Validation of Genome-Wide SSR Markers Developed for Genetic Diversity and Population Structure Study in Grain Amaranth (Amaranthus hypochondriacus)
Abstract
1. Introduction
2. Materials and Methods
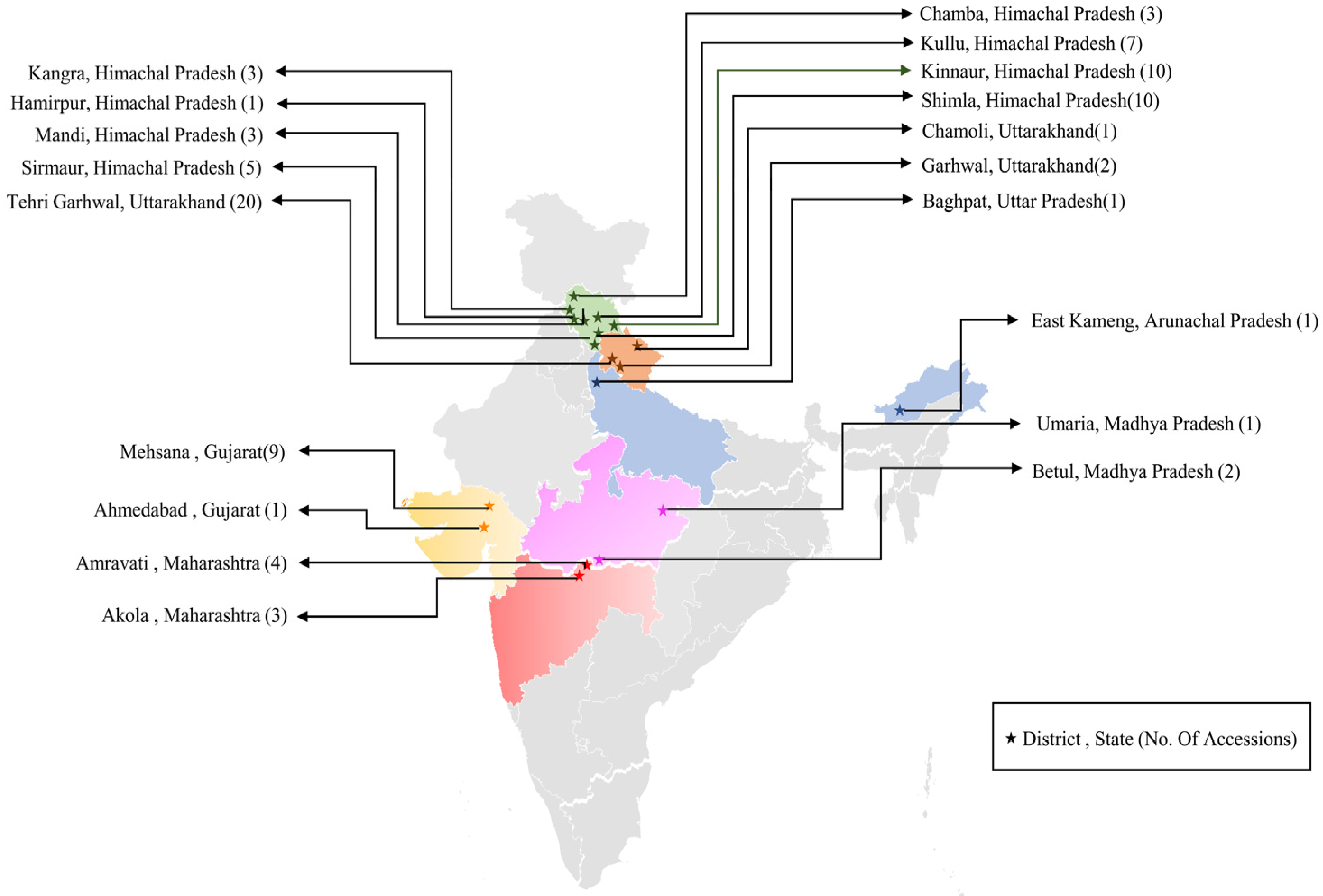
2.1. Plant Materials
2.2. Plant Genomic DNA Extraction
2.3. PCR Amplification and Gel Documentation
2.4. Scoring of Markers and Data Analysis
3. Results
3.1. Genetic Diversity Indices for g-SSR Markers
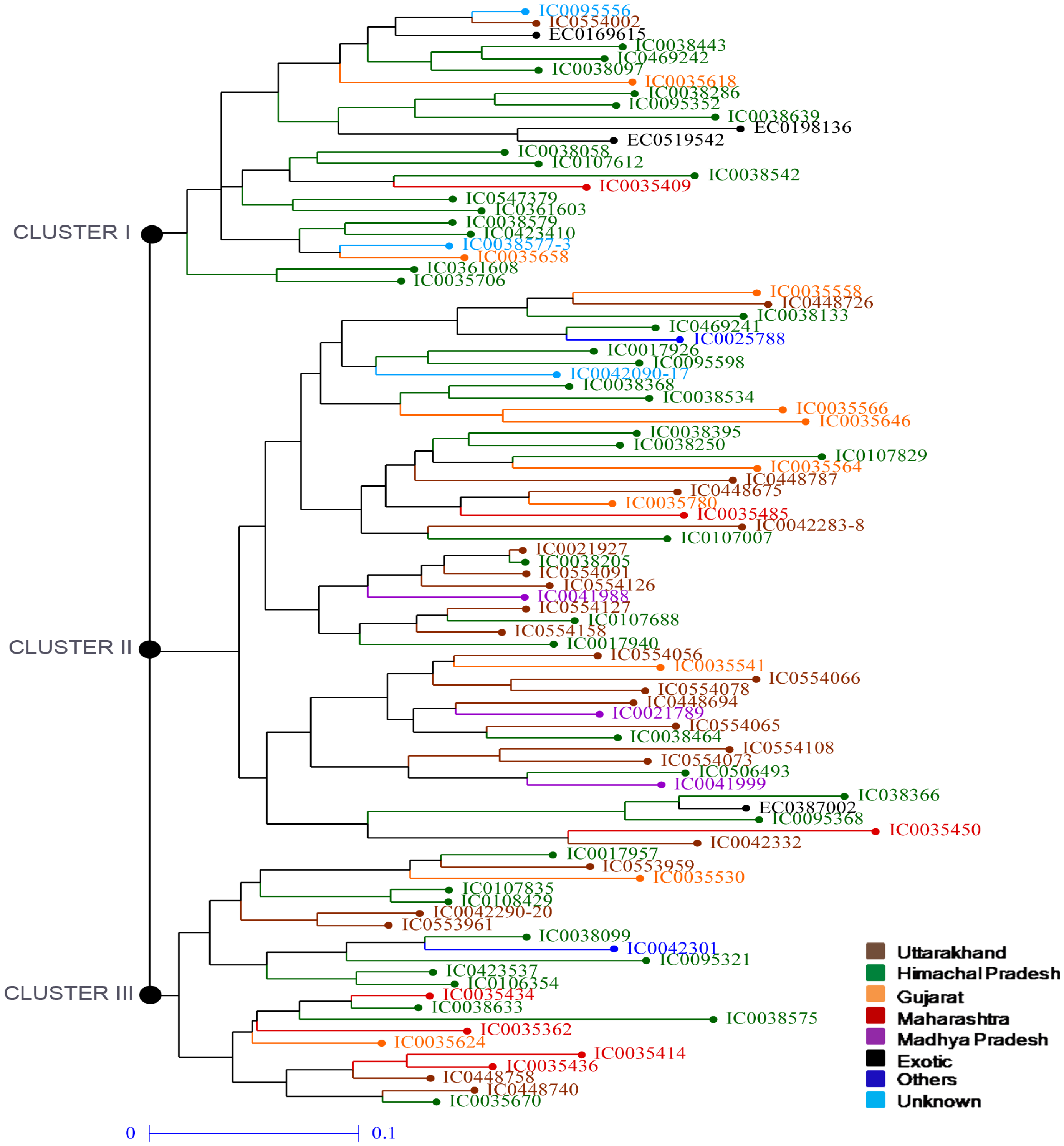
3.2. Phylogenetic Analysis of Amaranth Accessions
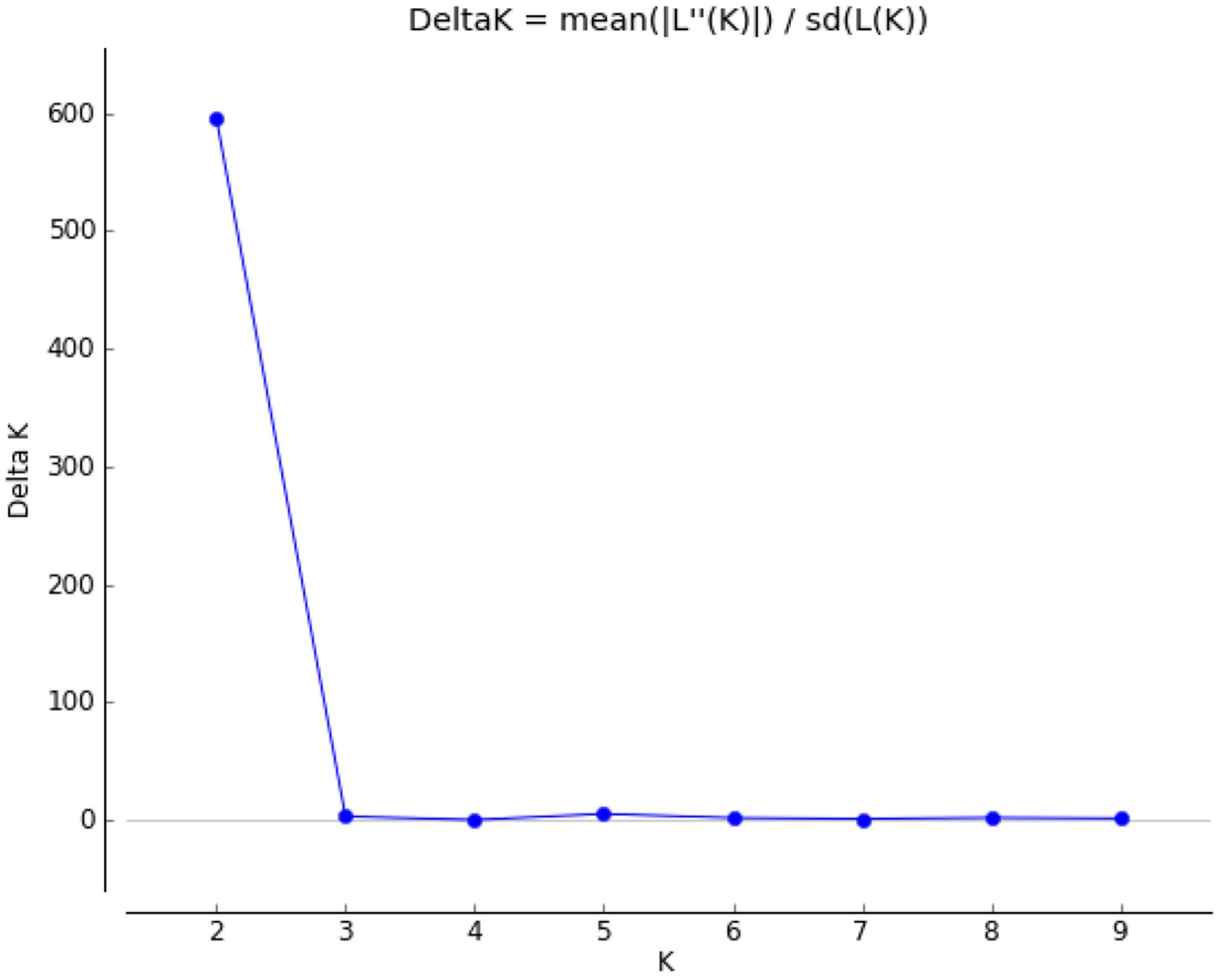
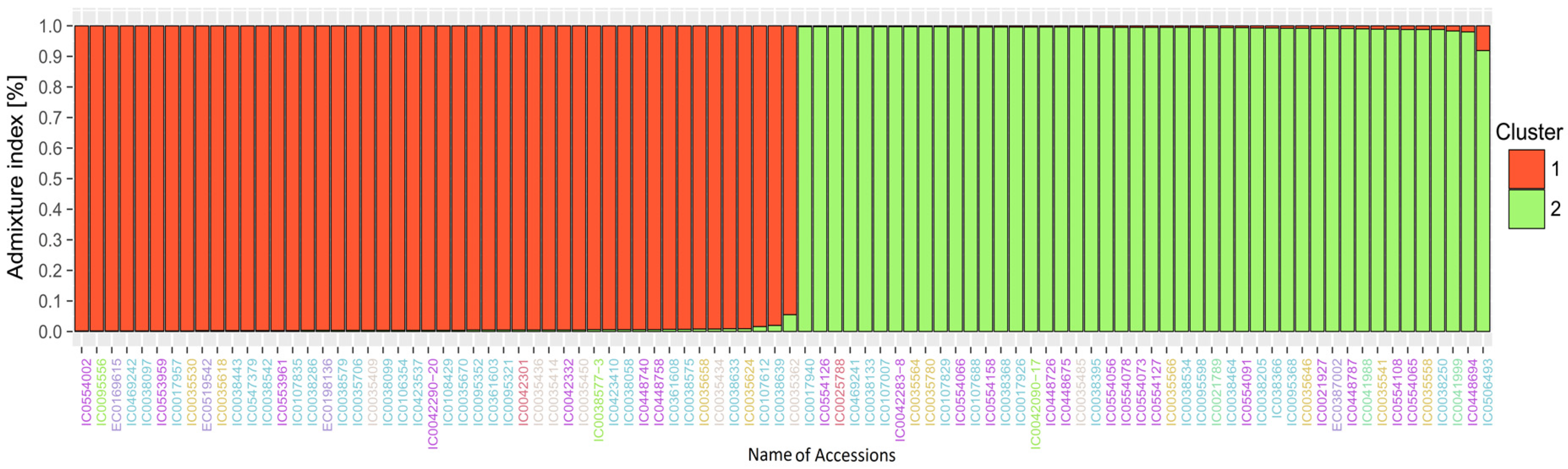
3.3. Analysis of Population Structure
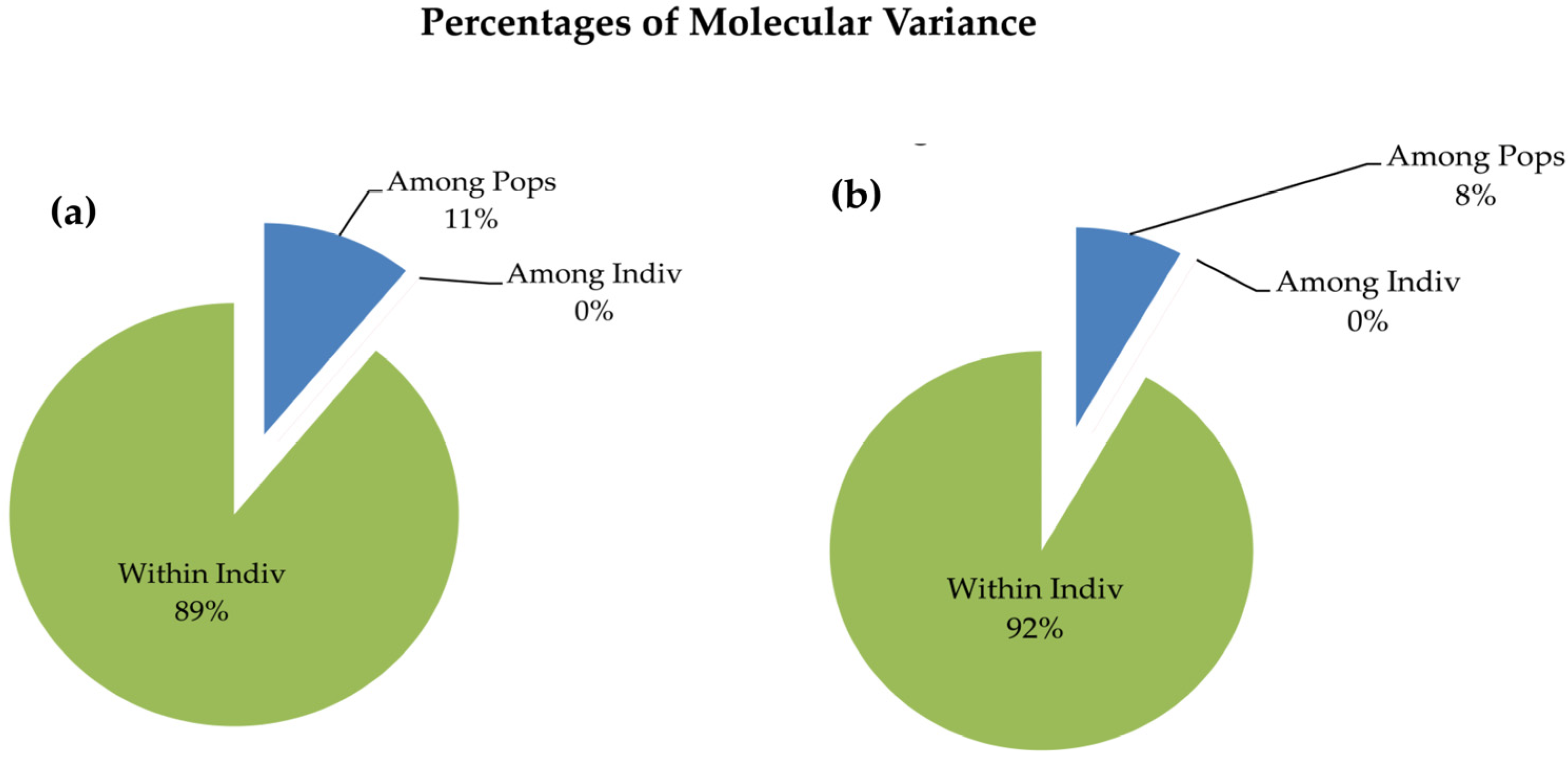
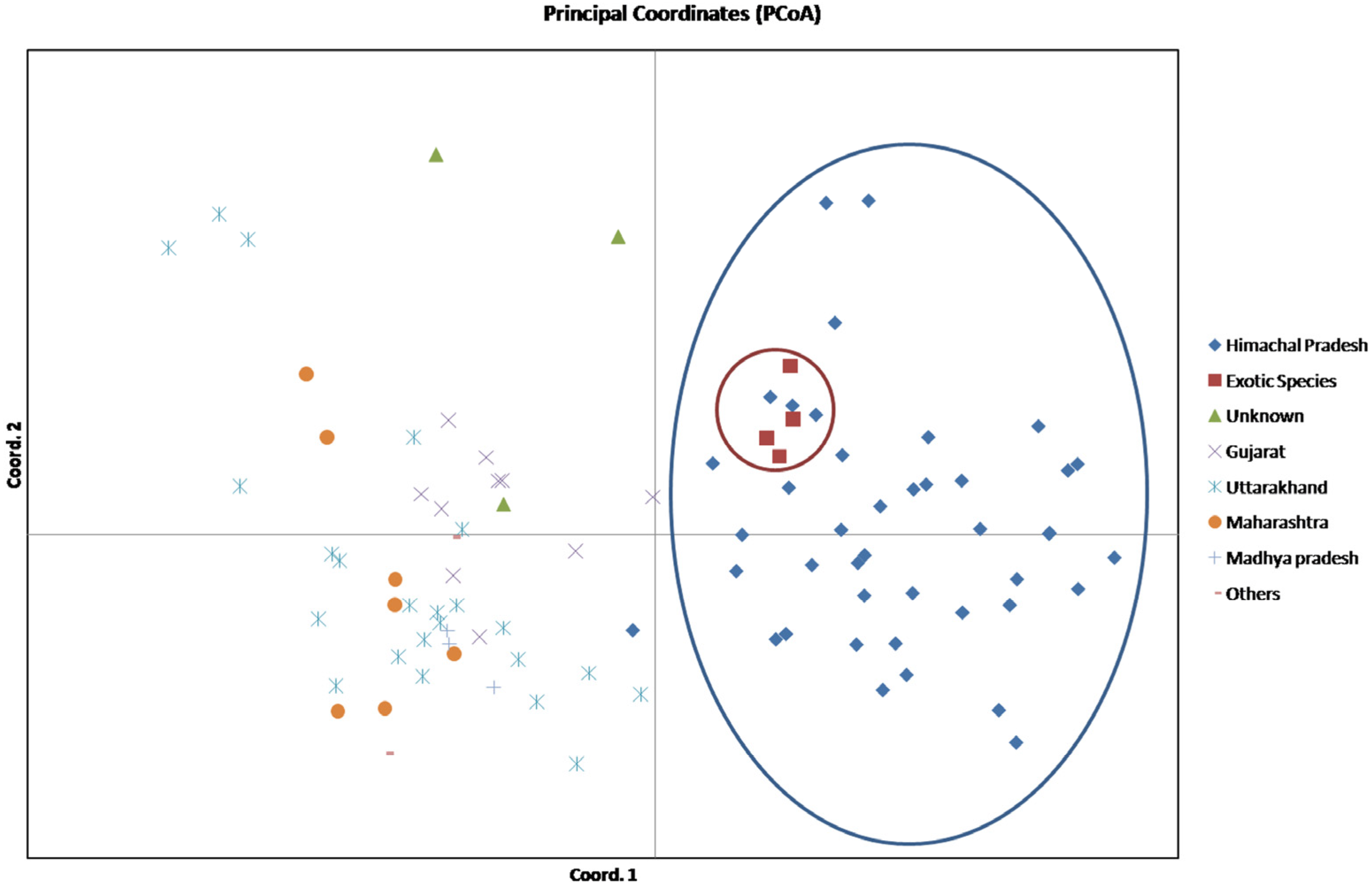
3.4. AMOVA and PCoA Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aderibigbe, O.R.; Ezekiel, O.O.; Owolade, S.O.; Korese, J.K.; Sturm, B.; Hensel, O. Exploring the potentials of underutilized grain amaranth (Amaranthus spp.) along the value chain for food and nutrition security: A review. Crit. Rev. Food Sci. Nutr. 2020, 62, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.M.; Baltensperger, D.D.; Kulakow, P.A.; Lehmann, J.W.; Myers, R.L.; Slabbert, M.M.; Sleugh, B.B. Genetic Resources and Breeding of Amaranthus. Plant Breed. Rev. 2010, 19, 227–285. [Google Scholar]
- Sauer, J.D. The Grain Amaranths and Their Relatives: A Revised Taxonomic and Geographic Survey. Ann. Mo. Bot. Gard. 1967, 54, 103. [Google Scholar] [CrossRef]
- Waselkov, K.E.; Boleda, A.S.; Olsen, K.M. A Phylogeny of the Genus Amaranthus (Amaranthaceae) Based on Several Low-Copy Nuclear Loci and Chloroplast Regions. Syst. Bot. 2018, 43, 439–458. [Google Scholar] [CrossRef]
- Nascimento, A.C.; Mota, C.; Coelho, I.; Gueifão, S.; Santos, M.; Matos, A.S.; Gimenez, A.; Lobo, M.; Samman, N.; Castanheira, I. Characterisation of nutrient profile of quinoa (Chenopodium quinoa), amaranth (Amaranthus caudatus), and purple corn (Zea mays L.) consumed in the North of Argentina: Proximates, minerals and trace elements. Food Chem. 2014, 148, 420–426. [Google Scholar] [CrossRef]
- Joshi, N.; Verma, K.C. A review on nutrition value of Amaranth (Amaranthus caudatus L.): The crop of future. J Pharm. Phytochem. 2020, 9, 317–319. [Google Scholar]
- Pedersen, B.; Hallgren, L.; Hansen, I.; Eggum, B.O. The nutritive value of amaranth grain (Amaranthus caudatus). Qual. Plant. Plant Foods Hum. Nutr. 1987, 36, 325–334. [Google Scholar] [CrossRef]
