Genome-Wide Association Analysis-Based Mining of Quality Genes Related to Linoleic and Linolenic Acids in Soybean
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials and Cultivation Management
2.2. Trait Identification and Statistical Analysis
2.3. SNP Genotyping
2.4. Genome-Wide Association Analysis
2.5. Screening and Annotation of Candidate Genes
2.6. Preliminary Identification of Candidate Genes
3. Results
3.1. Analysis of Phenotypic Data of Soybean Linoleic Acid and Linolenic Acid Content
3.2. Genotyping
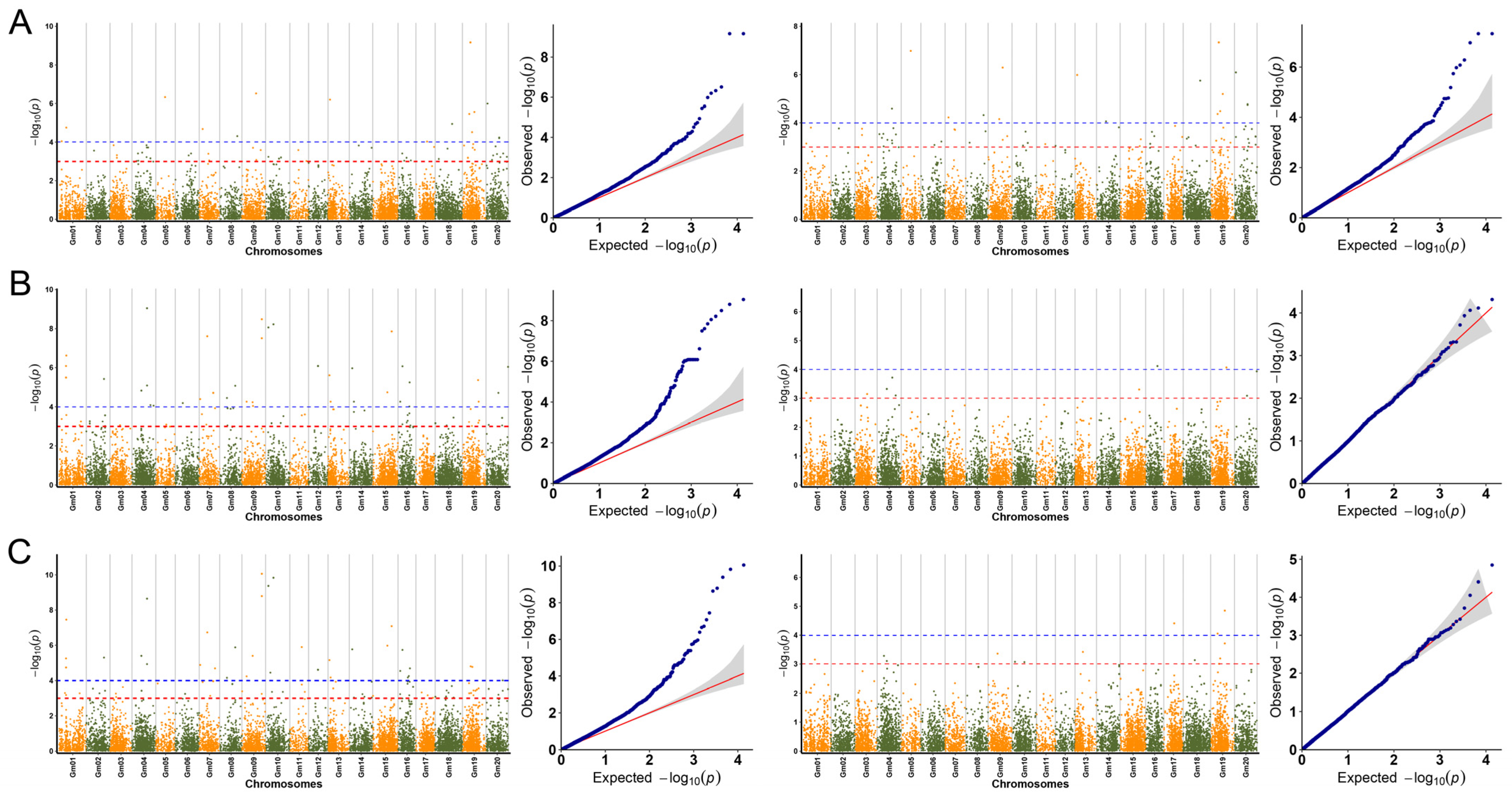
3.3. Genome-Wide Association Study of Linoleic Acid and Linolenic Acid Contents in Soybean Seeds
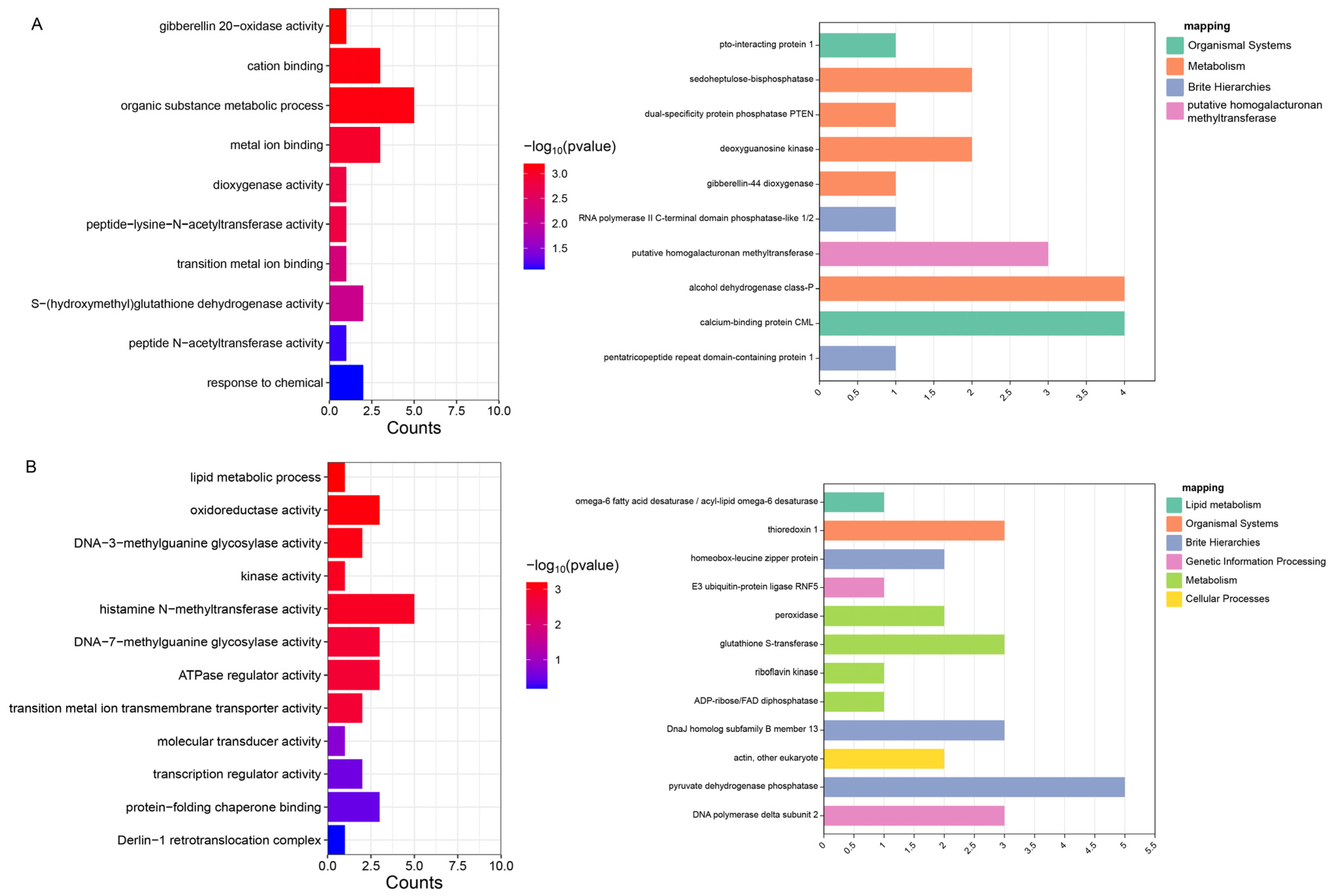
3.4. Screening and Annotation of Candidate Genes
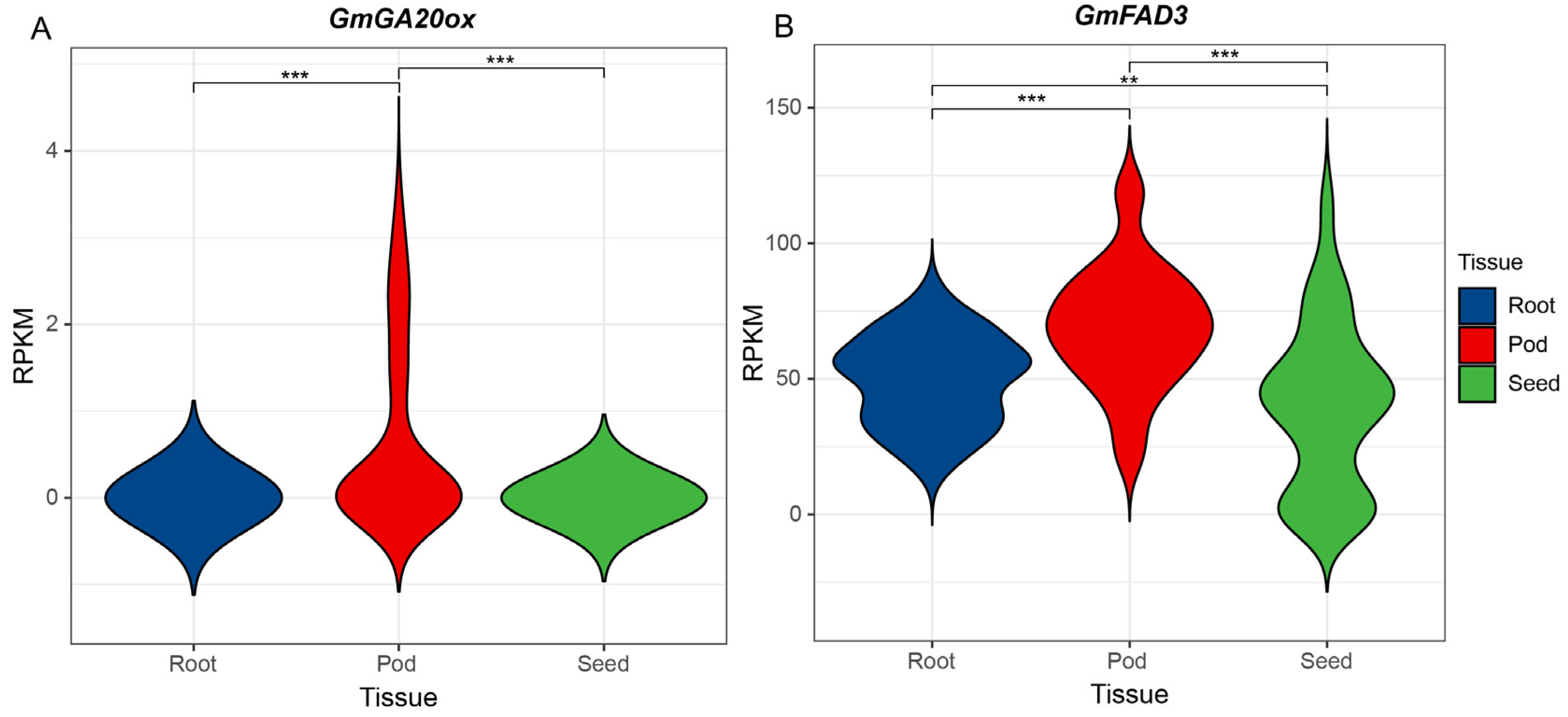
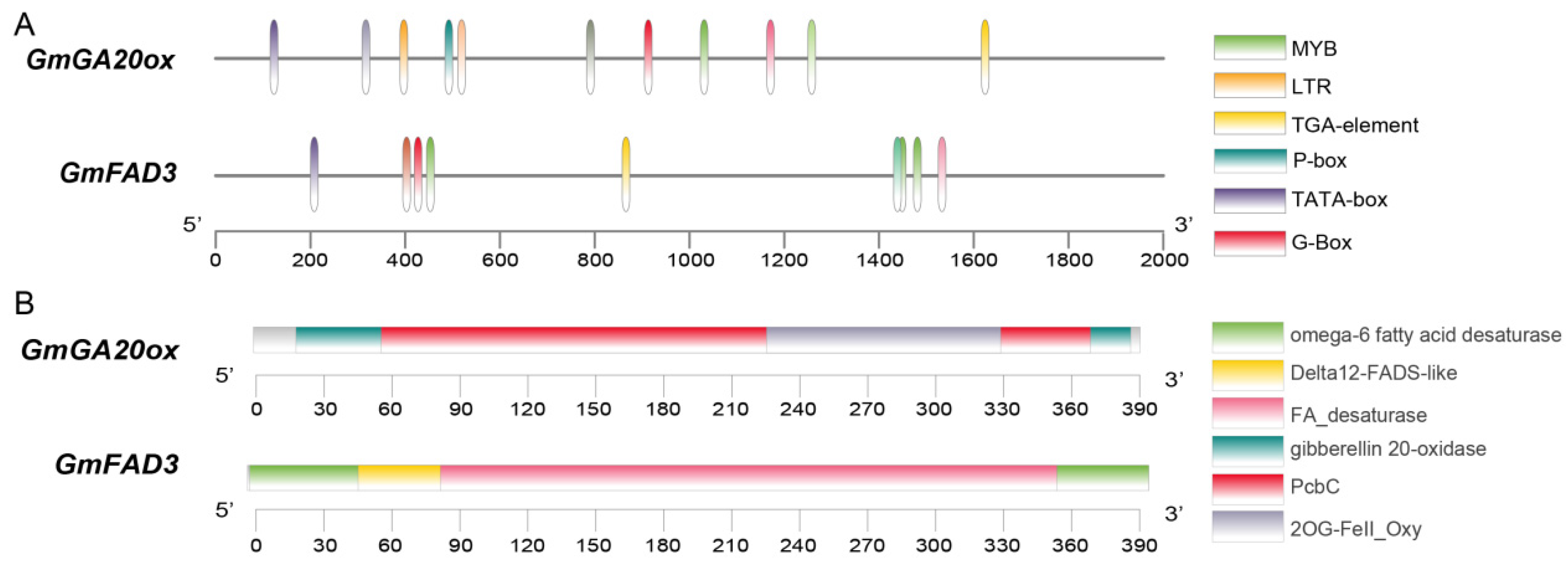
3.5. Preliminary Identification of Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ayilara, M.S.; Adeleke, B.S.; Babalola, O.O. Bioprospecting and Challenges of Plant Microbiome Research for Sustainable Agriculture, a Review on Soybean Endophytic Bacteria. Microb. Ecol. 2023, 85, 1113–1135. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M.; Yamada, T.; Hayashi, M.; Mano, S.; Nishimura, M. Soybean (Glycine Max L.) Triacylglycerol Lipase GmSDP1 Regulates the Quality and Quantity of Seed Oil. Sci. Rep. 2019, 9, 8924. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.K.; Achar, S.K.; Dasari, S.R.; Borugadda, V.B.; Goud, V.V. Analysis of Thermal, Oxidative and Cold Flow Properties of Methyl and Ethyl Esters Prepared from Soybean and Mustard Oils. J. Therm. Anal. Calorim. 2017, 130, 1501–1511. [Google Scholar] [CrossRef]
- Xu, K.; Saaoud, F.; Shao, Y.; Lu, Y.; Wu, S.; Zhao, H.; Chen, K.; Vazquez-Padron, R.; Jiang, X.; Wang, H.; et al. Early Hyperlipidemia Triggers Metabolomic Reprogramming with Increased SAH, Increased Acetyl-CoA-Cholesterol Synthesis, and Decreased Glycolysis. Redox Biol. 2023, 64, 102771. [Google Scholar] [CrossRef] [PubMed]
- Flock, M.R.; Green, M.H.; Kris-Etherton, P.M. Effects of Adiposity on Plasma Lipid Response to Reductions in Dietary Saturated Fatty Acids and Cholesterol. Adv. Nutr. 2011, 2, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-Y.; Franco, D.A.; Schwartz, E.; D’Souza, K.; Karnick, S.; Reaven, P.D. HDL Inhibits Saturated Fatty Acid Mediated Augmentation of Innate Immune Responses in Endothelial Cells by a Novel Pathway. Atherosclerosis 2017, 259, 83–96. [Google Scholar] [CrossRef]
