Exploration of Piezo Channels in Bread Wheat (Triticum aestivum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of TaPiezos and Their Chromosomal Distribution
2.2. Phylogenetic Analysis
2.3. Gene Structure Analysis
2.4. Cis-Regulatory Element Analysis
2.5. Physicochemical, Motif Analyses and Multiple Sequence Alignment
2.6. Expression Profiling of Piezo Genes
2.7. qRT-PCR Analysis
2.8. Protein–Protein and miRNA-Interaction Analysis
3. Results
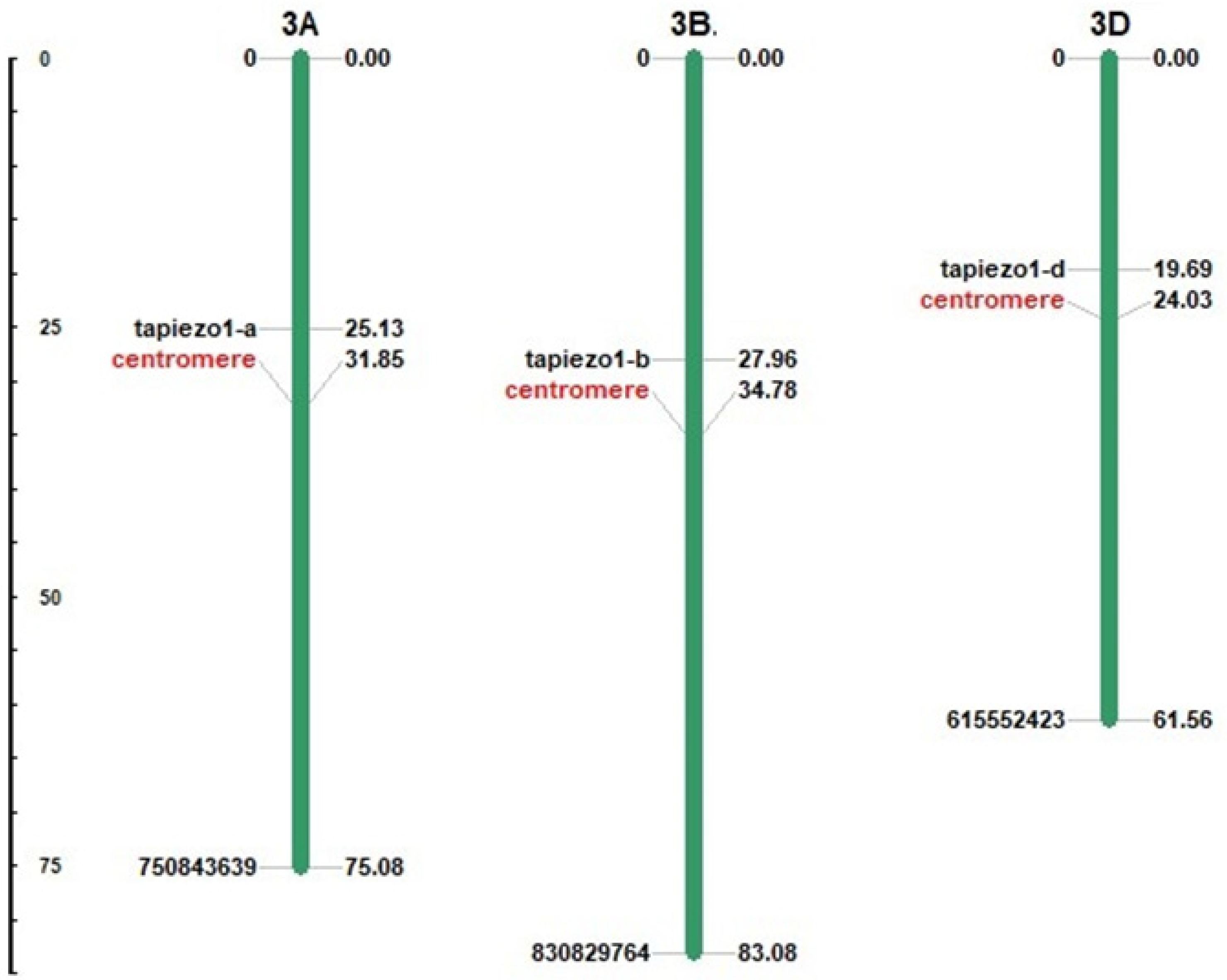
3.1. Identification and Chromosomal Localization of TaPiezo
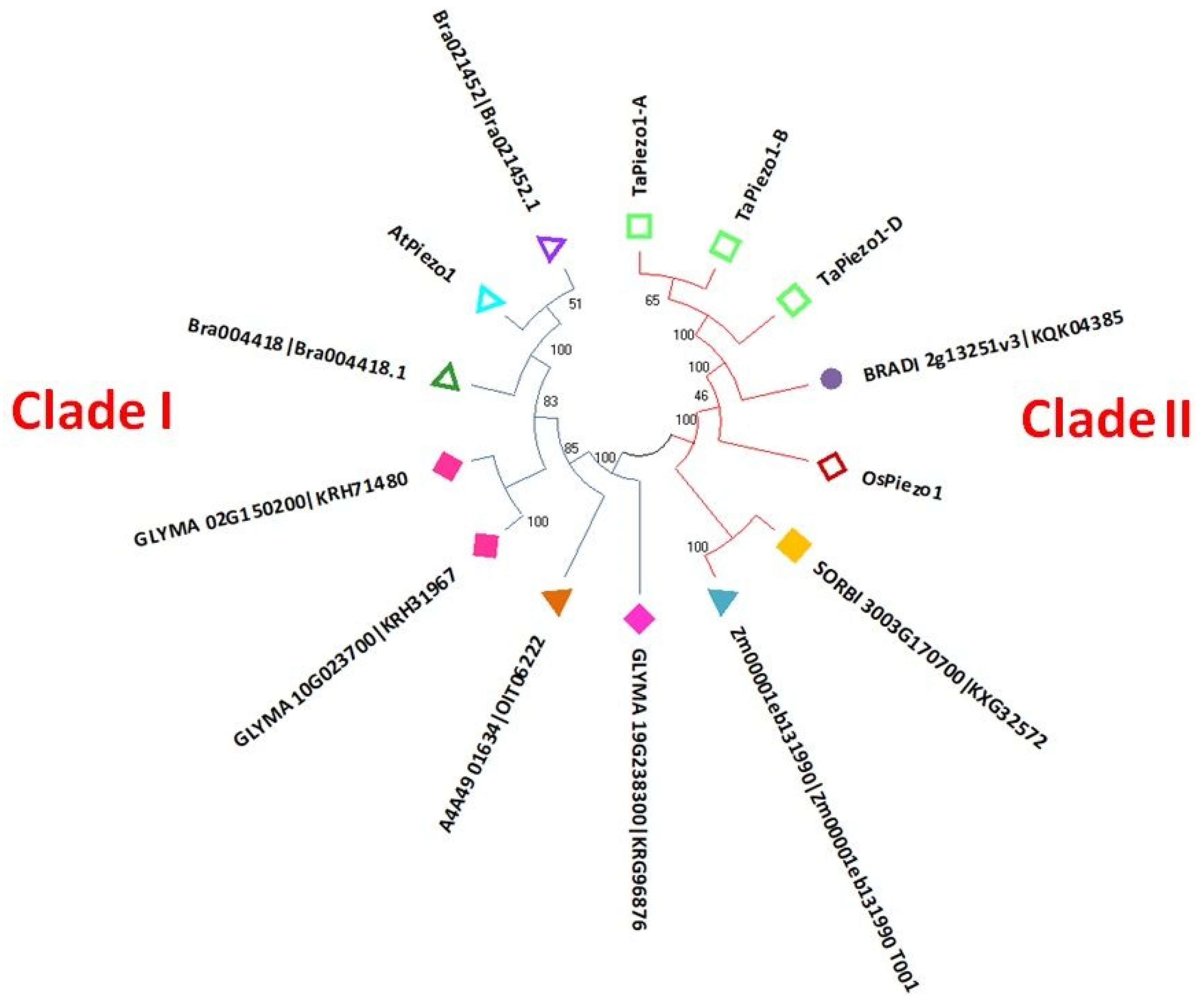
3.2. Phylogenetic Analysis
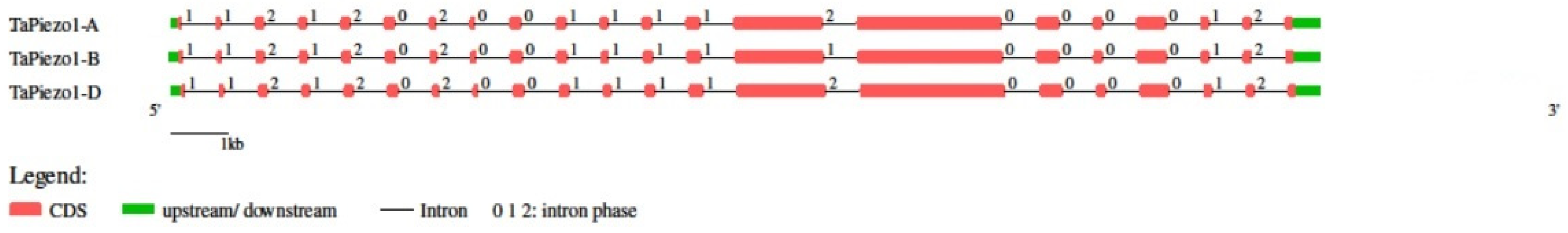
3.3. Gene Structure Analysis
3.4. Cis-Regulatory Element Analysis
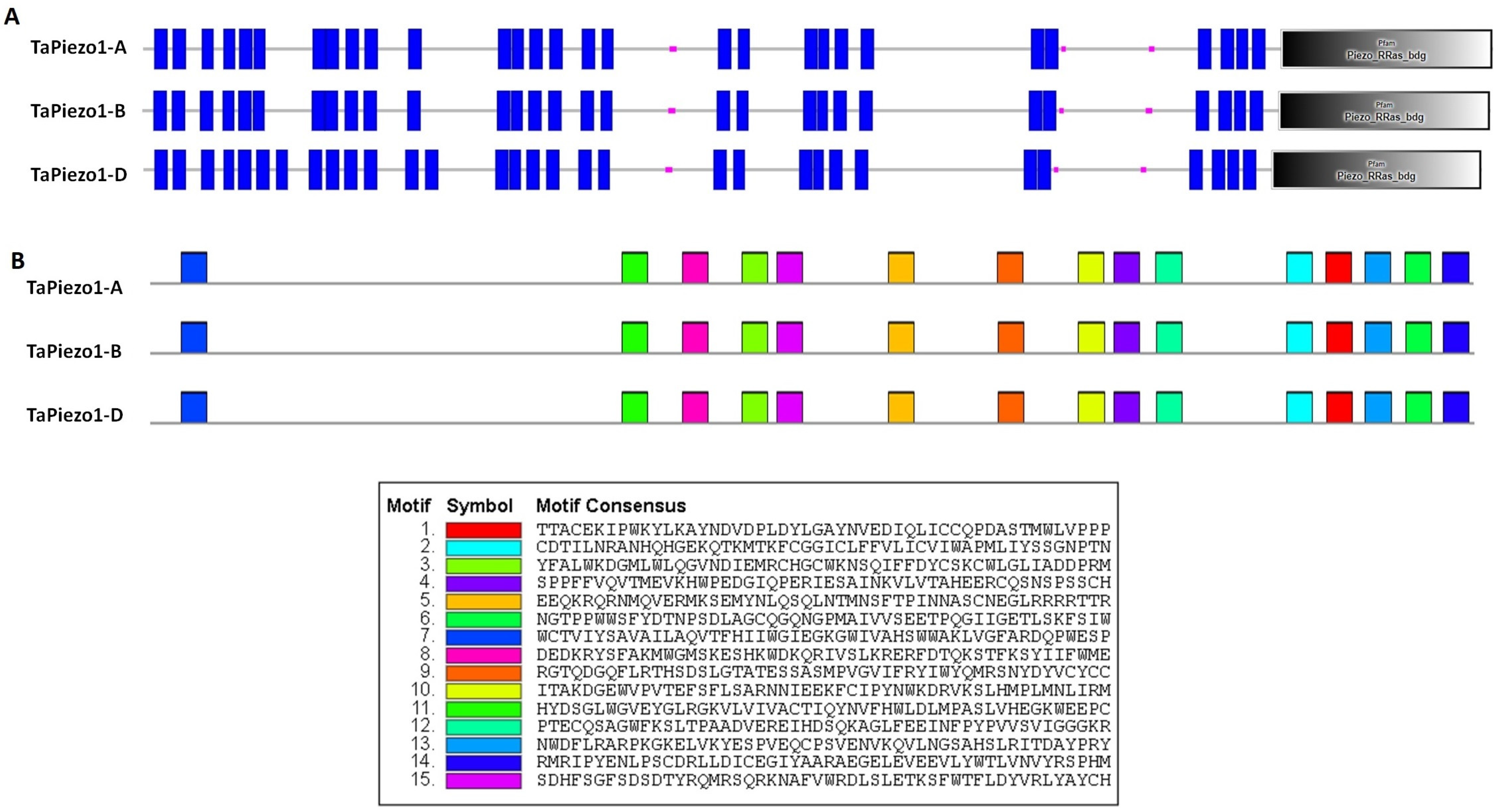
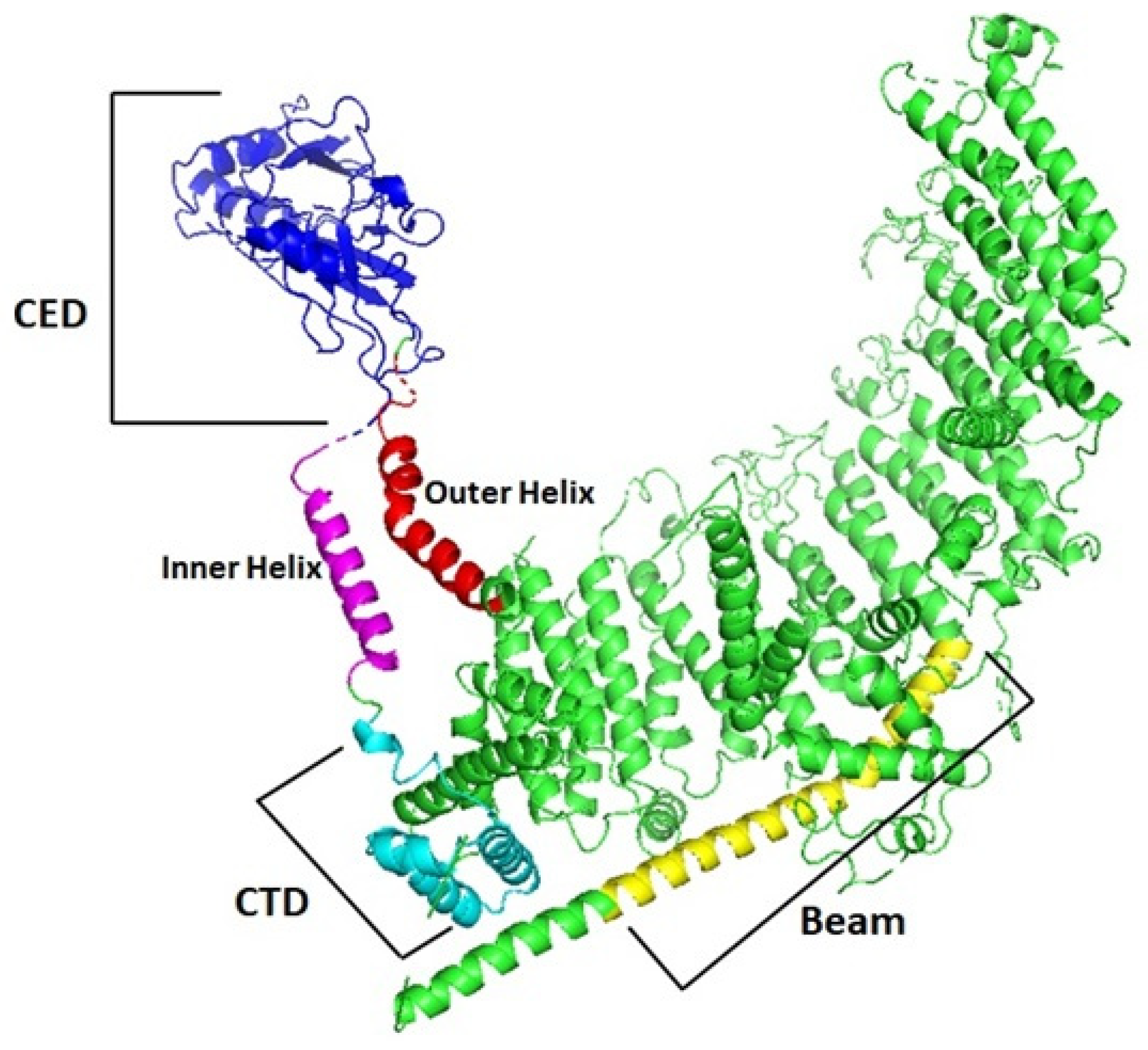
3.5. Protein Characterization
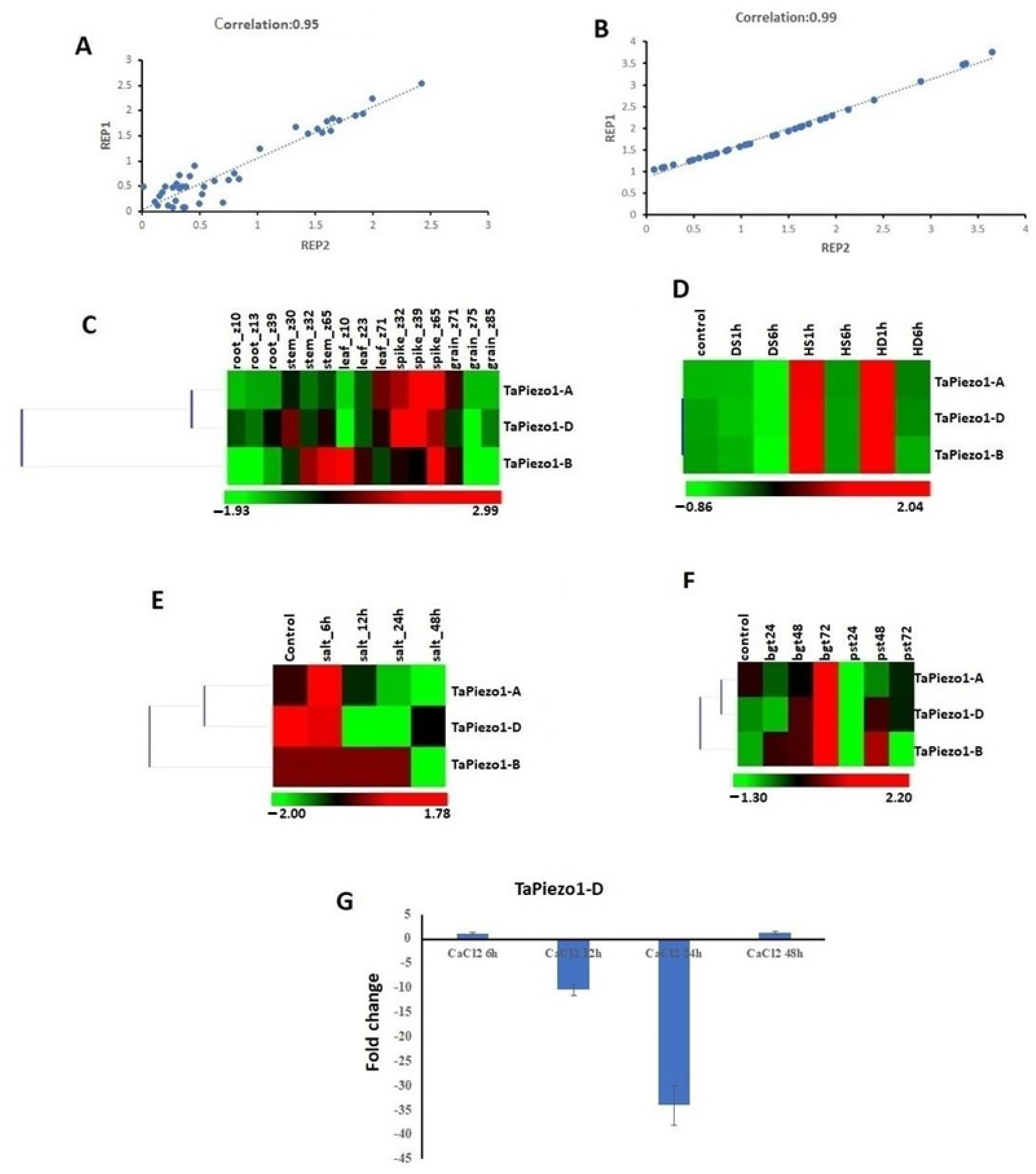
3.6. Expression Profiling of Piezo Genes in Tissue Developmental Stages
3.7. Expression Profiling of Piezo Genes in Abiotic and Biotic Stresses
3.8. qRT-PCR Analysis
3.9. Protein–Protein Interaction
3.10. miRNAs Interaction Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hamilton, E.S.; Schlegel, A.M.; Haswell, E.S. United in diversity: Mechanosensitive ion channels in plants. Annu. Rev. Plant Biol. 2015, 66, 113–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamant, O.; Haswell, E.S. Life behind the wall: Sensing mechanical cues in plants. BMC Biol. 2017, 15, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.E.; Coste, B.; Chadha, A.; Cook, B.; Patapoutian, A. The role of Drosophila Piezo in mechanical nociception. Nature 2012, 483, 209–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranade, S.S.; Woo, S.H.; Dubin, A.E.; Moshourab, R.A.; Wetzel, C.; Petrus, M.; Mathur, J.; Bégay, V.; Coste, B.; Mainquist, J.; et al. Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature 2014, 516, 121–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murthy, S.E.; Dubin, A.E.; Patapoutian, A. Piezos thrive under pressure: Mechanically activated ion channels in health and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Coste, B.; Mathur, J.; Schmidt, M.; Earley, T.J.; Ranade, S.; Petrus, M.J.; Dubin, A.E.; Patapoutian, A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 2010, 330, 55–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rode, B.; Shi, J.; Endesh, N.; Drinkhill, M.J.; Webster, P.J.; Lotteau, S.J.; Bailey, M.A.; Yuldasheva, N.Y.; Ludlow, M.J.; Cubbon, R.M.; et al. Piezo1 channels sense whole body physical activity to reset cardiovascular homeostasis and enhance performance. Nat. Commun. 2017, 8, 350. [Google Scholar] [CrossRef]
- Wu, J.; Lewis, A.H.; Grandl, J. Touch, tension, and transduction–the function and regulation of Piezo ion channels. Trends Biochem. Sci. 2017, 42, 57–71. [Google Scholar] [CrossRef] [Green Version]
- Ge, J.; Li, W.; Zhao, Q.; Li, N.; Chen, M.; Zhi, P.; Li, R.; Gao, N.; Xiao, B.; Yang, M. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature 2015, 527, 64–69. [Google Scholar] [CrossRef]
- Saotome, K.; Murthy, S.E.; Kefauver, J.M.; Whitwam, T.; Patapoutian, A.; Ward, A.B. Structure of the mechanically activated ion channel Piezo1. Nature 2018, 554, 481–486. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhou, H.; Chi, S.; Wang, Y.; Wang, J.; Geng, J.; Wu, K.; Liu, W.; Zhang, T.; Dong, M.Q.; et al. Structure and mechanogating mechanism of the Piezo1 channel. Nature 2018, 554, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Liu, B.; Shao, Q.; Huang, X.; Li, J.; Luan, S.; He, K. AtPiezo plays an important role in root cap mechanotransduction. Int. J. Mol. Sci. 2021, 22, 467. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.; Guoqiang, H.; Jin, S.; Fengli, Z.; Dabing, Z. Bioinformatics analysis for Piezo in rice. Reprod. Breed. 2021, 1, 108–113. [Google Scholar] [CrossRef]
