Evaluation of the Genetic Diversity of Greek Garlic (Allium sativum L.) Accessions Using DNA Markers and Association with Phenotypic and Chemical Variation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and Amplification
2.3. SSR Analysis
2.4. ISSR Analysis
2.5. Statistical Analysis
2.5.1. SSR Markers
2.5.2. ISSR Markers
2.5.3. Association Analysis
3. Results
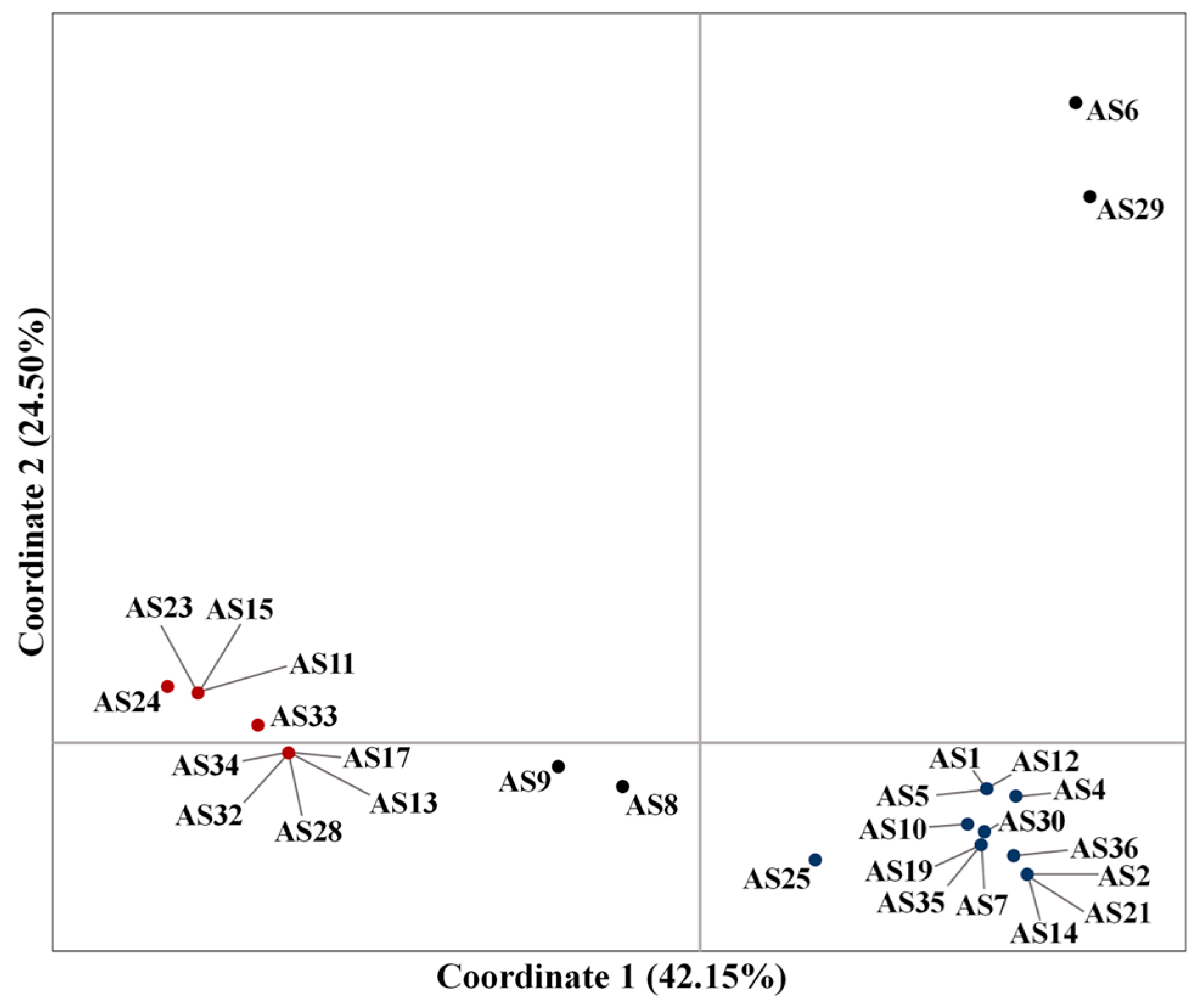
3.1. Genetic Relationships Based on the SSR Markers
3.2. Genetic Relationships Based on ISSR Analysis
3.3. Association of SSR Loci with Chemical Compounds and Bulb Morphological Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elsharkawy, G.A.; Hegazi, H.H.; Azab, E.; Gobouri, A.A.; Sayed, S.A. Assessment of Genetic Diversity among Egyptian Garlic Landraces Based on Morphological Characteristics and ISSR Markers. Eur. J. Hortic. Sci. 2021, 86, 579–589. [Google Scholar] [CrossRef]
- Karakan, F.Y. Relationship between Volatile Sulfur Compounds, Mineral Content, Morphological and Molecular Characterization of Local Garlic Genotypes. Bangladesh J. Bot. 2022, 51, 147–155. [Google Scholar] [CrossRef]
- Friesen, N.; Smirnov, S.V.; Shmakov, A.I.; Herden, T.; Oyuntsetseg, B.; Hurka, H. Allium Species of Section Rhizomatosa, Early Members of the Central Asian Steppe Vegetation. Flora Morphol. Distrib. Funct. Ecol. Plants 2020, 263, 151536. [Google Scholar] [CrossRef]
- Parreño, R.; Rodríguez-Alcocer, E.; Martínez-Guardiola, C.; Carrasco, L.; Castillo, P.; Arbona, V.; Jover-Gil, S.; Candela, H. Turning Garlic into a Modern Crop: State of the Art and Perspectives. Plants 2023, 12, 1212. [Google Scholar] [CrossRef]
- Benke, A.P.; Krishna, R.; Mahajan, V.; Ansari, W.A.; Gupta, A.J.; Khar, A.; Shelke, P.; Thangasamy, A.; Shabeer, T.P.A.; Singh, M.; et al. Genetic Diversity of Indian Garlic Core Germplasm Using Agro-Biochemical Traits and SRAP Markers. Saudi J. Biol. Sci. 2021, 28, 4833–4844. [Google Scholar] [CrossRef]
- Etoh, T.; Simon, P.W. Diversity, Fertility and Seed Production of Garlic. In Allium Crop Sciences: Recent Advances; Rabinowitch, H.D., Currah, L., Eds.; CABI Publishing: Wallingford, UK, 2002; pp. 101–117. ISBN 9780851995106. [Google Scholar]
- Shaaf, S.; Sharma, R.; Kilian, B.; Walther, A.; Özkan, H.; Karami, E.; Mohammadi, B. Genetic Structure and Eco-Geographical Adaptation of Garlic Landraces (Allium sativum L.) in Iran. Genet. Resour. Crop. Evol. 2014, 61, 1565–1580. [Google Scholar] [CrossRef]
- Fritsch, R.M.; Friesen, N. Evolution, Domestication and Taxonomy. In Allium Crop Science: Recent Advances; Rabinowitch, H., Currah, L., Eds.; CABI Publishing: Wallingford, UK, 2002; pp. 5–30. ISBN 9780851995106. [Google Scholar]