- He, M.; Qin, C.-X.; Wang, X.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Biosynthesis and Regulation. Front. Plant Sci. 2020, 11, 390. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, F.; Huang, H.; Mao, Y.; Ye, D. Biomarker of Dietary Linoleic Acid and Risk for Stroke: A Systematic Review and Meta-Analysis. Nutrition 2020, 79–80, 110953. [Google Scholar] [CrossRef]
- Badawy, S.; Liu, Y.; Guo, M.; Liu, Z.; Xie, C.; Marawan, M.A.; Ares, I.; Lopez-Torres, B.; Martínez, M.; Maximiliano, J.-E.; et al. Conjugated Linoleic Acid (CLA) as a Functional Food: Is It Beneficial or Not? Food Res. Int. 2023, 172, 113158. [Google Scholar] [CrossRef]
- Ghanem, A.K.; Ahmad, K.; Javier, D.A.; Rezvanizadeh, V.; Kinninger, A.; Hamal, S.A.; Flores, F.; Dailing, C.; Roy, S.K.; Budoff, M.J. Effects of Supplements Containing Curcumin, Omega Fatty Acids, Gamma Linolenic Acid, Vitamin E, Vitamin D, Hydroxytyrosol, And Astaxanthin On Cardiovascular Health: A Randomized, Double-Blind, Placebo-Controlled Clinical Trial. Am. J. Prev. Cardiol. 2023, 13, 100401. [Google Scholar] [CrossRef]
- Kim, Y.; Ilich, J.Z. Implications of Dietary α-Linolenic Acid in Bone Health. Nutrition 2011, 27, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Monnard, C.R.; Dulloo, A.G. Polyunsaturated Fatty Acids as Modulators of Fat Mass and Lean Mass in Human Body Composition Regulation and Cardiometabolic Health. Obes. Rev. 2021, 22, e13197. [Google Scholar] [CrossRef] [PubMed]
- Uprety, B.K.; Rakshit, S.K. Use of Essential Oils From Various Plants to Change the Fatty Acids Profiles of Lipids Obtained From Oleaginous Yeasts. J. Am. Oil Chem. Soc. 2018, 95, 135–148. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Rajion, M.A.; Meng, G.Y.; Boo, L.J.; Ebrahimi, M.; Royan, M.; Sahebi, M.; Azizi, P.; Abiri, R.; Jahromi, M.F. Conjugated Linoleic Acid: A Potent Fatty Acid Linked to Animal and Human Health. Crit. Rev. Food Sci. Nutr. 2017, 57, 2737–2748. [Google Scholar] [CrossRef] [PubMed]