- Mousavi, S.A.; Dubin, A.E.; Zeng, W.Z.; Coombs, A.M.; Do, K.; Ghadiri, D.A.; Keenan, W.T.; Ge, C.; Zhao, Y.; Patapoutian, A. PIEZO ion channel is required for root mechanotransduction in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2102188118. [Google Scholar] [CrossRef]
- Zhang, Z.; Tong, X.; Liu, S.Y.; Chai, L.X.; Zhu, F.F.; Zhang, X.P.; Zou, J.Z.; Wang, X.B. Genetic analysis of a Piezo-like protein suppressing systemic movement of plant viruses in Arabidopsis thaliana. Sci. Rep. 2019, 9, 3187. [Google Scholar] [CrossRef]
- Radin, I.; Richardson, R.A.; Weiner, E.R.; Bascom, C.S.; Bezanilla, M.; Haswell, E.S. Regulation of vacuole morphology by PIEZO channels in spreading earth moss. BioRxiv 2020, 2020-08. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Kaur, A.; Taneja, M.; Tyagi, S.; Sharma, A.; Singh, K.; Upadhyay, S.K. Genome-wide characterization and expression analysis suggested diverse functions of the mechanosensitive channel of small conductance-like (MSL) genes in cereal crops. Sci. Rep. 2020, 10, 16583. [Google Scholar] [CrossRef]
- Madhu; Kaur, A.; Tyagi, S.; Singh, K.; Upadhyay, S.K. Exploration of glutathione reductase for abiotic stress response in bread wheat (Triticum aestivum L.). Plant Cell Rep. 2022, 41, 639–654. [Google Scholar] [CrossRef]
- Kaur, A.; Sharma, A.; Dixit, S.; Singh, K.; Upadhyay, S.K. OSCA Genes in Bread Wheat: Molecular Characterization, Expression Profiling, and Interaction Analyses Indicated Their Diverse Roles during Development and Stress Response. Int. J. Mol. Sci. 2022, 23, 14867. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.; Biosciences, I.; Carlsbad, C.J.G.B.B. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E.; et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef]
