.; Koehler, A.-K.; Challinor, A.J. Assessing Uncertainty and Complexity in Regional-Scale Crop Model Simulations. Eur. J. Agron. 2017, 88, 84–95. [Google Scholar] [CrossRef]
- Chapagain, R.; Remenyi, T.A.; Harris, R.M.B.; Mohammed, C.L.; Huth, N.; Wallach, D.; Rezaei, E.E.; Ojeda, J.J. Decomposing Crop Model Uncertainty: A Systematic Review. Field Crops Res. 2022, 279, 108448. [Google Scholar] [CrossRef]
- Ma, L.; Malone, R.W.; Heilman, P.; Ahuja, L.R.; Meade, T.; Saseendran, S.A.; Ascough, J.C.; Kanwar, R.S. Sensitivity of Tile Drainage Flow and Crop Yield on Measured and Calibrated Soil Hydraulic Properties. Geoderma 2007, 140, 284–296. [Google Scholar] [CrossRef]
- Varella, H.; Buis, S.; Launay, M.; Guérif, M. Global Sensitivity Analysis for Choosing the Main Soil Parameters of a Crop Model to Be Determined. Agric. Sci. 2012, 3, 24661. [Google Scholar] [CrossRef]
- Rosenzweig, C.; Elliott, J.; Deryng, D.; Ruane, A.C.; Müller, C.; Arneth, A.; Boote, K.J.; Folberth, C.; Glotter, M.; Khabarov, N.; et al. Assessing Agricultural Risks of Climate Change in the 21st Century in a Global Gridded Crop Model Intercomparison. Proc. Natl. Acad. Sci. USA 2014, 111, 3268–3273. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.X.; He, J.R.; Williams, R.C.; Izaurralde, J.D. Atwood sensitivity and uncertainty analyses of crop yields and soil organic carbon simulated with epic. Trans. ASAE 2005, 48, 1041–1054. [Google Scholar] [CrossRef]
- Araya, A.; Hoogenboom, G.; Luedeling, E.; Hadgu, K.M.; Kisekka, I.; Martorano, L.G. Assessment of Maize Growth and Yield Using Crop Models under Present and Future Climate in Southwestern Ethiopia. Agric. For. Meteorol. 2015, 214–215, 252–265. [Google Scholar] [CrossRef]
- Deb, P.; Kiem, A.S.; Babel, M.S.; Chu, S.T.; Chakma, B. Evaluation of Climate Change Impacts and Adaptation Strategies for Maize Cultivation in the Himalayan Foothills of India. J. Water Clim. Chang. 2015, 6, 596–614. [Google Scholar] [CrossRef]
- Li, Z.; He, J.; Xu, X.; Jin, X.; Huang, W.; Clark, B.; Yang, G.; Li, Z. Estimating Genetic Parameters of DSSAT-CERES Model with the GLUE Method for Winter Wheat (Triticum aestivum L.) Production. Comput. Electron. Agric. 2018, 154, 213–221. [Google Scholar] [CrossRef]