- Bressani, R.; Gonzáles, J.M.; Zúñiga, J.; Breuner, M.; Elías, L.G. Yield, selected chemical composition and nutritive value of 14 selections of amaranth grain representing four species. J. Sci. Food Agric. 1987, 38, 347–356. [Google Scholar] [CrossRef]
- Rodriguez, J.P.; Rahman, H.; Thushar, S.; Singh, R.K. Healthy and Resilient Cereals and Pseudo-Cereals for Marginal Agriculture: Molecular Advances for Improving Nutrient Bioavailability. Front. Genet. 2020, 11, 49. [Google Scholar] [CrossRef]
- Martirosyan, D.M.; Miroshnichenko, L.A.; Kulakova, S.N.; Pogojeva, A.V.; Zoloedov, V.I. Amaranth oil application for coronary heart disease and hypertension. Lipids Health Dis. 2007, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.; Gremaud, G.; Baumgartner, M.; Rein, D.; Monnard, I.; Kratky, E.; Geiger, W.; Burri, J.; Dionisi, F.; Allan, M.; et al. Cholesterol-Lowering Properties of Amaranth Grain and Oil in Hamsters. Int. J. Vitam. Nutr. Res. 2003, 73, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.R.; Lin, Y.K.; Fang, J.Y. Biological and Pharmacological Activities of Squalene and Related Compounds: Potential Uses in Cosmetic Dermatology. Molecules 2009, 14, 540–554. [Google Scholar] [CrossRef]
- Achigan-Dako, E.G.; Sogbohossou, O.E.D.; Maundu, P. Current knowledge on Amaranthus spp.: Research avenues for improved nutritional value and yield in leafy amaranths in sub-Saharan Africa. Euphytica 2014, 197, 303–317. [Google Scholar] [CrossRef]
- Rastogi, A.; Shukla, S. Amaranth: A New Millennium Crop of Nutraceutical Values. Crit. Rev. Food Sci. Nutr. 2013, 53, 109–125. [Google Scholar] [CrossRef]
- Begna, T. Role and economic importance of crop genetic diversity in food security. J. Agric. Sc. Food Technol. 2021, 7, 164–169. [Google Scholar]
- EL-Mansy, A.; Abd El-Moneim, D.; ALshamrani, S.; Safhi, F.; Abdein, M.; Ibrahim, A. Genetic Diversity Analysis of Tomato (Solanum lycopersicum L.) with Morphological, Cytological, and Molecular Markers under Heat Stress. Horticulturae 2021, 7, 65. [Google Scholar] [CrossRef]
- Kumar, C.; Singh, S.K.; Pramanick, K.K.; Verma, M.K.; Srivastav, M.; Singh, R.; Bharadwaj, C.; Naga, K.C. Morphological and biochemical diversity among the Malus species including indigenous Himalayan wild apples. Sci. Hortic. 2018, 233, 204–219. [Google Scholar] [CrossRef]
- Bhandari, H.R.; Bhanu, A.N.; Srivastava, K.; Singh, M.N.; Shreya; Hemantaranjan, A. Assessment of genetic diversity in crop plants—An overview. Adv. Plants Agric. Res. 2017, 7, 279–286. [Google Scholar]
- Kumar, P.; Gupta, V.K.; Misra, A.K.; Modi, D.R.; Pandey, B.K. Potential of molecular markers in plant biotechnology. Plant Omics J. 2009, 2, 141–162. [Google Scholar]