- Pingault, L.; Choulet, F.; Alberti, A.; Glover, N.; Wincker, P.; Feuillet, C.; Paux, E. Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biol. 2015, 16, 29. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Papatheodorou, I.; Fonseca, N.A.; Keays, M.; Tang, Y.A.; Barrera, E.; Bazant, W.; Burke, M.; Füllgrabe, A.; Fuentes, A.M.P.; George, N.; et al. Expression Atlas: Gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018, 46, D246–D251. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Boil. 2015, 15, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, Z.; Khan, A.A.; Lin, Q.; Han, Y.; Mu, P.; Liu, Y.; Zhang, H.; Li, L.; Meng, X.; et al. Expression partitioning of homeologs and tandem duplications contribute to salt tolerance in wheat (Triticum aestivum L.). Sci. Rep. 2016, 6, 21476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Yang, Y.; Wang, C.; Liu, M.; Li, H.; Fu, Y.; Wang, Y.; Nie, Y.; Liu, X.; Ji, W. Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genom. 2014, 15, 898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, J.; Gordish-Dressman, H.; Hoffman, E.P. An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics 2006, 22, 808–814. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Tyagi, S.; Alok, A.; Singh, K.; Upadhyay, S.K. Thaumatin-like protein kinases: Molecular characterization and transcriptional profiling in five cereal crops. Plant Sci. 2020, 290, 110317. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, H.; Rajput, R.; Pandey, A.; Upadhyay, S.K. Molecular Characterization Revealed the Role of Thaumatin-Like Proteins of Bread Wheat in Stress Response. Front. Plant Sci. 2022, 12, 1664-462X. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; Von Mering, C.; et al. STRING v9. 1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [Green Version]