- Pathak, T.B.; Jones, J.W.; Fraisse, C.W.; Wright, D.; Hoogenboom, G. Uncertainty Analysis and Parameter Estimation for the CSM-CROPGRO-Cotton Model. Agron. J. 2012, 104, 1363–1373. [Google Scholar] [CrossRef]
- Sun, M.; Zhang, X.; Huo, Z.; Feng, S.; Huang, G.; Mao, X. Uncertainty and Sensitivity Assessments of an Agricultural–Hydrological Model (RZWQM2) Using the GLUE Method. J. Hydrol. 2016, 534, 19–30. [Google Scholar] [CrossRef]
- Jin, X.; Li, Z.; Nie, C.; Xu, X.; Feng, H.; Guo, W.; Wang, J. Parameter Sensitivity Analysis of the AquaCrop Model Based on Extended Fourier Amplitude Sensitivity under Different Agro-Meteorological Conditions and Application. Field Crops Res. 2018, 226, 1–15. [Google Scholar] [CrossRef]
- Tan, J.; Cao, J.; Cui, Y.; Duan, Q.; Gong, W. Comparison of the Generalized Likelihood Uncertainty Estimation and Markov Chain Monte Carlo Methods for Uncertainty Analysis of the ORYZA_V3 Model. Agron. J. 2019, 111, 555–564. [Google Scholar] [CrossRef]
- Cooman, A.; Schrevens, E. A Monte Carlo Approach for Estimating the Uncertainty of Predictions with the Tomato Plant Growth Model, Tomgro. Biosyst. Eng. 2006, 94, 517–524. [Google Scholar] [CrossRef]
- Dzotsi, K.A.; Basso, B.; Jones, J.W. Parameter and Uncertainty Estimation for Maize, Peanut and Cotton Using the SALUS Crop Model. Agric. Syst. 2015, 135, 31–47. [Google Scholar] [CrossRef]
- Wallach, D.; Thorburn, P.J. The Error in Agricultural Systems Model Prediction Depends on the Variable Being Predicted. Environ. Model. Softw. 2014, 62, 487–494. [Google Scholar] [CrossRef]
- Ines, A.V.M.; Honda, K.; Das Gupta, A.; Droogers, P.; Clemente, R.S. Combining Remote Sensing-Simulation Modeling and Genetic Algorithm Optimization to Explore Water Management Options in Irrigated Agriculture. Agric. Water Manag. 2006, 83, 221–232. [Google Scholar] [CrossRef]
- Dong, T.; Liu, J.; Qian, B.; Zhao, T.; Jing, Q.; Geng, X.; Wang, J.; Huffman, T.; Shang, J. Estimating Winter Wheat Biomass by Assimilating Leaf Area Index Derived from Fusion of Landsat-8 and MODIS Data. Int. J. Appl. Earth Obs. Geoinf. 2016, 49, 63–74. [Google Scholar] [CrossRef]
- Jin, X.; Li, Z.; Yang, G.; Yang, H.; Feng, H.; Xu, X.; Wang, J.; Li, X.; Luo, J. Winter Wheat Yield Estimation Based on Multi-Source Medium Resolution Optical and Radar Imaging Data and the AquaCrop Model Using the Particle Swarm Optimization Algorithm. ISPRS J. Photogramm. Remote Sens. 2017, 126, 24–37. [Google Scholar] [CrossRef]