- Zhang, Y.; He, Q.; Zhou, X.; Zheng, S.; Wang, Y.; Li, P.; Wang, Y. Genetic diversity and population structure of 93 rice cultivars (lines) (Oryza sativa Xian group) in Qinba in China by 3 types of genetic markers. BMC Genom. 2022, 23, 550. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kumar, C.; Paliwal, R.; Choudhury, D.R.; Singh, I.; Kumar, A.; Kumari, A.; Singh, R. Development of Novel Genomic Simple Sequence Repeat (g-SSR) Markers and Their Validation for Genetic Diversity Analyses in Kalmegh [Andrographis paniculata (Burm. F.) Nees]. Plants 2020, 9, 1734. [Google Scholar] [CrossRef]
- Paliwal, R.; Singh, R.; Choudhury, D.R.; Tiwari, G.; Kumar, A.; Bhat, K.C.; Singh, R. Molecular Characterization of Tinospora cordifolia (Willd.) Miers Using Novel g-SSR Markers and Their Comparison with EST-SSR and SCoT Markers for Genetic Diversity Study. Genes 2022, 13, 2042. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.; Yan, W.; Xu, J.; Duan, S.; Li, G.; Jin, L. Genome-wide simple sequence repeat markers in potato: Abundance, distribution, composition, and polymorphism. DNA Res. 2021, 28, dsab020. [Google Scholar] [CrossRef]
- Singh, R.; Narzary, D.; Bhardwaj, J.; Singh, A.K.; Kumar, S.; Kumar, A. Molecular diversity and SSR transferability studies in Vetiver grass (Vetiveria zizanioides L. Nash). Ind. Crops Prod. 2014, 53, 187–198. [Google Scholar] [CrossRef]
- Choudhury, D.R.; Singh, N.; Singh, A.K.; Kumar, S.; Srinivasan, K.; Tyagi, R.K.; Ahmad, A.; Singh, N.K.; Singh, R. Analysis of Genetic Diversity and Population Structure of Rice Germplasm from North-Eastern Region of India and Development of a Core Germplasm Set. PLoS ONE 2014, 9, e113094. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, S.K.; Singh, R.; Choudhury, D.R.; Bharadwaj, J.; Gupta, V.; Singode, A. Genetic diversity and population structure study of drumstick (Moringa oleifera Lam.) using morphological and SSR markers. Ind. Crops Prod. 2014, 60, 316–325. [Google Scholar] [CrossRef]
- Hipparagi, Y.; Singh, R.; Choudhury, D.R.; Gupta, V. Genetic diversity and population structure analysis of Kala bhat (Glycine max (L.) Merrill) genotypes using SSR markers. Hereditas 2017, 154, 9. [Google Scholar] [CrossRef]
- Paliwal, R.; Kumar, R.; Choudhury, D.R.; Singh, A.K.; Kumar, S.; Kumar, A.; Bhatt, K.C.; Singh, R.; Mahato, A.K.; Singh, N.K.; et al. Development of genomic simple sequence repeats (g-SSR) markers in Tinospora cordifolia and their application in diversity analyses. Plant Gene 2016, 5, 118–125. [Google Scholar] [CrossRef]