- Knight, H. Calcium signaling during abiotic stress in plants. In International Review of Cytology; Academic Press: Cambridge, MA, USA, 1999; Volume 195, pp. 269–324. [Google Scholar]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frachisse, J.M.; Thomine, S.; Allain, J.M. Calcium and plasma membrane force-gated ion channels behind development. Curr. Opin. Plant Biol. 2020, 53, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid ssignalling in guard cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef]
- Coste, B.; Xiao, B.; Santos, J.S.; Syeda, R.; Grandl, J.; Spencer, K.S.; Kim, S.E.; Schmidt, M.; Mathur, J.; Dubin, A.E.; et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 2012, 483, 176–181. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.R.; MacKinnon, R. Structure-based membrane dome mechanism for Piezo mechanosensitivity. Elife 2017, 6, e33660. [Google Scholar] [CrossRef]
- Woo, S.H.; Lukacs, V.; de Nooij, J.C.; Zaytseva, D.; Criddle, C.R.; Francisco, A.; Jessell, T.M.; Wilkinson, K.A.; Patapoutian, A. Piezo2 is the principal mechanotransduction channel for proprioception. Nat. Neurosci. 2015, 18, 1756–1762. [Google Scholar] [CrossRef] [Green Version]
- Rakhshandehroo, F.; Takeshita, M.; Squires, J.; Palukaitis, P. The influence of RNA-dependent RNA polymerase 1 on potato virus Y infection and on other antiviral response genes. Mol. Plant Microbe Interact. 2009, 22, 1312–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.W.; Sun, Z.H.; Zhou, Y.H.; Shi, K.; Li, X.; Zhang, G.Q.; Xia, X.J.; Chen, Z.X.; Yu, J.Q. The role of hydrogen peroxide and nitric oxide in the induction of plant-encoded RNA-dependent RNA polymerase 1 in the basal defense against Tobacco mosaic virus. PLoS ONE 2013, 8, e76090. [Google Scholar] [CrossRef]