- Silvestro, P.C.; Pignatti, S.; Pascucci, S.; Yang, H.; Li, Z.; Yang, G.; Huang, W.; Casa, R. Estimating Wheat Yield in China at the Field and District Scale from the Assimilation of Satellite Data into the Aquacrop and Simple Algorithm for Yield (SAFY) Models. Remote Sens. 2017, 9, 509. [Google Scholar] [CrossRef]
- Jégo, G.; Pattey, E.; Liu, J. Using Leaf Area Index, Retrieved from Optical Imagery, in the STICS Crop Model for Predicting Yield and Biomass of Field Crops. Field Crops Res. 2012, 131, 63–74. [Google Scholar] [CrossRef]
- Dente, L.; Satalino, G.; Mattia, F.; Rinaldi, M. Assimilation of Leaf Area Index Derived from ASAR and MERIS Data into CERES-Wheat Model to Map Wheat Yield. Remote Sens. Environ. 2008, 112, 1395–1407. [Google Scholar] [CrossRef]
- He, B.; Li, X.; Quan, X.; Qiu, S. Estimating the Aboveground Dry Biomass of Grass by Assimilation of Retrieved LAI Into a Crop Growth Model. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 550–561. [Google Scholar] [CrossRef]
- Jin, H.; Li, A.; Wang, J.; Bo, Y. Improvement of Spatially and Temporally Continuous Crop Leaf Area Index by Integration of CERES-Maize Model and MODIS Data. Eur. J. Agron. 2016, 78, 1–12. [Google Scholar] [CrossRef]
- Curnel, Y.; de Wit, A.J.W.; Duveiller, G.; Defourny, P. Potential Performances of Remotely Sensed LAI Assimilation in WOFOST Model Based on an OSS Experiment. Agric. For. Meteorol. 2011, 151, 1843–1855. [Google Scholar] [CrossRef]
- Roux, S.; Brun, F.; Wallach, D. Combining Input Uncertainty and Residual Error in Crop Model Predictions: A Case Study on Vineyards. Eur. J. Agron. 2014, 52, 191–197. [Google Scholar] [CrossRef]
- Ibrahim, O.M.; Gaafar, A.A.; Wali, A.M.; Tawfik, M.M.; El-Nahas, M.M. Estimating Cultivar Coefficients of a Spring Wheat Using GenCalc and GLUE in DSSAT. J. Agron. 2016, 15, 130–135. [Google Scholar] [CrossRef]
- Tao, F.; Zhang, Z.; Liu, J.; Yokozawa, M. Modelling the Impacts of Weather and Climate Variability on Crop Productivity over a Large Area: A New Super-Ensemble-Based Probabilistic Projection. Agric. For. Meteorol. 2009, 149, 1266–1278. [Google Scholar] [CrossRef]