- Abe, K.; Hori, Y.; Myoda, T. Volatile Compounds of Fresh and Processed Garlic (Review). Exp. Ther. Med. 2019, 19, 1585–1593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fesseha, H.; Goa, E. Therapeutic Value of Garlic (Allium sativum): A Review. Adv. Food Technol. Nutr. Sci.-Open J. 2019, 5, 107–117. [Google Scholar] [CrossRef]
- Avgeri, I.; Zeliou, K.; Petropoulos, S.A.; Bebeli, P.J.; Papasotiropoulos, V.; Lamari, F.N. Variability in Bulb Organosulfur Compounds, Sugars, Phenolics, and Pyruvate among Greek Garlic Genotypes: Association with Antioxidant Properties. Antioxidants 2020, 9, 967. [Google Scholar] [CrossRef]
- Batiha, G.E.S.; Beshbishy, A.M.; Wasef, L.G.; Elewa, Y.H.A.; Al-Sagan, A.A.; El-Hack, M.E.A.; Taha, A.E.; Abd-Elhakim, Y.M.; Devkota, H.P. Chemical Constituents and Pharmacological Activities of Garlic (Allium sativum L.): A Review. Nutrients 2020, 12, 872. [Google Scholar] [CrossRef] [Green Version]
- Morales-González, J.A.; Madrigal-Bujaidar, E.; Sánchez-Gutiérrez, M.; Izquierdo-Vega, J.A.; Del Carmen Valadez-Vega, M.; Álvarez-González, I.; Morales-González, Á.; Madrigal-Santillán, E. Garlic (Allium sativum L.): A Brief Review of Its Antigenotoxic Effects. Foods 2019, 8, 343. [Google Scholar] [CrossRef] [Green Version]
- Bradley, K.F.; Rieger, M.A.; Collins, G.G. Classification of Australian Garlic Cultivars by DNA Fingerprinting. Aust. J. Exp. Agric. 1996, 36, 613–618. [Google Scholar] [CrossRef]
- Poljuha, D.; Franić, M.; Kralj, I.; Weber, T.; Šatović, Z.; Ban, D.; Toth, N.; Dumičić, G.; Kereša, S.; da Cunha, C.P.; et al. Genetic Diversity and Structure Analysis of Croatian Garlic Collection Assessed by SSR Markers. Folia Hortic. 2021, 33, 157–171. [Google Scholar] [CrossRef]
- Volk, G.M.; Henk, A.D.; Richards, C.M. Genetic Diversity among U.S. Garlic Clones as Detected Using AFLP Methods. J. Am. Soc. Hortic. Sci. Jashs 2004, 129, 559–569. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Qiao, L.; Chen, B.; Zheng, Y.; Zhi, C.; Zhang, S.; Pan, Y.; Cheng, Z. SSR Markers Development and Their Application in Genetic Diversity Evaluation of Garlic (Allium sativum) Germplasm. Plant Divers 2022, 44, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Sun, X.; Cao, M.; Zhang, Y.; Hang, Y.; Chen, M. Molecular Markers for Authentification of Allium sativum L. Cultivar ‘Taicangbaisuan’ and Genetic Relationships among 9 Chinese Garlic Cultivars. Genet. Resour. Crop Evol. 2021, 68, 1961–1970. [Google Scholar] [CrossRef]
- Panthee, D.R.; Kc, R.B.; Regmi, H.N.; Subedi, P.P.; Bhattarai, S.; Dhakal, J. Diversity Analysis of Garlic (Allium sativum L.) Germplasms Available in Nepal Based on Morphological Characters. Genet. Resour. Crop Evol. 2006, 53, 205–212. [Google Scholar] [CrossRef]