- Priolli, R.H.G.; Carvalho, C.R.L.; Bajay, M.M.; Pinheiro, J.B.; Vello, N.A. Genome Analysis to Identify SNPs Associated with Oil Content and Fatty Acid Components in Soybean. Euphytica 2019, 215, 54. [Google Scholar] [CrossRef]
- Silva, L.C.C.; Bueno, R.D.; da Matta, L.B.; Pereira, P.H.S.; Mayrink, D.B.; Piovesan, N.D.; Sediyama, C.S.; Fontes, E.P.B.; Cardinal, A.J.; Dal-Bianco, M. Characterization of a New GmFAD3A Allele in Brazilian CS303TNKCA Soybean Cultivar. Theor. Appl. Genet. 2018, 131, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Byfield, G.E.; Upchurch, R.G. Effect of Temperature on Microsomal Omega-3 Linoleate Desaturase Gene Expression and Linolenic Acid Content in Developing Soybean Seeds. Crop Sci. 2007, 47, 2445–2452. [Google Scholar] [CrossRef]
- Panahabadi, R.; Ahmadikhah, A.; Farrokhi, N.; Bagheri, N. Genome-Wide Association Study (GWAS) of Germination and Post-Germination Related Seedling Traits in Rice. Euphytica 2022, 218, 112. [Google Scholar] [CrossRef]
- Gajardo Balboa, H.; Wittkop, B.; Soto-Cerda, B.; Higgins, E.; Parkin, I.; Snowdon, R.; Federico, M.; Iniguez-Luy, F. Association Mapping of Seed Quality Traits in Brassica Napus L. Using GWAS and Candidate QTL Approaches. Mol. Breed. 2015, 35, 143. [Google Scholar] [CrossRef]
- Cho, S.; Kim, D.; Lee, S. A Comparative Evaluation of a Single and Stereo Lighthouse Systems for 3-D Estimation. IEEE Sens. J. 2021, 21, 24791–24800. [Google Scholar] [CrossRef]
- Mandozai, A.; Moussa, A.A.; Zhang, Q.; Qu, J.; Du, Y.; Anwari, G.; Al Amin, N.; Wang, P. Genome-Wide Association Study of Root and Shoot Related Traits in Spring Soybean (Glycine max L.) at Seedling Stages Using SLAF-Seq. Front. Plant Sci. 2021, 12, 568995. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, Y.; Tao, Y.; Xu, C.; Li, X.; Zhang, X.; Han, Y.; Yang, X.; Sun, J.; Li, W.; et al. Identification of Genetic Loci and Candidate Genes Related to Soybean Flowering through Genome Wide Association Study. BMC Genom. 2019, 20, 987. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Li, D.; Zhao, X.; Zhan, Y.; Teng, W.; Qiu, L.; Zheng, H.; Li, W.; Han, Y. Identification of a Candidate Gene Associated with Isoflavone Content in Soybean Seeds Using Genome-Wide Association and Linkage Mapping. Plant J. 2020, 104, 950–963. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Qin, D.; Piersanti, A.; Zhang, Q.; Miceli, C.; Wang, P. Genome-Wide Association Study Identifies Candidate Genes Related to Oleic Acid Content in Soybean Seeds. BMC Plant Biol. 2020, 20, 399. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chang, H.; Feng, L.; Jing, Y.; Teng, W.; Qiu, L.; Zheng, H.; Han, Y.; Li, W. Genome-wide Association Mapping and Candidate Gene Analysis for Saturated Fatty Acid Content in Soybean Seed. Plant Breed. 2019, 138, 588–598. [Google Scholar] [CrossRef]
- Sun, X.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H.; et al. SLAF-Seq: An Efficient Method of Large-Scale De Novo SNP Discovery and Genotyping Using High-Throughput Sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef]
- Wu, S.; Alseekh, S.; Cuadros-Inostroza, Á.; Fusari, C.M.; Mutwil, M.; Kooke, R.; Keurentjes, J.B.; Fernie, A.R.; Willmitzer, L.; Brotman, Y. Combined Use of Genome-Wide Association Data and Correlation Networks Unravels Key Regulators of Primary Metabolism in Arabidopsis Thaliana. PLOS Genet. 2016, 12, e1006363. [Google Scholar] [CrossRef]
- Patil, I. Visualizations with Statistical Details: The “ggstatsplot” Approach. J. Open Source Softw. 2021, 6, 3167. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, S.; Li, W.; Liu, S.; Li, X.; Fang, Y.; Zhang, J.; Wang, Y.; Xu, S.; Zhang, J.; et al. Identification of QTNs Controlling Seed Protein Content in Soybean Using Multi-Locus Genome-Wide Association Studies. Front. Plant Sci. 2018, 9, 1690. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-Wide Efficient Mixed-Model Analysis for Association Studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.D. Qqman: An R Package for Visualizing GWAS Results Using Q-Q and Manhattan Plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Zhang, Q.; Sun, T.; Wang, J.; Fei, J.; Liu, Y.; Liu, L.; Wang, P. Genome-Wide Association Study and High-Quality Gene Mining Related to Soybean Protein and Fat. BMC Genom. 2023, 24, 596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, G.; Zhang, W.; Wang, Q.; Xu, W.; Liu, X.; Cui, X.; Chen, X.; Chen, H. Identification of Loci Governing Soybean Seed Protein Content via Genome-Wide Association Study and Selective Signature Analyses. Front. Plant Sci. 2022, 13, 1045953. [Google Scholar] [CrossRef] [PubMed]