- Kumar, C.; Kumar, R.; Singh, S.K.; Goswami, A.K.; Nagaraja, A.; Paliwal, R.; Singh, R. Development of novel g-SSR markers in guava (Psidium guajava L.) cv. Allahabad Safeda and their application in genetic diversity, population structure and cross species transferability studies. PLoS ONE 2020, 15, e0237538. [Google Scholar] [CrossRef]
- Das, M.; Banerjee, S.; Dhariwal, R.; Vyas, S.; Mir, R.R.; Topdar, N.; Kundu, A.; Khurana, J.P.; Tyagi, A.K.; Sarkar, D.; et al. Development of SSR markers and construction of a linkage map in jute. J. Genet. 2012, 91, 21–31. [Google Scholar] [CrossRef]
- Kaur, G.; Joshi, A.; Jain, D. SSR-Marker assisted evaluation of Genetic Diversity in Mungbean (Vigna radiata (L.) Wilcezk) genotypes. Braz. Arch. Biol. Technol. 2018, 61. [Google Scholar] [CrossRef]
- Moe, K.T.; Zhao, W.; Song, H.S.; Kim, Y.H.; Chung, J.W.; Cho, Y.I.; Park, P.H.; Park, H.S.; Chae, S.C.; Park, Y.J. Development of SSR markers to study diversity in the genus Cymbidium. Biochem. Syst. Ecol. 2010, 38, 585–594. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of Genetic Diversity Assessment in Crop Plants and Its Recent Advances: An Overview of Its Analytical Perspectives. Genet. Res. Int. 2015, 2015, 431487. [Google Scholar] [CrossRef]
- Lee, J.R.; Hong, G.Y.; Dixit, A.; Chung, J.W.; Ma, K.H.; Lee, J.H.; Kang, H.K.; Cho, Y.H.; Gwag, J.G.; Park, Y.J. Characterization of microsatellite loci developed for Amaranthus hypochondriacus and their cross-amplifications in wild species. Conserv. Genet. 2007, 9, 243–246. [Google Scholar] [CrossRef]
- Faseela, K.V.; Salikutty, J. Molecular characterization of amaranth landraces and assessment of interspecific relationships among Amaranthus spp. (L.) using RAPD markers. Indian J. Genet. Plant Breed. 2007, 67, 12–17. [Google Scholar]
- Park, Y.J.; Nishikawa, T.; Matsushima, K.; Minami, M.; Nemoto, K. A rapid and reliable PCR-restriction fragment length polymorphism (RFLP) marker for the identification of Amaranthus cruentus species. Breed Sci. 2014, 64, 422–426. [Google Scholar] [CrossRef]
- Chandi, A.; Milla-Lewis, S.R.; Jordan, D.L.; York, A.C.; Burton, J.D.; Zuleta, M.C.; Whitaker, J.R.; Culpepper, A.S. Use of AFLP Markers to Assess Genetic Diversity in Palmer Amaranth (Amaranthus palmeri) Populations from North Carolina and Georgia. Weed Sci. 2013, 61, 136–145. [Google Scholar] [CrossRef]
