Impact of Allelic Variation in Maturity Genes E1–E4 on Soybean Adaptation to Central and West Siberian Regions of Russia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Field Experiments
2.3. Genomic DNA Extraction PCR Amplification and Restriction
2.4. Statistical Analyses
3. Results
3.1. Genotyping of E1–E4
3.1.1. Distribution of E1–E4 Alleles in Different Origin Groups
3.1.2. Established E Genotypes and Their Distribution in Russian Accessions
3.2. Field Experiments
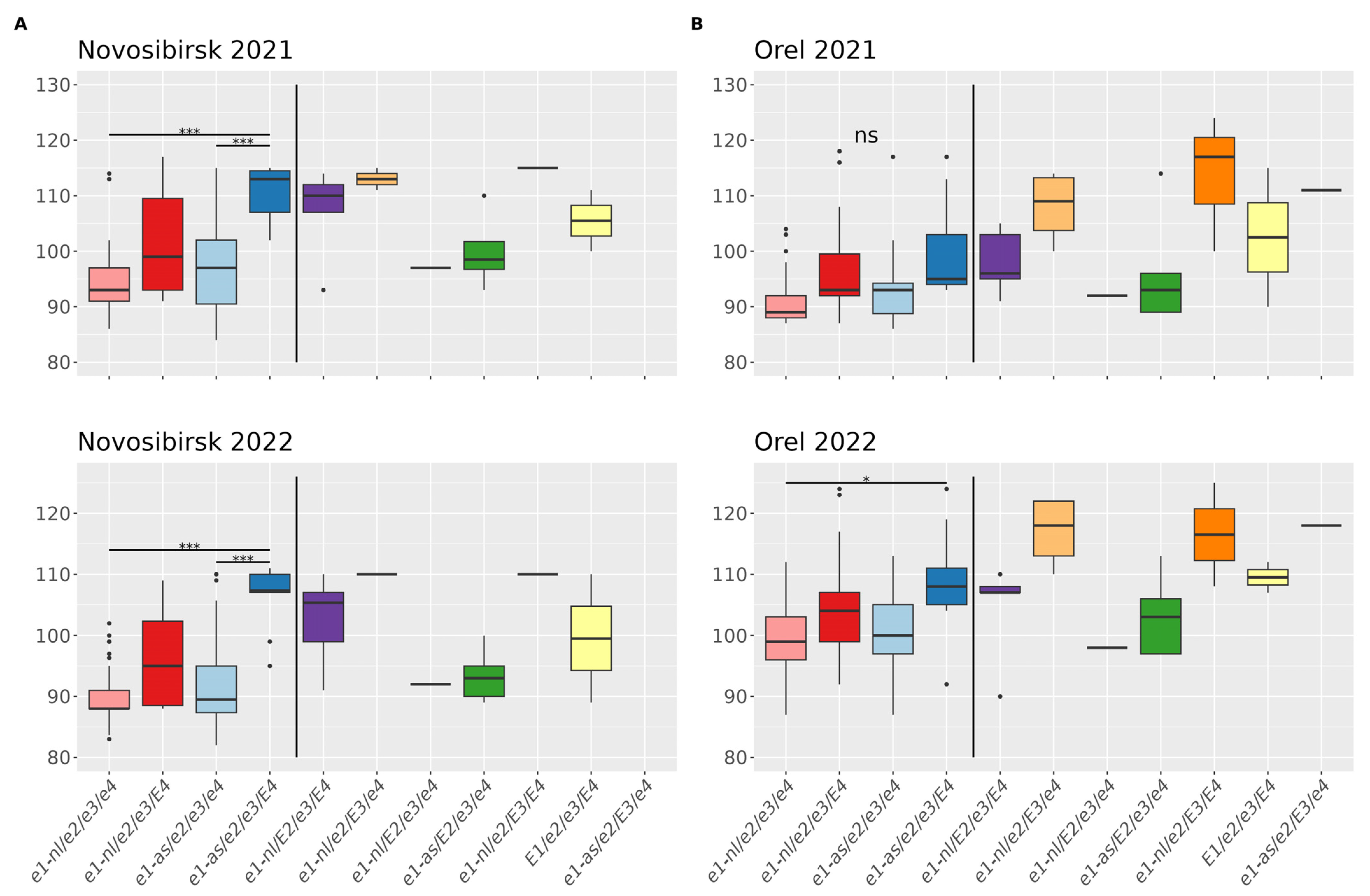
3.2.1. Association of E Genotypes with the Flowering Time
3.2.2. Association of E Genotypes with the Maturity Time
3.3. Statistical Analysis
4. Discussion
4.1. Distribution of E1–E4 Alleles among Russian Accessions
4.2. The Effect of Established E Genotypes on Flowering and Maturity
4.3. Optimal Genotypes for the Studied Regions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kurasch, A.K.; Hahn, V.; Leiser, W.L.; Vollmann, J.; Schori, A.; Bétrix, C.; Mayr, B.; Winkler, J.; Mechtler, K.; Aper, J.; et al. Identification of Mega-environments in Europe and Effect of Allelic Variation at Maturity E Loci on Adaptation of European Soybean. Plant Cell Environ. 2017, 40, 765–778. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Song, W.; Wang, L.; Sun, X.; Qi, Y.; Wu, T.; Sun, S.; Jiang, B.; Wu, C.; Hou, W.; et al. Allele Combinations of Maturity Genes E1–E4 Affect Adaptation of Soybean to Diverse Geographic Regions and Farming Systems in China. PLoS ONE 2020, 15, e0235397. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, T.; Wen, H.; Song, W.; Xu, C.; Han, T.; Sun, S.; Wu, C. Allelic Variation of Soybean Maturity Genes E1–E4 in the Huang-Huai-Hai River Valley and the Northwest China. Agriculture 2021, 11, 478. [Google Scholar] [CrossRef]
- Liu, B.; Kanazawa, A.; Matsumura, H.; Takahashi, R.; Harada, K.; Abe, J. Genetic Redundancy in Soybean Photoresponses Associated With Duplication of the Phytochrome A Gene. Genetics 2008, 180, 995–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, S.; Hideshima, R.; Xia, Z.; Tsubokura, Y.; Sato, S.; Nakamoto, Y.; Yamanaka, N.; Takahashi, R.; Ishimoto, M.; Anai, T.; et al. Map-Based Cloning of the Gene Associated with the Soybean Maturity Locus E3. Genetics 2009, 182, 1251–1262. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, S.; Xia, Z.; Hideshima, R.; Tsubokura, Y.; Sato, S.; Yamanaka, N.; Takahashi, R.; Anai, T.; Tabata, S.; Kitamura, K.; et al. A Map-Based Cloning Strategy Employing a Residual Heterozygous Line Reveals That the GIGANTEA Gene Is Involved in Soybean Maturity and Flowering. Genetics 2011, 188, 395–407. [Google Scholar] [CrossRef] [Green Version]