- Shook, J.M.; Zhang, J.; Jones, S.E.; Singh, A.; Diers, B.W.; Singh, A.K. Meta-GWAS for Quantitative Trait Loci Identification in Soybean. G3 GenesGenomesGenetics 2021, 11, jkab117. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, F.; Yu, Y.; Feng, L.; Jia, J.; Liu, B.; Li, B.; Guo, H.; Zhai, J. A Comprehensive Online Database for Exploring ∼20,000 Public Arabidopsis RNA-Seq Libraries. Mol. Plant 2020, 13, 1231–1233. [Google Scholar] [CrossRef]
- Karikari, B.; Wang, Z.; Zhou, Y.; Yan, W.; Feng, J.; Zhao, T. Identification of Quantitative Trait Nucleotides and Candidate Genes for Soybean Seed Weight by Multiple Models of Genome-Wide Association Study. BMC Plant Biol. 2020, 20, 404. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Valliyodan, B.; Cannon, S.B.; Bayer, P.E.; Shu, S.; Brown, A.V.; Ren, L.; Jenkins, J.; Chung, C.Y.-L.; Chan, T.-F.; Daum, C.G.; et al. Construction and Comparison of Three Reference-Quality Genome Assemblies for Soybean. Plant J. 2019, 100, 1066–1082. [Google Scholar] [CrossRef] [PubMed]
- Carrera, C.; Martínez, M.J.; Dardanelli, J.; Balzarini, M. Environmental Variation and Correlation of Seed Components in Nontransgenic Soybeans: Protein, Oil, Unsaturated Fatty Acids, Tocopherols, and Isoflavones. Crop Sci. 2011, 51, 800–809. [Google Scholar] [CrossRef]
- Li, X.; Tian, R.; Shao, Z.; Zhang, H.; Chu, J.; Li, W.; Kong, Y.; Du, H.; Zhang, C. Genetic Loci and Causal Genes for Seed Fatty Acids Accumulation across Multiple Environments and Genetic Backgrounds in Soybean. Mol. Breed. 2021, 41, 31. [Google Scholar] [CrossRef] [PubMed]
- Sritongtae, C.; Monkham, T.; Sanitchon, J.; Lodthong, S.; Srisawangwong, S.; Chankaew, S. Identification of Superior Soybean Cultivars through the Indication of Specific Adaptabilities within Duo-Environments for Year-Round Soybean Production in Northeast Thailand. Agronomy 2021, 11, 585. [Google Scholar] [CrossRef]
- Bachlava, E.; Burton, J.W.; Brownie, C.; Wang, S.; Auclair, J.; Cardinal, A.J. Heritability of Oleic Acid Content in Soybean Seed Oil and Its Genetic Correlation with Fatty Acid and Agronomic Traits. Crop Sci. 2008, 48, 1764–1772. [Google Scholar] [CrossRef]