- Trnka, M.; Dubrovský, M.; Semerádová, D.; Žalud, Z. Projections of Uncertainties in Climate Change Scenarios into Expected Winter Wheat Yields. Theor Appl Clim. 2004, 77, 229–249. [Google Scholar] [CrossRef]
- Wang, Z. Potato Processing Industry in China: Current Scenario, Future Trends and Global Impact. Potato Res. 2023, 66, 543–562. [Google Scholar] [CrossRef]
- Na, T. Analysis of Accuracy and Stability in Regional Trial of Potato Varieties. Chin. Acad. Agric. Sci. 2011. Available online: https://www.cnki.net/KCMS/detail/detail.aspx?dbcode=CMFD&dbname=CMFD2012&filename=1012318224.nh&uniplatform=OVERSEA&v=3HNO6lOT58LlKVk4qABr8jWx3PCs11KNuQbKz2gjn0g-iepASYD5CjSi8R6hXY7p (accessed on 31 October 2023).
- Duan, D.; YingBin, H.; JinKuan, Y.; Li, L.; RuiYang, X.; WenJuan, L. Parameter Sensitivity Analysis and Suitability Evaluation of DSSAT-SUBSTOR Potato Model. J. Anhui Agric. Univ. 2019, 46, 521–527. [Google Scholar]
- Gao, F.; Luan, X.; Yin, Y.; Sun, S.; Li, Y.; Mo, F.; Wang, J. Exploring Long-Term Impacts of Different Crop Rotation Systems on Sustainable Use of Groundwater Resources Using DSSAT Model. J. Clean. Prod. 2022, 336, 130377. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, M.; Liao, Z.; Guo, J.; Zhang, F.; Fan, J.; Feng, H.; Yang, Q.; Wu, L.; Wang, X. Performance Evaluation of AquaCrop and DSSAT-SUBSTOR-Potato Models in Simulating Potato Growth, Yield and Water Productivity under Various Drip Fertigation Regimes. Agric. Water Manag. 2023, 276, 108076. [Google Scholar] [CrossRef]
- Fleisher, D.H.; Cavazzoni, J.; Giacomelli, G.A.; Ting, K.C. Adaptation of SUBSTOR for Controlled-Environment Potato Production with Elevated Carbon Dioxide. Trans. ASAE 2003, 46, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Griffin, T.S.; Johnson, B.S.; Ritchie, J.T.; IBSNAT Project; University of Hawaii (Honolulu); Department of Agronomy and Soil Science; Michigan State University, Department of Crop and Soil Science. A Simulation Model for Potato Growth and Development: SUBSTOR-Potato Version 2.0; IBSNAT Research Report Series; Michigan State University, Department of Crop and Soil Sciences: East Lansing, MI, USA, 1993. [Google Scholar]
- Zhou, Z. Machine Learning; Tsinghua University Press: Beijing, China, 2016. [Google Scholar]