- Ovesná, J.; Leišová-Svobodová, L.; Kučera, L. Microsatellite Analysis Indicates the Specific Genetic Basis of Czech Bolting Garlic. Czech J. Genet. Plant Breed. 2014, 50, 226–234. [Google Scholar] [CrossRef] [Green Version]
- Hoogerheide, E.S.S.; Azevedo Filho, J.A.; Vencovsky, R.; Zucchi, M.I.; Zago, B.W.; Pinheiro, J.B. Genetic Variability of Garlic Accessions as Revealed by Agro-Morphological Traits Evaluated under Different Environments. Genet. Mol. Res. 2017, 16, gmr16029612. [Google Scholar] [CrossRef]
- Kıraç, H.; Dalda Şekerci, A.; Coşkun, Ö.F.; Gülşen, O. Morphological and Molecular Characterization of Garlic (Allium sativum L.) Genotypes Sampled from Turkey. Genet. Resour. Crop Evol. 2022, 69, 1833–1841. [Google Scholar] [CrossRef]
- Stavělíková, H. Morphological Characteristics of Garlic (Allium sativum L.) Genetic Resources Collection–Information. Hortic. Sci. 2008, 35, 130–135. [Google Scholar] [CrossRef] [Green Version]
- Hirata, S.; Abdelrahman, M.; Yamauchi, N.; Shigyo, M. Diversity Evaluation Based on Morphological, Physiological and Isozyme Variation in Genetic Resources of Garlic (Allium sativum L.) Collected Worldwide. Genes Genet. Syst. 2016, 91, 161–173. [Google Scholar] [CrossRef] [Green Version]
- Bonasia, A.; Conversa, G.; Lazzizera, C.; Loizzo, P.; Gambacorta, G.; Elia, A. Evaluation of Garlic Landraces from Foggia Province (Puglia Region; Italy). Foods 2020, 9, 850. [Google Scholar] [CrossRef]
- Chen, S.; Chen, W.; Shen, X.; Yang, Y.; Qi, F.; Liu, Y.; Meng, H. Analysis of the Genetic Diversity of Garlic (Allium sativum L.) by Simple Sequence Repeat and Inter Simple Sequence Repeat Analysis and Agro-Morphological Traits. Biochem. Syst. Ecol. 2014, 55, 260–267. [Google Scholar] [CrossRef]
- Rakesh Sharma, V.; Malik, S.; Kumar, M.; Sirohi, A.; Nagaraju, K. Assessment of Genetic Diversity in Garlic (Allium sativum L.) Genotypes Based on ISSR Markers. Plant Arch. 2016, 16, 88–95. [Google Scholar]
- Mohammadi, B.; Khodadadi, M.; Karami, E.; Shaaf, S. Variation in Agro-Morphological Characters in Iranian Garlic Landraces. Int. J. Veg. Sci. 2014, 20, 202–215. [Google Scholar] [CrossRef]
- Wang, H.; Li, X.; Shen, D.; Oiu, Y.; Song, J. Diversity Evaluation of Morphological Traits and Allicin Content in Garlic (Allium sativum L.) from China. Euphytica 2014, 198, 243–254. [Google Scholar] [CrossRef]