- Sung, M.; Van, K.; Lee, S.; Nelson, R.; LaMantia, J.; Taliercio, E.; McHale, L.K.; Mian, M.A.R. Identification of SNP Markers Associated with Soybean Fatty Acids Contents by Genome-Wide Association Analyses. Mol. Breed. 2021, 41, 27. [Google Scholar] [CrossRef]
- Gupta, S.K.; Manjaya, J.G. Advances in Improvement of Soybean Seed Composition Traits Using Genetic, Genomic and Biotechnological Approaches. Euphytica 2022, 218, 99. [Google Scholar] [CrossRef]
- Talukdar, A.; Shivakumar, M.; Chandra, S. Recent Advances in Breeding for Modified Fatty Acid Profile in Soybean Oil. In Quality Breeding in Field Crops; Qureshi, A.M.I., Dar, Z.A., Wani, S.H., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 159–172. ISBN 978-3-030-04609-5. [Google Scholar]
- Ning, L.; Kan, G.; Du, W.; Guo, S.; Wang, Q.; Zhang, G.; Cheng, H.; Yu, D. Association Analysis for Detecting Significant Single Nucleotide Polymorphisms for Phosphorus-Deficiency Tolerance at the Seedling Stage in Soybean [Glycine Max (L) Merr.]. Breed. Sci. 2016, 66, 191–203. [Google Scholar] [CrossRef]
- Fulton, T.M.; Beck-Bunn, T.; Emmatty, D.; Eshed, Y.; Lopez, J.; Petiard, V.; Uhlig, J.; Zamir, D.; Tanksley, S.D. QTL Analysis of an Advanced Backcross of Lycopersicon Peruvianum to the Cultivated Tomato and Comparisons with QTLs Found in Other Wild Species. Theor. Appl. Genet. 1997, 95, 881–894. [Google Scholar] [CrossRef]
- Hyten, D.L.; Pantalone, V.R.; Saxton, A.M.; Schmidt, M.E.; Sams, C.E. Molecular Mapping and Identification of Soybean Fatty Acid Modifier Quantitative Trait Loci. J. Am. Oil Chem. Soc. 2004, 81, 1115–1118. [Google Scholar] [CrossRef]
- Kim, H.-K.; Kim, Y.; Kim, S.T.; Son, B.G.; Choi, Y.W.; Kang, J.S.; Lee, Y.J.; Cho, Y.-S.; Choi, I.S. Analysis of Quantitative Trait Loci (QTLs) for Seed Size and Fatty Acid Composition Using Recombinant Inbred Lines in Soybean. J. Life Sci. 2010, 20, 1186–1192. [Google Scholar] [CrossRef]
- Peng, F.Y.; Weselake, R.J. Gene Coexpression Clusters and Putative Regulatory Elements Underlying Seed Storage Reserve Accumulation in Arabidopsis. BMC Genom. 2011, 12, 286. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Kapoor, S.; Tyagi, A.K. Transcription Factors Regulating the Progression of Monocot and Dicot Seed Development. BioEssays News Rev. Mol. Cell. Dev. Biol. 2011, 33, 189–202. [Google Scholar] [CrossRef]