- Dietterich, T.G. Approximate Statistical Tests for Comparing Supervised Classification Learning Algorithms. Neural Comput 1998, 10, 1895–1923. [Google Scholar] [CrossRef] [PubMed]
- Efron, B.; Tibshirani, R.J. An Introduction to the Bootstrap; Chapman and Hall/CRC: Broken Sound Parkway, NW, USA, 1994; ISBN 978-0-429-24659-3. [Google Scholar]
- Jones, J.W.; He, J.; Boote, K.J.; Wilkens, P.; Porter, C.H.; Hu, Z. Estimating DSSAT Cropping System Cultivar-Specific Parameters Using Bayesian Techniques. In Advances in Agricultural Systems Modeling; Ahuja, L.R., Ma, L., Eds.; American Society of Agronomy and Soil Science Society of America: Madison, WI, USA, 2015; pp. 365–393. ISBN 978-0-89118-196-5. [Google Scholar]
- Raymundo, R. Performance of the SUBSTOR-Potato Model across Contrasting Growing Conditions. Field Crops Res. 2017, 202, 57–76. [Google Scholar] [CrossRef]
- Yang, Y.; Wilson, L.T.; Li, T.; Paleari, L.; Confalonieri, R.; Zhu, Y.; Tang, L.; Qiu, X.; Tao, F.; Chen, Y.; et al. Integration of Genomics with Crop Modeling for Predicting Rice Days to Flowering: A Multi-Model Analysis. Field Crops Res. 2022, 276, 108394. [Google Scholar] [CrossRef]
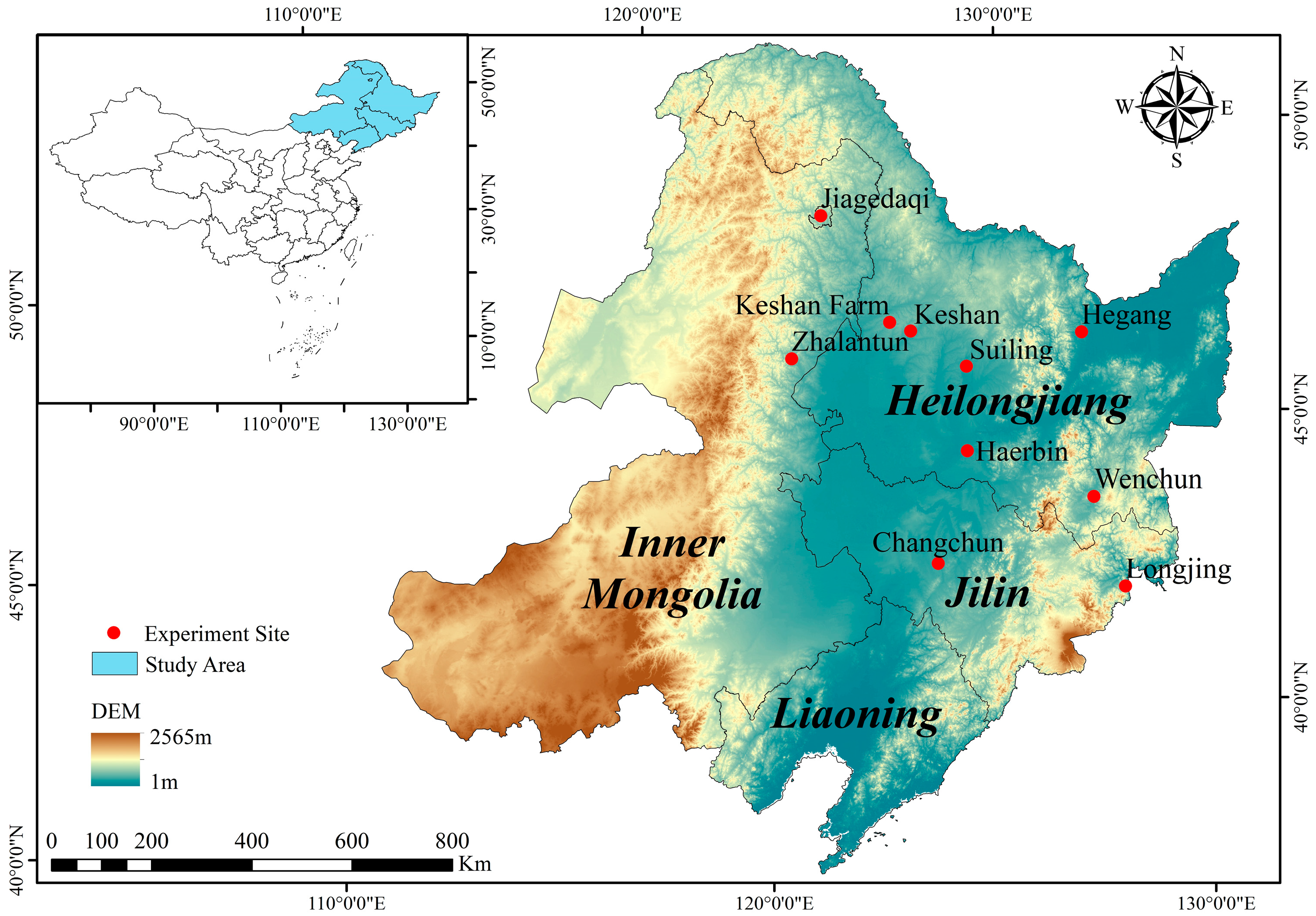

| Experimental Site | Year | Longitude (°) | Latitude (°) | Altitude (m) | Observed Yield (t/ha) | GSL * (d) | ≥10 °C GDD * | Soil Type |
|---|---|---|---|---|---|---|---|---|
| Haerbin | 2016–2020 | 126.63 | 45.75 | 155 | 32.8 ± 13.2 * | 114 ± 7 | 2243 ± 186 | SiLo * |
| Wenchun | 2016–2020 | 129.50 | 44.42 | 251 | 44.2 ± 3.9 | 124 ± 4 | 2259 ± 85 | SiLo |
| Zhalantun | 2016–2020 | 122.74 | 48.03 | 307 | 37.4 ± 5.4 | 131 ± 11 | 2240 ± 142 | ClLo * |
| Keshan | 2016–2020 | 125.87 | 48.07 | 236 | 27.6 ± 2.7 | 113 ± 4 | 2252 ± 86 | SiClLo * |
| Keshan Farm | 2016–2020 | 125.37 | 48.30 | 315 | 47.1 ± 8 | 121 ± 10 | 2251 ± 182 | SiClLo |
| Suiling | 2016–2020 | 127.10 | 47.23 | 212 | 35.9 ± 4.4 | 107 ± 9 | 2254 ± 214 | SiClLo |
| Hegang | 2016–2020 | 130.27 | 47.33 | 228 | 31.1 ± 1.8 | 123 ± 11 | 2258 ± 169 | SiLo |
| Jiagedaqi | 2016–2020 | 124.12 | 50.40 | 372 | 28.7 ± 6 | 117 ± 7 | 2237 ± 105 | Lo * |
| Changchun | 2016, 2018–2020 | 125.32 | 43.83 | 237 | 34.7 ± 13.5 | 124 ± 6 | 2266 ± 106 | SiLo |
| Longjing | 2016, 2018–2020 | 129.70 | 42.70 | 242 | 33.5 ± 9.2 | 122 ± 5 | 2263 ± 77 | Lo |
| Genotypic Parameters | Definition | Type |