- Yebirzaf, Y.; Belete, N.; Tegibew, W.; Yohaness, G.; Abayneh, M.; Kassahun, Y. Collection and Characterization of Garlic (Allium sativm L.) Germplasm for Growth and Bulb Yield at Debre Markos, Ethiopia. J. Hortic. For. 2018, 10, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Barboza, K.; Salinas, M.C.; Acuña, C.V.; Bannoud, F.; Beretta, V.; García-Lampasona, S.; Burba, J.L.; Galmarini, C.R.; Cavagnaro, P.F. Assessment of Genetic Diversity and Population Structure in a Garlic (Allium sativum L.) Germplasm Collection Varying in Bulb Content of Pyruvate, Phenolics, and Solids. Sci. Hortic. 2020, 261, 108900. [Google Scholar] [CrossRef]
- Benke, A.P.; Khar, A.; Mahajan, V.; Gupta, A.; Singh, M. Study on Dispersion of Genetic Variation among Indian Garlic Ecotypes Using Agro Morphological Traits. Indian J. Genet. Plant Breed. 2020, 80, 94–102. [Google Scholar] [CrossRef]
- Petropoulos, S.A.; Fernandes, Â.; Ntatsi, G.; Petrotos, K.; Barros, L.; Ferreira, I.C.F.R. Nutritional Value, Chemical Characterization and Bulb Morphology of Greek Garlic Landraces. Molecules 2018, 23, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polyzos, N.; Papasotiropoulos, V.; Lamari, F.N.; Petropoulos, S.A.; Bebeli, P.J. Phenotypic Characterization and Quality Traits of Greek Garlic (Allium sativum L.) Germplasm Cultivated at Two Different Locations. Genet. Resour. Crop Evol. 2019, 66, 1671–1689. [Google Scholar] [CrossRef]
- Ma, K.H.; Kwag, J.G.; Zhao, W.; Dixit, A.; Lee, G.A.; Kim, H.H.; Chung, I.M.; Kim, N.S.; Lee, J.S.; Ji, J.J.; et al. Isolation and Characteristics of Eight Novel Polymorphic Microsatellite Loci from the Genome of Garlic (Allium sativum L.). Sci. Hortic. 2009, 122, 355–361. [Google Scholar] [CrossRef]
- Kaushik, S.; Kumar, M.; Prakash, S.; Kumar, V.; Singh, M.K.; Singh, B.; Malik, S.; Singh, K. Study of Genetic Diversity in Garlic (Allium sativum L.) by Using Morphological Characters. Progress. Agric. 2016, 16, 204. [Google Scholar] [CrossRef]
- García Lampasona, S.; Martínez, L.; Burba, J.L. Genetic diversity among selected Argentinean garlic clones (Allium sativum L.) using AFLP (Amplified Fragment Length Polymorphism). Euphytica 2003, 132, 115–119. [Google Scholar] [CrossRef]
- Ipek, M.; Ipek, A.; Simon, P.W. Molecular Characterization of Kastamonu Garlic: An Economically Important Garlic Clone in Turkey. Sci. Hortic. 2008, 115, 203–208. [Google Scholar] [CrossRef]
- Wang, H.; Li, X.; Liu, X.; Oiu, Y.; Song, J.; Zhang, X. Genetic Diversity of Garlic (Allium sativum L.) Germplasm from China by Fluorescent-Based AFLP, SSR and InDel Markers. Plant Breed. 2016, 135, 743–750. [Google Scholar] [CrossRef]
- Ipek, M.; Ipek, A.; Simon, P.W. Rapid Characterization of Garlic Clones with Locus-Specific DNA Markers. Turk. J. Agric. For. 2008, 32, 357–362. [Google Scholar]