- Lu, L.; Wei, W.; Li, Q.-T.; Bian, X.-H.; Lu, X.; Hu, Y.; Cheng, T.; Wang, Z.-Y.; Jin, M.; Tao, J.-J.; et al. A Transcriptional Regulatory Module Controls Lipid Accumulation in Soybean. New Phytol. 2021, 231, 661–678. [Google Scholar] [CrossRef] [PubMed]
- Reszczyńska, E.; Hanaka, A. Lipids Composition in Plant Membranes. Cell Biochem. Biophys. 2020, 78, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Pollard, M.; Shachar-Hill, Y. Kinetic Complexities of Triacylglycerol Accumulation in Developing Embryos from Camelina Sativa Provide Evidence for Multiple Biosynthetic Systems. J. Biol. Chem. 2022, 298, 101396. [Google Scholar] [CrossRef]
- Cao, D.; Cheng, H.; Wu, W.; Soo, H.M.; Peng, J. Gibberellin Mobilizes Distinct DELLA-Dependent Transcriptomes to Regulate Seed Germination and Floral Development in Arabidopsis. Plant Physiol. 2006, 142, 509–525. [Google Scholar] [CrossRef]
- Chen, M.; Du, X.; Zhu, Y.; Wang, Z.; Hua, S.; Li, Z.; Guo, W.; Zhang, G.; Peng, J.; Jiang, L. Seed Fatty Acid Reducer Acts Downstream of Gibberellin Signalling Pathway to Lower Seed Fatty Acid Storage in Arabidopsis. Plant Cell Environ. 2012, 35, 2155–2169. [Google Scholar] [CrossRef]
- Do, P.T.; Nguyen, C.X.; Bui, H.T.; Tran, L.T.N.; Stacey, G.; Gillman, J.D.; Zhang, Z.J.; Stacey, M.G. Demonstration of Highly Efficient Dual gRNA CRISPR/Cas9 Editing of the Homeologous GmFAD2–1A and GmFAD2–1B Genes to Yield a High Oleic, Low Linoleic and α-Linolenic Acid Phenotype in Soybean. BMC Plant Biol. 2019, 19, 311. [Google Scholar] [CrossRef]
- Kumar, V.; Vats, S.; Kumawat, S.; Bisht, A.; Bhatt, V.; Shivaraj, S.M.; Padalkar, G.; Goyal, V.; Zargar, S.; Gupta, S.; et al. Omics Advances and Integrative Approaches for the Simultaneous Improvement of Seed Oil and Protein Content in Soybean (Glycine max L.). Crit. Rev. Plant Sci. 2021, 40, 398–421. [Google Scholar] [CrossRef]
- Wu, N.; Lu, Q.; Wang, P.; Zhang, Q.; Zhang, J.; Qu, J.; Wang, N. Construction and Analysis of GmFAD2-1A and GmFAD2-2A Soybean Fatty Acid Desaturase Mutants Based on CRISPR/Cas9 Technology. Int. J. Mol. Sci. 2020, 21, 1104. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Liu, H.; Peng, D.; Zhang, J.; Chen, M. Linum Usitatissimum FAD2A and FAD3A Enhance Seed Polyunsaturated Fatty Acid Accumulation and Seedling Cold Tolerance in Arabidopsis Thaliana. Plant Sci. 2021, 311, 111014. [Google Scholar] [CrossRef] [PubMed]