|---|---|---|
| G2 (cm2·m−2·d−1) | Leaf expansion rate | For yield |
| G3 (g·m−2·d−1) | Tuber growth rate | For yield |
| PD | Determinacy | For yield |
| P2 | Sensitivity of tuber initiation to photoperiod | For phenology |
| TC (°C) | Coefficient for critical temperature | For phenology |
| Sampling Sequence Number | Sampling Method | Calibration Year | Validation Year |
|---|---|---|---|
| 1 | HO | 2017, 2018, 2019 | 2016, 2020 |
| 2 | 2016, 2018, 2019 | 2017, 2020 | |
| 3 | 2016, 2017, 2019 | 2018, 2020 | |
| 4 | 2016, 2017, 2018 | 2019, 2020 | |
| 5 | 2017, 2018, 2020 | 2016, 2019 | |
| 6 | CA | 2017, 2018, 2019, 2020 | 2016 |
| 7 | 2016, 2018, 2019, 2020 | 2017 | |
| 8 | 2016, 2017, 2019, 2020 | 2018 | |
| 9 | 2016, 2017, 2018, 2020 | 2019 | |
| 10 | 2016, 2017, 2018, 2019 | 2020 | |
| 11 | BS | 2017, 2018, 2020, 2018, 2020 | 2016, 2019 |
| 12 | 2017, 2019, 2020, 2019, 2020 | 2016, 2018 | |
| 13 | 2017, 2018, 2019, 2020, 2020 | 2016 | |
| 14 | 2016, 2018, 2019, 2016, 2018 | 2017, 2020 | |
| 15 | 2016, 2018, 2019, 2020, 2018 | 2017 |
| Sampling Sequence Number | Sampling Methods | Calibration Set | Validation Set |
|---|---|---|---|
| 1 | HO | 29 * | 19 ** |
| 2 | 34 * | 14 ** | |
| 3 | 33 * | 15 ** | |
| 4 | 34 * | 14 ** | |
| 5 | 31 * | 17 ** | |
| 6 | CA | 2017, 2018, 2019, 2020 *** | 2016 **** |
| 7 | 2016, 2018, 2019, 2020 *** | 2017 **** | |
| 8 | 2016, 2017, 2019, 2020 *** | 2018 **** | |
| 9 | 2016, 2017, 2018, 2020 *** | 2019 **** | |
| 10 | 2016, 2017, 2018, 2019 *** | 2020 **** | |
| 11 | BS | 30 * | 18 ** |
| 12 | 31 * | 17 ** | |
| 13 | 34 * | 14 ** | |
| 14 | 31 * | 17 ** | |
| 15 | 30 * | 18 ** |
| Observation Station | Sampling Methods | Mean of RMSE | CV of RMSE | Mean of RRMSE | CV of RRMSE |
|---|---|---|---|---|---|
| Zhalantun | HO | 8.70 | 20.86% | 23.09% | 21.84% |
| CA | 5.74 | 15.34% | |||
| BS | 8.12 | 23.11% | |||
| Wenchun | HO | 6.35 | 20.04% | 14.93% | 22.99% |
| CA | 4.58 | 10.35% | |||
| BS | 4.54 | 10.12% | |||
| Hegang | HO | 3.01 | 26.63% | 9.45% | 25.04% |
| CA | 1.84 | 5.93% | |||
| BS | 2.09 | 6.73% | |||
| Suiling | HO | 10.06 | 15.17% | 31.87% | 18.32% |
| CA | 7.73 | 24.40% | |||
| BS | 7.90 | 22.83% | |||
| Keshan | HO | 3.82 | 37.45% | 13.55% | 37.41% |
| CA | 2.89 | 10.48% | |||