- Mukherjee, A.; Sikdar, B.; Ghosh, B.; Banerjee, A.; Ghosh, E.; Bhattacharya, M.; Roy, S.C. RAPD and ISSR Analysis of Some Economically Important Species, Varieties, and Cultivars of the Genus Allium (Alliaceae). Turk. J. Botany 2013, 37, 605–618. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, J.; Chen, Q.; Chang, Y.; Du, J.; Meng, H. Analysis of the Genetic Diversity of Garlic (Allium sativum L.) Germplasm by SRAP. Biochem. Syst. Ecol. 2013, 50, 139–146. [Google Scholar] [CrossRef]
- Cunha, C.P.; Hoogerheide, E.S.S.; Zucchi, M.I.; Monteiro, M.; Pinheiro, J.B. New Microsatellite Markers for Garlic, Allium sativum (Alliaceae). Am. J. Bot. 2012, 99, e17–e19. [Google Scholar] [CrossRef]
- da Cunha, C.P.; Resende, F.V.; Zucchi, M.I.; Pinheiro, J.B. SSR-Based Genetic Diversity and Structure of Garlic Accessions from Brazil. Genetica 2014, 142, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Rakesh Sharma, V.; Kumar, V.; Sirohi, U.; Chaudhary, V.; Sharma, S.; Saripalli, G.; Naresh, R.K.; Yadav, H.K.; Sharma, S. Genetic Diversity and Population Structure Analysis of Indian Garlic (Allium sativum L.) Collection Using SSR Markers. Physiol. Mol. Biol. Plants 2019, 25, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A Rapid Total DNA Preparation Procedure for Fresh Plant Tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Lee, G.A.; Kwon, S.J.; Park, Y.J.; Lee, M.C.; Kim, H.H.; Lee, J.S.; Lee, S.Y.; Gwag, J.G.; Kim, C.K.; Ma, K.H. Cross-Amplification of SSR Markers Developed from Allium sativum to Other Allium Species. Sci. Hortic. 2011, 128, 401–407. [Google Scholar] [CrossRef]
- Yeh, F.; Yang, R.; Boyle, T.; Ye, Z.; Mao, J. POPGENE, the User-Friendly Shareware for Population Genetic Analysis. Mol. Biol. Biotechnol. Cent. Univ. Alta. Can. 1997, 10, 295–301. [Google Scholar]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising How the Computer Program CERVUS Accommodates Genotyping Error Increases Success in Paternity Assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis4, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenALEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research-an Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Hampl, V.; Pavlíček, A.; Flegr, J. Construction and Bootstrap Analysis of DNA Fingerprinting-Based Phylogenetic Trees with the Freeware Program FreeTree: Application to Trichomonad Parasites. Int. J. Syst. Evol. Microbiol. 2001, 51, 731–735. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.3.1; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, Scotland, 2010; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 12 March 2023).