- Xia, Z.; Watanabe, S.; Yamada, T.; Tsubokura, Y.; Nakashima, H.; Zhai, H.; Anai, T.; Sato, S.; Yamazaki, T.; Lü, S.; et al. Positional Cloning and Characterization Reveal the Molecular Basis for Soybean Maturity Locus E1 That Regulates Photoperiodic Flowering. Proc. Natl. Acad. Sci. USA 2012, 109, E2155–E2164. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Xu, Z.; Liu, B.; Kong, F.; Tsubokura, Y.; Watanabe, S.; Xia, Z.; Harada, K.; Kanazawa, A.; Yamada, T.; et al. Genetic Variation in Four Maturity Genes Affects Photoperiod Insensitivity and PHYA-Regulated Post-Flowering Responses of Soybean. BMC Plant Biol. 2013, 13, 91. [Google Scholar] [CrossRef] [Green Version]
- Tsubokura, Y.; Matsumura, H.; Xu, M.; Liu, B.; Nakashima, H.; Anai, T.; Kong, F.; Yuan, X.; Kanamori, H.; Katayose, Y.; et al. Genetic Variation in Soybean at the Maturity Locus E4 Is Involved in Adaptation to Long Days at High Latitudes. Agronomy 2013, 3, 117–134. [Google Scholar] [CrossRef] [Green Version]
- Langewisch, T.; Lenis, J.; Jiang, G.-L.; Wang, D.; Pantalone, V.; Bilyeu, K. The Development and Use of a Molecular Model for Soybean Maturity Groups. BMC Plant Biol. 2017, 17, 91. [Google Scholar] [CrossRef] [Green Version]
- Soybeans: Improvement, Production, and Uses; Shibles, R.M.; Harper, J.E.; Wilson, R.F.; Shoemaker, R.C. (Eds.) Agronomy Monographs; American Society of Agronomy, Crop Science Society of America, and Soil Science Society of America: Madison, WI, USA, 2004; ISBN 978-0-89118-266-5. [Google Scholar]
- Deriglazova, G. Current Trends in Soybean Cultivation in Russia. Agric. Lifestock Technol. Aгpoзoomexнuкa 2022, 5, 1–10. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from Milligram Amounts of Fresh, Herbarium and Mummified Plant Tissues. Plant Mol. Biol. 1985, 5, 69–76. [Google Scholar] [CrossRef]
- Perfil’ev, R.N.; Shcherban, A.B.; Salina, E.A. Development of a Marker Panel for Genotyping of Domestic Soybean Cultivars for Genes Controlling the Duration of Vegetation and Response to Photoperiod. Vavilov J. Genet. Breed. 2021, 25, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Patil, I. Visualizations with Statistical Details: The “ggstatsplot” Approach. J. Open Source Softw. 2021, 6, 3167. [Google Scholar] [CrossRef]