- Jimenez, F.R.; Maughan, P.J.; Alvarez, A.; Kietlinski, K.D.; Smith, S.M.; Pratt, D.B.; Elzinga, D.B.; Jellen, E.N. Assessment of Genetic Diversity in Peruvian Amaranth (Amaranthus caudatus and A. hybridus) Germplasm using Single Nucleotide Polymorphism Markers. Crop Sci. 2013, 53, 532–541. [Google Scholar] [CrossRef]
- Wang, X.Q.; Park, Y.J. Comparison of Genetic Diversity among Amaranth Accessions from South and Southeast Asia using SSR Markers. Korean J. Med. Crop Sci. 2013, 21, 220–228. [Google Scholar] [CrossRef]
- Suresh, S.; Chung, J.W.; Cho, G.T.; Sung, J.S.; Park, J.H.; Gwag, J.G.; Baek, H.J. Analysis of molecular genetic diversity and population structure in Amaranthus germplasm using SSR markers. Plant Biosyst.—Int. J. Deal. All Asp. Plant Biol. 2014, 148, 635–644. [Google Scholar]
- Nguyen, D.C.; Tran, D.S.; Tran, T.T.H.; Ohsawa, R.; Yoshioka, Y. Genetic diversity of leafy amaranth (Amaranthus tricolor L.) resources in Vietnam. Breed. Sci. 2019, 69, 640–650. [Google Scholar] [CrossRef]
- Sahu, S.K.; Thangaraj, M.; Kathiresan, K. DNA Extraction Protocol for Plants with High Levels of Secondary Metabolites and Polysaccharides without Using Liquid Nitrogen and Phenol. ISRN Mol. Biol. 2012, 2012, 205049. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Perrier, X.; Flori, A.; Bonnot, F. Data Analysis Methods. In Genetic Diversity of Cultivated Tropical Plants; Hamon, P., Seguin, M., Perrier, X., Glaszmann, J.C., Eds.; Science Publishers: Enfield, UK, 2003; pp. 43–76. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Barba de la Rosa, A.; Fomsgaard, I.S.; Laursen, B.; Mortensen, A.G.; Olvera-Martínez, L.; Silva-Sánchez, C.; Mendoza-Herrera, A.; Gonzalez-Castaneda, J.; De León-Rodríguez, A. Amaranth (Amaranthus hypochondriacus) as an alternative crop for sustainable food production: Phenolic acids and flavonoids with potential impact on its nutraceutical quality. J. Cereal Sci. 2009, 49, 117–121. [Google Scholar] [CrossRef]
- Brown, A.H.D. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Fatinah, A.A.; Arumingtyas, E.L.; Mastuti, R. Morphological and genetic variation of Amaranthus spinosus L.: An adaptation evidence of climate differences and gene interaction. Int. J. Biosci. 2013, 11, 205–212. [Google Scholar]
- Miah, G.; Rafii, M.; Ismail, M.; Puteh, A.; Rahim, H.; Islam, K.; Latif, M. A Review of Microsatellite Markers and Their Applications in Rice Breeding Programs to Improve Blast Disease Resistance. Int. J. Mol. Sci. 2013, 14, 22499–22528. [Google Scholar] [CrossRef]