- Prichard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, D.A.; vonHoldt, B.M. Structure Harvester: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Remington, D.L.; Thornsberry, J.M.; Matsuoka, Y.; Wilson, L.M.; Whitt, S.R.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S., IV. Structure of Linkage Disequilibrium and Phenotypic Associations in the Maize Genome. Proc. Natl. Acad. Sci. USA 2001, 98, 11479–11484. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A Program for Identifying Clustering Modes and Packaging Population Structure Inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [Green Version]
- IBM Corp. IBM SPSS Statistics for Macintosh, Version 28.0; IBM Corp: Armonk, NY, USA, 2021. [Google Scholar]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Figliuolo, G.; Candido, V.; Logozzo, G.; Miccolis, V.; Zeuli, P.L.S. Genetic Evaluation of Cultivated Garlic Germplasm (Allium sativum L. and A. ampeloprasum L.). Euphytica 2001, 121, 325–334. [Google Scholar] [CrossRef]
- Ipek, M.; Sahin, N.; Ipek, A.; Cansev, A.; Simon, P.W. Development and Validation of New SSR Markers from Expressed Regions in the Garlic Genome. Sci. Agric. 2015, 72, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Egea, L.A.; Mérida-García, R.; Kilian, A.; Hernandez, P.; Dorado, G. Assessment of Genetic Diversity and Structure of Large Garlic (Allium sativum) Germplasm Bank, by Diversity Arrays Technology “Genotyping-by-Sequencing” Platform (DArTseq). Front. Genet. 2017, 8, 98. [Google Scholar] [CrossRef] [Green Version]
- Gimenez, M.D.; Yañez-Santos, A.M.; Paz, R.C.; Quiroga, M.P.; Marfil, C.F.; Conci, V.C.; García-Lampasona, S.C. Assessment of genetic and epigenetic changes in virus-free garlic (Allium sativum L.) plants obtained by meristem culture followed by in vitro propagation. Plant Cell Rep. 2016, 35, 129–141. [Google Scholar] [CrossRef]
- Gimenez, M.D.; García-Lampasona, S. Before-after analysis of genetic and epigenetic markers in garlic: A 13-year experiment. Sci. Hortic. 2018, 240, 23–28. [Google Scholar] [CrossRef]
- Zhao, W.G.; Chung, J.W.; Lee, G.A.; Ma, K.H.; Kim, H.H.; Kim, K.T.; Chung, I.M.; Lee, J.K.; Kim, N.S.; Kim, S.M.; et al. Molecular Genetic Diversity and Population Structure of a Selected Core Set in Garlic and Its Relatives Using Novel SSR Markers. Plant Breed. 2011, 130, 46–54. [Google Scholar] [CrossRef]
- Lazaridi, E.; Ntatsi, G.; Savvas, D.; Bebeli, P.J. Diversity in cowpea (Vigna unguiculata (L.) Walp.) local populations from Greece. Genet. Resour. Crop Evol. 2017, 64, 1529–1551. [Google Scholar] [CrossRef]
- Frankham, R. Do Island Populations Have Less Genetic Variation than Mainland Populations? Heredity 1997, 78, 311–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Lampasona, S.; Asprelli, P.; Burba, J.L. Genetic Analysis of a Garlic (Allium sativum L.) Germplasm Collection from Argentina. Sci. Hortic. 2012, 138, 183–189. [Google Scholar] [CrossRef]