- Islam, N.; Bates, P.D.; Maria John, K.M.; Krishnan, H.B.; Zhang, Z.J.; Luthria, D.L.; Natarajan, S.S. Quantitative Proteomic Analysis of Low Linolenic Acid Transgenic Soybean Reveals Perturbations of Fatty Acid Metabolic Pathways. Proteomics 2019, 19, 1800379. [Google Scholar] [CrossRef] [PubMed]
- Abdelghany, A.M.; Zhang, S.; Azam, M.; Shaibu, A.S.; Feng, Y.; Qi, J.; Li, Y.; Tian, Y.; Hong, H.; Li, B.; et al. Natural Variation in Fatty Acid Composition of Diverse World Soybean Germplasms Grown in China. Agronomy 2020, 10, 24. [Google Scholar] [CrossRef]




| Trait | Years | Mean | Range | SD | CV(%) | H2 (%) | F Values from ANOVA | ||
|---|---|---|---|---|---|---|---|---|---|
| Line | Env. | Line × Env. | |||||||
| Linoleic Acid | 2019 | 58.42 ± 0.28 | 46.07–70.73 | 4.93 | 0.084 | 67.00 | 2.00 *** | 0.15 | 1 |
| 2020 | 58.40 ± 0.29 | 45.07–70.52 | 4.88 | 0.083 | |||||
| 2021 | 56.62 ± 0.23 | 47.02–67.98 | 3.88 | 0.068 | |||||
| Linolenic Acid | 2019 | 8.01 ± 0.16 | 1.39–15.77 | 2.66 | 0.332 | 73.20 | 2.00 *** | 2.00 *** | 0.41 |
| 2020 | 7.91 ± 0.15 | 2.39–15.46 | 2.59 | 0.327 | |||||
| 2021 | 6.72 ± 0.11 | 2.16–12.27 | 1.91 | 0.285 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Liu, L.; Zhang, Q.; Sun, T.; Wang, P. Genome-Wide Association Analysis-Based Mining of Quality Genes Related to Linoleic and Linolenic Acids in Soybean. Agriculture 2023, 13, 2250. https://doi.org/10.3390/agriculture13122250
Wang J, Liu L, Zhang Q, Sun T, Wang P. Genome-Wide Association Analysis-Based Mining of Quality Genes Related to Linoleic and Linolenic Acids in Soybean. Agriculture. 2023; 13(12):2250. https://doi.org/10.3390/agriculture13122250
Chicago/Turabian StyleWang, Jiabao, Lu Liu, Qi Zhang, Tingting Sun, and Piwu Wang. 2023. "Genome-Wide Association Analysis-Based Mining of Quality Genes Related to Linoleic and Linolenic Acids in Soybean" Agriculture 13, no. 12: 2250. https://doi.org/10.3390/agriculture13122250
APA StyleWang, J., Liu, L., Zhang, Q., Sun, T., & Wang, P. (2023). Genome-Wide Association Analysis-Based Mining of Quality Genes Related to Linoleic and Linolenic Acids in Soybean. Agriculture, 13(12), 2250. https://doi.org/10.3390/agriculture13122250