- Khaing, A.A.; Moe, K.T.; Chung, J.W.; Baek, H.J.; Park, Y.J. Genetic diversity and population structure of the selected core set inAmaranthus using SSR markers. Plant Breed. 2013, 132, 165–173. [Google Scholar] [CrossRef]
- Mallory, M.A.; Hall, R.V.; McNabb, A.R.; Pratt, D.B.; Jellen, E.N.; Maughan, P.J. Development and Characterization of Microsatellite Markers for the Grain Amaranths. Crop Sci. 2008, 48, 1098. [Google Scholar] [CrossRef]
- Rae, S.J.; Aldam, C.; Dominguez, I.; Hoebrechts, H.; Barnes, S.R.; Edwards, K.J. Development and incorporation of microsatellite markers into the linkage map of sugar beet (Beta vulgaris spp.). Appl Genet. 2000, 100, 1240–1248. [Google Scholar] [CrossRef]
- Mason, S.L.; Stevens, M.R.; Jellen, E.N.; Bonifacio, A.; Fairbanks, D.J.; Coleman, C.E.; McCarty, R.R.; Rasmussen, A.G.; Maughan, P.J. Development and Use of Microsatellite Markers for Germplasm Characterization in Quinoa (Chenopodium quinoa Willd.). Crop Sci. 2005, 45, 1618–1630. [Google Scholar] [CrossRef]
- Tiwari, K.K.; Thakkar, N.J.; Dharajiya, D.T.; Bhilocha, H.L.; Barvaliya, P.P.; Galvadiya, B.P.; Prajapati, N.N.; Patel, M.P.; Solanki, S.D. Genome-wide microsatellites in amaranth: Development, characterization, and cross-species transferability. 3 Biotech 2021, 11, 395. [Google Scholar] [CrossRef]
- Gupta, S.; Kumari, K.; Muthamilarasan, M.; Subramanian, A.; Prasad, M. Development and utilization of novel SSRs in foxtail millet [Setaria italica (L.) P. Beauv.]. Plant Breed. 2013, 132, 367–374. [Google Scholar] [CrossRef]
- Patil, P.G.; Singh, N.V.; Parashuram, S.; Bohra, A.; Sowjanya, R.; Gaikwad, N.; Mundewadikar, D.M.; Sangnure, V.R.; Jamma, S.M.; Injak, A.S.; et al. Genome-wide characterization and development of simple sequence repeat markers for genetic studies in pomegranate (Punica granatum L.). Trees 2020, 34, 987–998. [Google Scholar] [CrossRef]
| S.No. | Marker Name | Scaffold Number | g-SSR Start Position | g-SSR End Position | MAF | Allele No | GD | Ho | PIC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | AhySSR0595 | Scaffolds_1 | 5688613 | 5688666 | 0.4679 | 3 | 0.6304 | 0.9103 | 0.5557 |
| 2 | AhySSR3176 | Scaffolds_1 | 23101765 | 23101806 | 0.5054 | 2 | 0.4999 | 0.9891 | 0.375 |
| 3 | AhySSR4597 | Scaffolds_1 | 35176079 | 35176178 | 0.5915 | 4 | 0.4977 | 0.7683 | 0.391 |
| 4 | AhySSR0261 | Scaffolds_1 | 2618226 | 2618270 | 0.5068 | 2 | 0.4999 | 0.9315 | 0.375 |
| 5 | AhySSR1363 | Scaffolds_10 | 22467372 | 22467407 | 0.6902 | 2 | 0.4276 | 0.5761 | 0.3362 |
| 6 | AhySSR33457 | Scaffolds_10 | 39900 | 39915 | 0.7321 | 2 | 0.3922 | 0.5119 | 0.3153 |