- Morales, R.G.F.; Resende, J.T.V.; Resende, F.V.; Delatorre, C.A.; Figueiredo, A.S.T.; Da-Silva, P.R. Genetic Divergence among Brazilian Garlic Cultivars Based on Morphological Characters and AFLP Markers. Genet. Mol. Res. 2013, 12, 270–281. [Google Scholar] [CrossRef]
- Gomes Viana, J.P.; de Pires, C.J.; Bajay, M.M.; dos Santos Valente, S.E.; Pinheiro, J.B.; Zucchi, M.I.; de Almeida Lopes, Â.C.; Ferreira Gomes, R.L. Do the Importations of Crop Products Affect the Genetic Diversity from Landraces? A Study Case in Garlic (Allium sativum L.). Genet. Resour. Crop Evol. 2021, 68, 1199–1211. [Google Scholar] [CrossRef]
- Longmei, N.; Gill, G.K.; Zaidi, P.H.; Kumar, R.; Nair, S.K.; Hindu, V.; Vinayan, M.T.; Vikal, Y. Genome Wide Association Mapping for Heat Tolerance in Sub-Tropical Maize. BMC Genom. 2021, 22, 154. [Google Scholar] [CrossRef]
- Ghomi, K.; Rabiei, B.; Sabouri, H.; Alamdari, E.G. Association between SSR Markers and Phenologic Plus Agronomic Traits in Barley (Hordeum valgare L.) Under Cold Stress Conditions. Plant Mol. Biol. Report. 2023, 41, 164–184. [Google Scholar] [CrossRef]
- González, R.E.; Burba, J.L.; Camargo, A.B. A Physiological Indicator to Estimate Allicin Content in Garlic during Storage. J. Food Biochem. 2013, 37, 449–455. [Google Scholar] [CrossRef]
- Wall, M.M.; Corgan, J.N. Relationship between Pyruvate Analysis and Flavor Perception for Onion Pungency Determination. HortScience 1992, 27, 1029–1030. [Google Scholar] [CrossRef] [Green Version]
- Cavagnaro, P.F.; Camargo, A.; Galmarini, C.R.; Simon, P.W. Effect of Cooking on Garlic (Allium sativum L.) Antiplatelet Activity and Thiosulfinates Content. J. Agric. Food Chem. 2007, 55, 1280–1288. [Google Scholar] [CrossRef] [PubMed]

| Accessions | Collection Site | Prefecture | Latitude | Longitude | Altitude (m) |
|---|---|---|---|---|---|
| AS1 | Agios Petros | Lefkada | 38°40′ Ν | 20°36′ Ε | 328 |
| AS2 | Nea Vyssa (market) | Evros | 41°35′ Ν | 26°32′ Ε | 31 |
| AS4 | Polichni | Messinia | 37°16′ N | 21°56′ Ε | 432 |
| AS5 | Κarya | Lefkada | 38°45′ Ν | 20°38′ Ε | 510 |
| AS6 | Katouna | Lefkada | 38°46′ N | 20°42′ Ε | 165 |
| AS7 | Tripoli | Arkadia | 37°30′ N | 22°22′ Ε | 662 |
| AS8 | Manasi | Lefkada | 38°41′ Ν | 20°36′ Ε | 557 |
| AS9 | Vrysoula | Ioannina | 39°40′ Ν | 20°32′ Ε | 220 |
| AS10 | Trachy, Skyros Island | Evia | 38°57′ Ν | 24°30′ Ε | 10 |
| AS11 | Tsoureki | Messinia | 37°19′ Ν | 21°57′ Ε | 467 |
| AS12 | Κefalonia (market) | Kefalonia | 38°17′ Ν | 20°31′ Ε | 500 |
| AS13 | Andania | Messinia | 37°15′ Ν | 21°59′ Ε | 85 |
| AS14 | Komotini | Rodopi | 41°05′ Ν | 25°24′ Ε | 42 |
| AS15 | Altomira | Messinia | 36°58′ Ν | 22°13′ Ε | 827 |
| AS17 | Mavriki(i) | Arkadia | 37°23′ Ν | 22°27′ Ε | 950 |
| AS19 | Lithovouni | Arkadia | 37°28′ Ν | 22°27′ Ε | 676 |
| AS21 | Stadio Tripoleos | Arkadia | 37°27′ N | 22°26′ Ε | 675 |
| AS23 | Kakaletri | Messinia | 37°24′ Ν | 22°55′ Ε | 607 |
| AS24 | Dermatianika | Lakonia | 36°54′ Ν | 23°02′ Ε | 35 |
| AS25 | Mesa Vouni, Andros Island | Cyclades | 37°47′ Ν | 24°55′ Ε | 585 |
| AS28 | Kitries | Messinia | 36°55′ Ν | 22°08′ Ε | 3 |
| AS30 | Agios Theodoros | Kefalonia | 38°11′ Ν | 20°28′ Ε | 2 |