- Liao, Y.W.; Liu, Y.R.; Liang, J.Y.; Wang, W.P.; Zhou, J.; Xia, X.J.; Zhou, Y.H.; Yu, J.Q.; Shi, K. The relationship between the plant-encoded RNA-dependent RNA polymerase 1 and alternative oxidase in tomato basal defense against Tobacco mosaic virus. Planta 2015, 241, 641–650. [Google Scholar] [CrossRef]
- Cao, J.Y.; Xu, Y.P.; Li, W.; Li, S.S.; Rahman, H.; Cai, X.Z. Genome-wide identification of Dicer-like, Argonaute, and RNA-dependent RNA polymerase gene families in Brassica species and functional analyses of their Arabidopsis homologs in resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 2016, 7, 1614. [Google Scholar] [CrossRef] [Green Version]
- Koiwa, H.; Li, F.; McCully, M.G.; Mendoza, I.; Koizumi, N.; Manabe, Y.; Nakagawa, Y.; Zhu, J.; Rus, A.; Pardo, J.M.; et al. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell. 2003, 15, 2273–2284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.S.; Frank, J.; Kang, C.H.; Kajiura, H.; Vikram, M.; Ueda, A.; Kim, S.; Bahk, J.D.; Triplett, B.; Fujiyama, K.; et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 2008, 105, 5933–5938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Németh, K.; Salchert, K.; Putnoky, P.; Bhalerao, R.; Koncz-Kálmán, Z.; Stankovic-Stangeland, B.; Bakó, L.; Mathur, J.; Ökrész, L.; Stabel, S.; et al. Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 1998, 12, 3059–3073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baruah, A.; Šimková, K.; Hincha, D.K.; Apel, K.; Laloi, C. Modulation of 1O2-mediated retrograde signaling by the PLEIOTROPIC RESPONSE LOCUS 1 (PRL1) protein, a central integrator of stress and energy signaling. Plant J. 2009, 60, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Flores-Pérez, Ú.; Pérez-Gil, J.; Closa, M.; Wright, L.P.; Botella-Pavía, P.; Phillips, M.A.; Ferrer, A.; Gershenzon, J.; Rodríguez-Concepción, M. Pleiotropic regulatory locus 1 (PRL1) integrates the regulation of sugar responses with isoprenoid metabolism in Arabidopsis. Mol. Plant. 2010, 3, 101–112. [Google Scholar] [CrossRef]