- Tsubokura, Y.; Watanabe, S.; Xia, Z.; Kanamori, H.; Yamagata, H.; Kaga, A.; Katayose, Y.; Abe, J.; Ishimoto, M.; Harada, K. Natural Variation in the Genes Responsible for Maturity Loci E1, E2, E3 and E4 in Soybean. Ann. Bot. 2014, 113, 429–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tardivel, A.; Sonah, H.; Belzile, F.; O’Donoughue, L.S. Rapid Identification of Alleles at the Soybean Maturity Gene E3 Using Genotyping by Sequencing and a Haplotype-Based Approach. Plant Genome 2014, 7, 1–9. [Google Scholar] [CrossRef]
- Lin, X.; Dong, L.; Tang, Y.; Li, H.; Cheng, Q.; Li, H.; Zhang, T.; Ma, L.; Xiang, H.; Chen, L.; et al. Novel and Multifaceted Regulations of Photoperiodic Flowering by Phytochrome A in Soybean. Proc. Natl. Acad. Sci. USA 2022, 119, e2208708119. [Google Scholar] [CrossRef]
- Zhai, H.; Wan, Z.; Jiao, S.; Zhou, J.; Xu, K.; Nan, H.; Liu, Y.; Xiong, S.; Fan, R.; Zhu, J.; et al. GmMDE Genes Bridge the Maturity Gene E1 and Florigens in Photoperiodic Regulation of Flowering in Soybean. Plant Physiol. 2022, 189, 1021–1036. [Google Scholar] [CrossRef]
- Qin, C.; Li, H.; Zhang, S.; Lin, X.; Jia, Z.; Zhao, F.; Wei, X.; Jiao, Y.; Li, Z.; Niu, Z.; et al. GmEID1 Modulates Light Signaling through the Evening Complex to Control Flowering Time and Yield in Soybean. Proc. Natl. Acad. Sci. USA 2023, 120, e2212468120. [Google Scholar] [CrossRef]
- Dong, L.; Cheng, Q.; Fang, C.; Kong, L.; Yang, H.; Hou, Z.; Li, Y.; Nan, H.; Zhang, Y.; Chen, Q.; et al. Parallel Selection of Distinct Tof5 Alleles Drove the Adaptation of Cultivated and Wild Soybean to High Latitudes. Mol. Plant 2022, 15, 308–321. [Google Scholar] [CrossRef]
- Li, H.; Du, H.; He, M.; Wang, J.; Wang, F.; Yuan, W.; Huang, Z.; Cheng, Q.; Gou, C.; Chen, Z.; et al. Natural Variation of FKF1 Controls Flowering and Adaptation during Soybean Domestication and Improvement. New Phytol. 2023, 238, 1671–1684. [Google Scholar] [CrossRef] [PubMed]
- Kou, K.; Yang, H.; Li, H.; Fang, C.; Chen, L.; Yue, L.; Nan, H.; Kong, L.; Li, X.; Wang, F.; et al. A Functionally Divergent SOC1 Homolog Improves Soybean Yield and Latitudinal Adaptation. Curr. Biol. 2022, 32, 1728–1742.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Takeshima, R.; Harigai, K.; Xu, M.; Kong, F.; Liu, B.; Kanazawa, A.; Yamada, T.; Abe, J. Loss of Function of the E1-Like-b Gene Associates With Early Flowering Under Long-Day Conditions in Soybean. Front. Plant Sci. 2019, 9, 1867. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Wang, Y.; Li, S.; Wang, L.; Kou, K.; Kong, L.; Cheng, Q.; Dong, L.; Liu, B.; Kong, F.; et al. A Flowering Time Locus Dependent on E2 in Soybean. Mol. Breed. 2021, 41, 35. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Kim, E.-S.; Kim, K.H.; Jeong, N.; Lee, J.S.; Kang, S. Genetic Variance for Flowering Time Conferring E2 Gene in Photoperiod-Insensitive Early-Maturing Soybean Accessions and Topological Distribution in Korea Peninsula. Mol. Breed. 2018, 38, 148. [Google Scholar] [CrossRef]