| 7 | AhySSR37960 | Scaffolds_11 | 9628916 | 9628945 | 0.764 | 3 | 0.3656 | 0.427 | 0.3065 |
| 8 | AhySSR39451 | Scaffolds_11 | 22234840 | 22234866 | 0.2865 | 8 | 0.8102 | 0.8539 | 0.7845 |
| 9 | AhySSR38506 | Scaffolds_11 | 14445677 | 14445715 | 0.978 | 2 | 0.043 | 0.044 | 0.0421 |
| 10 | AhySSR0471 | Scaffolds_11 | 18505355 | 18505416 | 0.9773 | 2 | 0.0444 | 0.0455 | 0.0434 |
| 11 | AhySSR41001 | Scaffolds_12 | 12761843 | 12761877 | 0.8807 | 2 | 0.2102 | 0.0795 | 0.1881 |
| 12 | AhySSR39465 | Scaffolds_12 | 40173 | 40211 | 0.539 | 2 | 0.497 | 0.2987 | 0.3735 |
| 13 | AhySSR12555 | Scaffolds_13 | 146746 | 146817 | 0.7987 | 2 | 0.3216 | 0.4026 | 0.2699 |
| 14 | AhySSR43989 | Scaffolds_13 | 9524194 | 9524211 | 0.4839 | 5 | 0.5797 | 0.9355 | 0.4904 |
| 15 | AhySSR43465 | Scaffolds_13 | 6175139 | 6175180 | 0.5342 | 8 | 0.6538 | 0.4384 | 0.6173 |
| 16 | AhySSR44477 | Scaffolds_13 | 13640854 | 13640877 | 0.9477 | 2 | 0.0992 | 0.0814 | 0.0943 |
| 17 | AhySSR46467 | Scaffolds_14 | 7295227 | 7295270 | 0.5053 | 2 | 0.4999 | 0.9255 | 0.375 |
| 18 | AhySSR48966 | Scaffolds_15 | 6889536 | 6889637 | 0.5988 | 3 | 0.5349 | 0.8025 | 0.457 |
| 19 | AhySSR49996 | Scaffolds_15 | 13907952 | 13908026 | 0.5325 | 8 | 0.6273 | 0.9351 | 0.5736 |
| 20 | AhySSR0542 | Scaffolds_15 | 11049934 | 11050033 | 0.5495 | 2 | 0.4951 | 0.8352 | 0.3725 |
| 21 | AhySSR51527 | Scaffolds_16 | 8353654 | 8353737 | 0.7443 | 6 | 0.4301 | 0.5114 | 0.4124 |
| 22 | AhySSR51061 | Scaffolds_16 | 4284470 | 4284489 | 0.7527 | 2 | 0.3723 | 0.4946 | 0.303 |
| 23 | AhySSR5081 | Scaffolds_2 | 1020604 | 1020639 | 0.3444 | 10 | 0.7533 | 1 | 0.7189 |
| 24 | AhySSR8059 | Scaffolds_2 | 22784760 | 22784831 | 0.5774 | 2 | 0.488 | 0.0119 | 0.3689 |
| 25 | AhySSR11576 | Scaffolds_3 | 16269717 | 16269792 | 0.633 | 2 | 0.4646 | 0.6702 | 0.3567 |
| 26 | AhySSR12579 | Scaffolds_3 | 23384578 | 23384688 | 0.7926 | 2 | 0.3288 | 0.4149 | 0.2748 |
| 27 | AhySSR16096 | Scaffolds_4 | 18892950 | 18892976 | 0.5722 | 4 | 0.5566 | 0.8556 | 0.4796 |
| 28 | AhySSR18586 | Scaffolds_5 | 10835301 | 10835348 | 0.4471 | 6 | 0.6828 | 1 | 0.6324 |
| 29 | AhySSR19060 | Scaffolds_5 | 14161515 | 14161619 | 0.4718 | 10 | 0.6646 | 0.9577 | 0.6119 |
| 30 | AhySSR18091 | Scaffolds_5 | 7461113 | 7461163 | 0.6193 | 2 | 0.4715 | 0.7614 | 0.3604 |
| 31 | AhySSR23586 | Scaffolds_6 | 20936577 | 20936639 | 0.3673 | 5 | 0.7097 | 0.9388 | 0.6602 |
| 32 | AhySSR21572 | Scaffolds_6 | 6051583 | 6051604 | 0.9506 | 2 | 0.0939 | 0.0741 | 0.0895 |
| 33 | AhySSR28598 | Scaffolds_8 | 10843033 | 10843092 | 0.6744 | 2 | 0.4392 | 0.6512 | 0.3427 |