| BS | 1.72 | 6.08% | |||
| Keshan Farm | HO | 6.40 | 6.07% | 12.87% | 7.85% |
| CA | 5.96 | 12.67% | |||
| BS | 6.73 | 14.58% | |||
| Jiagedaqi | HO | 7.60 | 16.94% | 25.22% | 18.35% |
| CA | 10.25 | 35.70% | |||
| BS | 7.89 | 27.96% | |||
| Haerbin | HO | 14.52 | 11.22% | 38.59% | 8.79% |
| CA | 11.69 | 35.64% | |||
| BS | 13.98 | 42.46% |
| Observation Station | Sampling Method | G2 | G3 | PD | P2 | TC |
|---|---|---|---|---|---|---|
| Zhalantun | HO | 1772.54 | 24.46 | 0.78 | 0.48 | 16.98 |
| CA | 1683.80 | 23.62 | 0.68 | 0.50 | 16.48 | |
| BS | 1779.36 | 22.38 | 0.76 | 0.42 | 16.44 | |
| CV | 3.05% | 4.46% | 7.15% | 8.92% | 1.81% | |
| Wenchun | HO | 1384.06 | 23.74 | 0.76 | 0.40 | 17.50 |
| CA | 1585.56 | 23.52 | 1.00 | 0.40 | 18.30 | |
| BS | 1329.64 | 23.96 | 1.00 | 0.40 | 17.70 | |
| CV | 9.41% | 0.93% | 15.06% | 0.00% | 2.33% | |
| Hegang | HO | 1548.48 | 23.28 | 0.84 | 0.64 | 15.96 |
| CA | 1266.10 | 23.60 | 0.84 | 0.80 | 17.56 | |
| BS | 1476.32 | 23.02 | 0.96 | 0.80 | 18.44 | |
| CV | 10.26% | 1.25% | 7.87% | 12.37% | 7.26% | |
| Suiling | HO | 1262.18 | 22.94 | 0.60 | 0.80 | 18.58 |
| CA | 1576.18 | 21.70 | 0.76 | 0.74 | 19.44 | |
| BS | 1544.84 | 22.96 | 0.68 | 0.82 | 19.34 | |
| CV | 11.84% | 3.20% | 11.76% | 5.29% | 2.46% | |
| Keshan | HO | 1683.14 | 23.72 | 0.88 | 0.86 | 19.78 |
| CA | 1835.72 | 23.38 | 0.88 | 0.84 | 19.10 | |
| BS | 1619.58 | 23.00 | 1.00 | 0.90 | 20.58 | |
| CV | 6.49% | 1.54% | 7.53% | 3.53% | 3.74% | |
| Keshan Farm | HO | 1597.34 | 23.18 | 0.78 | 0.34 | 18.86 |
| CA | 1729.30 | 23.80 | 0.60 | 0.34 | 16.58 | |
| BS | 1810.84 | 23.88 | 0.66 | 0.36 | 16.88 | |
| CV | 6.29% | 1.62% | 13.48% | 3.33% | 7.10% | |
| Jiagedaqi | HO | 1756.38 | 21.98 | 0.92 | 0.72 | 18.92 |
| CA | 1460.30 | 23.22 | 0.82 | 0.72 | 18.80 | |
| BS | 1559.70 | 22.64 | 0.78 | 0.72 | 20.24 | |
| CV | 9.46% | 2.74% | 8.58% | 0.00% | 4.14% | |
| Haerbin | HO | 1633.28 | 23.20 | 0.78 | 0.72 | 16.84 |
| CA | 1348.58 | 23.30 | 0.92 | 0.54 | 15.82 | |
| BS | 1764.50 | 23.28 | 0.82 | 0.56 | 15.18 | |
| CV | 13.44% | 0.23% | 8.58% | 16.26% | 5.25% |
| Sampling Method | G2 | G3 | PD | P2 | TC |
|---|---|---|---|---|---|
| HO | 11.28% | 3.08% | 12.14% | 30.89% | 7.26% |
| CA | 12.39% | 2.84% | 15.95% | 31.10% | 7.62% |
| BS | 10.40% | 2.39% | 16.59% | 34.37% | 10.51% |