- Sun, M.; Shen, Y.; Li, H.; Yang, J.; Cai, X.; Zheng, G.; Zhu, Y.; Jia, B.; Sun, X. The multiple roles of OsmiR535 in modulating plant height, panicle branching and grain shape. Plant Sci. 2019, 283, 60–69. [Google Scholar] [CrossRef]
- Zhang, L.L.; Huang, Y.Y.; Zheng, Y.P.; Liu, X.X.; Zhou, S.X.; Yang, X.M.; Liu, S.L.; Li, Y.; Li, J.L.; Zhao, S.L.; et al. Osa-miR535 targets SQUAMOSA promoter binding protein-like 4 to regulate blast disease resistance in rice. Plant J. 2022, 110, 166–178. [Google Scholar] [CrossRef]
- Li, W.; Cui, X.; Meng, Z.; Huang, X.; Xie, Q.; Wu, H.; Jin, H.; Zhang, D.; Liang, W. Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012, 158, 1279–1292. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Jiang, N.; Hou, X.; Wu, S.; Zhang, Q.; Meng, J.; Luan, Y. Genome-wide identification of lncRNAs and analysis of ceRNA networks during tomato resistance to Phytophthora infestans. Phytopathology 2020, 110, 456–464. [Google Scholar] [CrossRef]
- Hou, X.; Cui, J.; Liu, W.; Jiang, N.; Zhou, X.; Qi, H.; Meng, J.; Luan, Y. LncRNA39026 enhances tomato resistance to Phytophthora infestans by decoying miR168a and inducing PR gene expression. Phytopathology 2020, 110, 873–880. [Google Scholar] [CrossRef]
- Wu, J.; Wang, D.; Liu, Y.; Wang, L.; Qiao, X.; Zhang, S. Identification of miRNAs involved in pear fruit development and quality. BMC Genom. 2014, 15, 953. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene Name | 5′-3′ |
|---|---|
| TaARF_Forward primer | TGATAGGGAACGTGTTGTTGAGGC |
| TaARF_Reverse primer | AGCCAGTCAAGACCCTCGTACAAC |
| TaPiezo1-D_Forward primer | AGGAGAGGATTTCACAATTGGAGGCTG |
| TaPiezo1-D_Reverse primer | CTTCAACCAAAGAAAGGACAGCAGCAG |