| 34 | AhySSR29093 | Scaffolds_8 | 14673069 | 14673119 | 0.4902 | 6 | 0.682 | 0.7255 | 0.6407 |
| 35 | AhySSR29571 | Scaffolds_8 | 17923890 | 17923961 | 0.3404 | 9 | 0.8077 | 1 | 0.7855 |
| 36 | AhySSR31577 | Scaffolds_9 | 7849897 | 7849947 | 0.9828 | 2 | 0.0339 | 0.0115 | 0.0333 |
| Mean | 0.6286 | 3.8333 | 0.4641 | 0.6075 | 0.4002 |
| Population | Percent Polymorphism | I | Ho | He |
|---|---|---|---|---|
| Himachal Pradesh | 100.00% | 0.883 | 0.555 | 0.497 |
| Uttarakhand | 97.22% | 0.82 | 0.527 | 0.48 |
| Maharashtra | 91.67% | 0.751 | 0.484 | 0.459 |
| Gujarat | 91.67% | 0.715 | 0.547 | 0.444 |
| Madhya Pradesh | 75.00% | 0.587 | 0.583 | 0.39 |
| Exotic species | 80.56% | 0.548 | 0.583 | 0.368 |
| Unknown | 83.33% | 0.651 | 0.435 | 0.412 |
| Other | 63.89% | 0.49 | 0.431 | 0.33 |
| Mean | 85.42% | 0.681 | 0.518 | 0.423 |
| Axis | Axis-1 | Axis-2 | Axis-3 |
|---|---|---|---|
| Variation (%) | 11.72 | 8.51 | 7.48 |
| Cumulative variation (%) | 11.72 | 20.23 | 27.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vats, G.; Das, D.; Gupta, R.; Singh, A.; Maurya, A.; Rajkumar, S.; Singh, A.K.; Bharadwaj, R.; Kumar, S.; Kaushik, S.K.; et al. Validation of Genome-Wide SSR Markers Developed for Genetic Diversity and Population Structure Study in Grain Amaranth (Amaranthus hypochondriacus). Agriculture 2023, 13, 431. https://doi.org/10.3390/agriculture13020431
Vats G, Das D, Gupta R, Singh A, Maurya A, Rajkumar S, Singh AK, Bharadwaj R, Kumar S, Kaushik SK, et al. Validation of Genome-Wide SSR Markers Developed for Genetic Diversity and Population Structure Study in Grain Amaranth (Amaranthus hypochondriacus). Agriculture. 2023; 13(2):431. https://doi.org/10.3390/agriculture13020431
Chicago/Turabian StyleVats, Gautam, Dimpi Das, Rajat Gupta, Akshay Singh, Avantika Maurya, S. Rajkumar, Amit Kumar Singh, Rakesh Bharadwaj, Sandeep Kumar, Surinder Kumar Kaushik, and et al. 2023. "Validation of Genome-Wide SSR Markers Developed for Genetic Diversity and Population Structure Study in Grain Amaranth (Amaranthus hypochondriacus)" Agriculture 13, no. 2: 431. https://doi.org/10.3390/agriculture13020431
APA StyleVats, G., Das, D., Gupta, R., Singh, A., Maurya, A., Rajkumar, S., Singh, A. K., Bharadwaj, R., Kumar, S., Kaushik, S. K., Gupta, V., Singh, K., & Singh, R. (2023). Validation of Genome-Wide SSR Markers Developed for Genetic Diversity and Population Structure Study in Grain Amaranth (Amaranthus hypochondriacus). Agriculture, 13(2), 431. https://doi.org/10.3390/agriculture13020431