- Funatsuki, H.; Kawaguchi, K.; Matsuba, S.; Sato, Y.; Ishimoto, M. Mapping of QTL Associated with Chilling Tolerance during Reproductive Growth in Soybean. Theor. Appl. Genet. 2005, 111, 851–861. [Google Scholar] [CrossRef]
- Tsegaw, M.; Zegeye, W.A.; Jiang, B.; Sun, S.; Yuan, S.; Han, T.; Wu, T. Progress and Prospects of the Molecular Basis of Soybean Cold Tolerance. Plants 2023, 12, 459. [Google Scholar] [CrossRef]
- Takahashi, R.; Abe, J. Soybean Maturity Genes Associated with Seed Coat Pigmentation and Cracking in Response to Low Temperatures. Crop Sci. 1999, 39, 1657–1662. [Google Scholar] [CrossRef]
- Cober, E.R.; Tanner, J.W.; Voldeng, H.D. Genetic Control of Photoperiod Response in Early-Maturing, Near-Isogenic Soybean Lines. Crop Sci. 1996, 36, 601–605. [Google Scholar] [CrossRef]
- Cober, E.R.; Tanner, J.W.; Voldeng, H.D. Soybean Photoperiod-Sensitivity Loci Respond Differentially to Light Quality. Crop Sci. 1996, 36, 606–610. [Google Scholar] [CrossRef]


| Genotype | E1 | E2 | E3 | E4 | N | % |
|---|---|---|---|---|---|---|
| e1/e2/e3/e4 | e1-nl | e2-ns | e3-fs | e4-SORE-1 | 54 | 30.7 |
| e1-as | e2-ns | e3-fs | e4-kes | 16 | 9.1 | |
| e1-nl | e2-ns | e3-fs | e4-kes | 15 | 8.5 | |
| e1-nl | e2-ns | e3-tr | e4-SORE-1 | 10 | 5.7 | |
| e1-as | e2-ns | e3-fs | e4-SORE-1 | 9 | 5.1 | |
| e1-as | e2-ns | e3-tr | e4-SORE-1 | 6 | 3.4 | |
| e1-nl | e2-ns | e3-ns | e4-SORE-1 | 6 | 3.4 | |
| e1-as | e2-ns | e3-ns | e4-kes | 2 | 1.1 | |
| e1-as | e2-ns | e3-tr | e4-kes | 2 | 1.1 | |
| e1-as | e2-ns | e3-ns | e4-SORE-1 | 1 | 0.6 | |
| e1-fs | e2-ns | e3-fs | e4-kes | 1 | 0.6 | |
| e1-nl | e2-ns | e3-ns | e4-kes | 1 | 0.6 | |
| e1/e2/e3/E4 | e1-nl | e2-ns | e3-fs | E4 | 9 | 5.1 |
| e1-nl | e2-ns | e3-tr | E4 | 9 | 5.1 | |
| e1-as | e2-ns | e3-fs | E4 | 8 | 4.5 | |
| e1-as | e2-ns | e3-tr | E4 | 4 | 2.3 | |
| e1-as | e2-ns | e3-mix * | E4 | 1 | 0.6 | |
| e1-nl | e2-ns | e3-mix * | E4 | 1 | 0.6 | |
| e1/e2/E3/e4 | e1-nl | e2-ns | E3-Ha | e4-SORE-1 | 4 | 2.3 |
| e1-as | e2-ns | E3-Ha | e4-SORE-1 | 1 | 0.6 | |
| e1/e2/E3/E4 | e1-nl | e2-ns | E3-Ha | E4 | 3 | 1.7 |
| e1/E2/e3/e4 | e1-as | E2 | e3-fs | e4-kes | 5 | 2.8 |
| e1-nl | E2 | e3-fs | e4-kes | 1 | 0.6 | |
| e1/E2/e3/E4 | e1-nl | E2 | e3-tr | E4 | 5 | 2.8 |