| Gene Name | Light Response | Growth and Development | Stress Response | Hormone Response |
|---|---|---|---|---|
| TaPiezo1-A | G-box, TCT-motif, I-box, TCCC-motif, G-Box, Box 4 | CCGTCC-box, GCN4_motif, CCGTCC motif, AAGAA-motif | ARE, as-1, LTR, STRE, WRE3, A-box, W box, MYB, GC-motif, Myb-binding site, MYB-like sequence, TC-rich repeats | TCA-element, ABRE, CGTCA-motif, TGA-element, TGACG-motif |
| TaPiezo1-B | AE-box, G-box, G-Box, | O2-site, RY-element, CCGTCC-box, CCGTCC motif | MYC, Myb-binding site, STRE, MYB recognition site, MYB, WRE3, LTR, ARE, Myb, as-1, A-box | TGACG-motif, ABRE, CGTCA-motif |
| TaPiezo1-D | Sp1, G-box, TCT-motif, ACE | CCGTCC motif, O2-site, CCGTCC-box, CAT-box | W box, LTR, MYB-like sequence, WRE3, GC-motif, TC-rich repeats, MYC, A-box, box S, Myb, MBS, STRE, as-1, Myb-binding site, DRE core, MYB recognition site, MYB | CGTCA-motif, AT~ABRE, ABRE4, TGACG-motif, TATC-box, P-box, ABRE3a, ABRE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaur, A.; Madhu; Sharma, A.; Singh, K.; Upadhyay, S.K. Exploration of Piezo Channels in Bread Wheat (Triticum aestivum L.). Agriculture 2023, 13, 783. https://doi.org/10.3390/agriculture13040783
Kaur A, Madhu, Sharma A, Singh K, Upadhyay SK. Exploration of Piezo Channels in Bread Wheat (Triticum aestivum L.). Agriculture. 2023; 13(4):783. https://doi.org/10.3390/agriculture13040783
Chicago/Turabian StyleKaur, Amandeep, Madhu, Alok Sharma, Kashmir Singh, and Santosh Kumar Upadhyay. 2023. "Exploration of Piezo Channels in Bread Wheat (Triticum aestivum L.)" Agriculture 13, no. 4: 783. https://doi.org/10.3390/agriculture13040783
APA StyleKaur, A., Madhu, Sharma, A., Singh, K., & Upadhyay, S. K. (2023). Exploration of Piezo Channels in Bread Wheat (Triticum aestivum L.). Agriculture, 13(4), 783. https://doi.org/10.3390/agriculture13040783