| CV of CV | 8.82% | 12.75% | 16.17% | 6.06% | 21.04% |
| Sampling Sequence Number | Sampling Method | G2 | G3 | PD | P2 | TC | RMSE | RRMSE |
|---|---|---|---|---|---|---|---|---|
| 1 | HO | 2044.40 | 23.10 | 0.90 | 0.70 | 18.20 | 8.85 | 25.57% |
| 2 | HO | 1230.40 | 22.70 | 0.90 | 0.70 | 18.30 | 11.45 | 33.32% |
| 3 | HO | 1962.30 | 25.00 | 0.80 | 0.70 | 18.40 | 8.54 | 25.00% |
| 4 | HO | 2091.20 | 25.80 | 1.00 | 0.70 | 19.60 | 10.82 | 32.21% |
| 5 | HO | 1729.40 | 23.30 | 0.70 | 0.80 | 20.10 | 9.58 | 28.82% |
| Average | 1811.54 | 23.98 | 0.86 | 0.72 | 18.92 | 9.85 | 28.98% | |
| 6 | CA | 1960.00 | 24.00 | 0.60 | 0.70 | 19.70 | 11.16 | 31.85% |
| 7 | CA | 992.00 | 21.90 | 0.90 | 0.70 | 21.10 | 9.19 | 26.28% |
| 8 | CA | 1813.20 | 23.40 | 1.00 | 0.50 | 16.40 | 12.82 | 35.83% |
| 9 | CA | 1902.10 | 24.30 | 1.00 | 0.60 | 17.70 | 8.29 | 23.30% |
| 10 | CA | 1790.00 | 22.70 | 0.80 | 0.70 | 17.20 | 17.04 | 50.11% |
| Average | 1691.46 | 23.26 | 0.86 | 0.64 | 18.42 | 11.70 | 33.48% | |
| 11 | BS | 990.70 | 24.00 | 1.00 | 0.60 | 18.30 | 7.00 | 19.61% |
| 12 | BS | 2193.30 | 24.90 | 0.80 | 0.70 | 18.30 | 7.91 | 21.76% |
| 13 | BS | 1702.00 | 22.40 | 0.80 | 0.70 | 16.90 | 16.75 | 45.88% |
| 14 | BS | 1750.30 | 21.80 | 0.60 | 0.80 | 21.50 | 9.60 | 26.95% |
| 15 | BS | 1104.20 | 25.50 | 0.80 | 0.80 | 21.30 | 10.71 | 28.86% |
| Average | 1548.10 | 23.72 | 0.80 | 0.72 | 19.26 | 10.39 | 28.61% | |
| CV | 23.22% | 5.17% | 15.55% | 11.13% | 8.25% | 26.80% | 26.89% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Wang, X.; He, Y.; Zha, Y.; Chen, H.; Han, S. Variability in Estimating Crop Model Genotypic Parameters: The Impact of Different Sampling Methods and Sizes. Agriculture 2023, 13, 2207. https://doi.org/10.3390/agriculture13122207
Ma X, Wang X, He Y, Zha Y, Chen H, Han S. Variability in Estimating Crop Model Genotypic Parameters: The Impact of Different Sampling Methods and Sizes. Agriculture. 2023; 13(12):2207. https://doi.org/10.3390/agriculture13122207
Chicago/Turabian StyleMa, Xintian, Xiangyi Wang, Yingbin He, Yan Zha, Huicong Chen, and Shengnan Han. 2023. "Variability in Estimating Crop Model Genotypic Parameters: The Impact of Different Sampling Methods and Sizes" Agriculture 13, no. 12: 2207. https://doi.org/10.3390/agriculture13122207