| E1/e2/e3/E4 | E1 | e2-ns | e3-tr | E4 | 2 | 1.1 |
| Trait | Location | Factor | df | Sum Sq | F Value |
|---|---|---|---|---|---|
| DTF | Novosibirsk | E1 | 1 | 411.9 | 59.6 *** |
| E4 | 1 | 527.6 | 76.4 *** | ||
| year | 1 | 235.9 | 34.1 *** | ||
| residuals | 312 | 2155.5 | |||
| Orel | E1 | 1 | 252.5 | 50.3 *** | |
| E4 | 1 | 82.4 | 16.4 *** | ||
| year | 1 | 126.6 | 25.2 *** | ||
| residuals | 312 | 1567.4 | |||
| DTM | Novosibirsk | E1 | 1 | 1215.8 | 34.6 *** |
| E4 | 1 | 2995.2 | 85.3 *** | ||
| year | 1 | 1557.1 | 44.3 *** | ||
| residuals | 284 | 9975.4 | |||
| Orel | E1 | 1 | 454.5 | 12.4 ** | |
| E4 | 1 | 2588.8 | 70.4 *** | ||
| year | 1 | 5258 | 143 *** | ||
| residuals | 312 | 11469.2 | |||
| DTF | Novosibirsk and Orel | E1 | 1 | 606.6 | 99.3 *** |
| E4 | 1 | 533.9 | 87.4 *** | ||
| year | 1 | 336.9 | 55.1 *** | ||
| location | 1 | 2308.4 | 377.7 *** | ||
| residuals | 615 | 3758.8 | |||
| DTM | E1 | 1 | 1355.7 | 28 *** | |
| E4 | 1 | 6137 | 126.7 *** | ||
| year | 1 | 506.5 | 10.5 ** | ||
| location | 1 | 541.8 | 11.2 *** | ||
| residuals | 597 | 28918.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perfil’ev, R.; Shcherban, A.; Potapov, D.; Maksimenko, K.; Kiryukhin, S.; Gurinovich, S.; Panarina, V.; Polyudina, R.; Salina, E. Impact of Allelic Variation in Maturity Genes E1–E4 on Soybean Adaptation to Central and West Siberian Regions of Russia. Agriculture 2023, 13, 1251. https://doi.org/10.3390/agriculture13061251
Perfil’ev R, Shcherban A, Potapov D, Maksimenko K, Kiryukhin S, Gurinovich S, Panarina V, Polyudina R, Salina E. Impact of Allelic Variation in Maturity Genes E1–E4 on Soybean Adaptation to Central and West Siberian Regions of Russia. Agriculture. 2023; 13(6):1251. https://doi.org/10.3390/agriculture13061251
Chicago/Turabian StylePerfil’ev, Roman, Andrey Shcherban, Dmitriy Potapov, Konstantin Maksimenko, Sergey Kiryukhin, Sergey Gurinovich, Veronika Panarina, Revmira Polyudina, and Elena Salina. 2023. "Impact of Allelic Variation in Maturity Genes E1–E4 on Soybean Adaptation to Central and West Siberian Regions of Russia" Agriculture 13, no. 6: 1251. https://doi.org/10.3390/agriculture13061251
APA StylePerfil’ev, R., Shcherban, A., Potapov, D., Maksimenko, K., Kiryukhin, S., Gurinovich, S., Panarina, V., Polyudina, R., & Salina, E. (2023). Impact of Allelic Variation in Maturity Genes E1–E4 on Soybean Adaptation to Central and West Siberian Regions of Russia. Agriculture, 13(6), 1251. https://doi.org/10.3390/agriculture13061251