| AS32 | Megali Mantineia | Messinia | 36°57′ Ν | 22°09′ Ε | 207 |
| AS33 | Kato Doloi | Messinia | 36°93′ Ν | 22°17′ Ε | 315 |
| AS34 | Milos Island | Cyclades | 36°40′ Ν | 24°23′ Ε | 153 |
| AS35 | Manthurea | Arkadia | 37°24′ Ν | 22°23′ Ε | 750 |
| AS36 | Mavriki(ii) | Arkadia | 37°23′ Ν | 22°27′ Ε | 950 |
| SSR | Repeat Motif | Forward Primer | Labelling | Reverse Primer | Tm (°C) |
|---|---|---|---|---|---|
| Asa08 1 | (GT)8 | TGATTGAAACGAATCCCACA | 5′ FAM | GGGGGTTACCTGAACCTGTTA | 54 |
| Asa10 1 | (AC)7 | TTGTTGTTCTGCCATTTT | 5′ HEX | GATCTAAGCCGAGAGAAA | 48 |
| Asa24 1 | (GT)4(GT)3(GT)5 | TTGTTGTGCCGAGTTCCATA | 5′ FAM | CAGCAATTTACCAAAGCCAAG | 57 |
| GB-AS-076 2 | (GA)7 | CGGCGGGTTTAGTGATTT | 5′ HEX | TTCGTTGGGTTCGATTTG | 52 |
| GB-AS-102 2 | (AAAT)3 | AATCATCTTCGGGCCACT | 5′ TAM | CCTAGAACGAGTGTGAAGGG | 52 |
| Primer ID | Primer Sequence (5′–3′) | Tm (°C) | No of Bands Scored | Range (bp) | PIC |
|---|---|---|---|---|---|
| UBC-811 | (GA)8C | 51 °C | 19 | 590–1875 | 0.500 |
| UBC-812 | (GA)8A | 51 °C | 16 | 400–2700 | 0.371 |
| UBC-818 | (CA)8G | 51 °C | 18 | 490–1800 | 0.499 |
| UBC-826 | (AC)8C | 51 °C | 14 | 650–2700 | 0.456 |
| UBC-846 | (CA)8RT | 51 °C | 10 | 680–1300 | 0.500 |
| UBC-880 | (GGAGA)3 | 51 °C | 7 | 550–1390 | 0.499 |
| Locus | Na | Ne | Nm | I | PIC | Obs. Ho | Exp. Ho | Obs. He | Exp. He | Fst |
|---|---|---|---|---|---|---|---|---|---|---|
| Asa08 | 9 | 3.358 | 0.317 | 1.548 | 0.669 | 0.214 | 0.295 | 0.786 | 0.705 | 0.440 |
| Asa10 | 5 | 3.609 | 0.079 | 1.436 | 0.683 | 0.630 | 0.274 | 0.370 | 0.726 | 0.759 |
| Asa24 | 4 | 1.873 | 0.338 | 0.807 | 0.398 | 0.464 | 0.532 | 0.536 | 0.468 | 0.425 |
| GB-AS-076 | 4 | 2.302 | 0.936 | 0.941 | 0.470 | 0.107 | 0.432 | 0.893 | 0.568 | 0.211 |
| GB-AS-102 | 4 | 1.157 | 0.038 | 0.332 | 0.133 | 0.964 | 0.864 | 0.036 | 0.136 | 0.868 |
| Mean | 5.2 | 2.460 | 0.251 | 1.013 | 0.471 | 0.476 | 0.480 | 0.524 | 0.520 | 0.500 |
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among Pops | 27 | 183.036 | 6.779 | 0.678 | 34% |
| Among Indiv | 112 | 0.000 | 0.000 | 0.000 | 0% |
| Within Indiv | 140 | 182.500 | 1.304 | 1.304 | 66% |
| Total | 279 | 365.536 | 1.981 | 100% |
| Genotype | Trait | Fisher’s Exact Test (p) | Logistic Regression (p) |
|---|---|---|---|
| SSR | |||
| Asa08-Locus 7 | Allylprop1enyldisulfide | 0.037 | 0.037 |
| Asa10-Locus 4 | Pyruvate | 0.036 | 0.05 |
| Asa10-Locus 2 | Bulb position of root disc (BPRD) | <0.001 | 0.024 |
| Asa10-Locus 5 | Shape of bulb base (SBB) | 0.006 | 0.045 |
| Asa10-Locus 2 | 0.002 | 0.024 | |
| ISSR | |||
| 811-Locus 8 | 4H-1,2,3-trithiine | 0.041 | 0.007 |
| 826-Locus 11 | Anthocyanin stripes on clove scales (ASCS) | 0.003 | 0.05 |
| 812-Locus 14 | Bulb compactness of cloves (BCC) | 0.004 | 0.006 |
| 818-Locus 9 | Bulb position of root disc (BPRD) | 0.050 | 0.05 |
| 846-Locus 6 | Bulb structure type (BST) | 0.010 | 0.008 |
| 880-Locus 2 | Carbohydrates | 0.008 | 0.032 |
| 811-Locus 3 | Diallyl trisulfide (DATS) | 0.025 | 0.02 |
| 811-Locus 2 | Number of cloves per compound bulb (NC/B) | 0.010 | 0.01 |
| 818-Locus 18 | 0.034 | 0.035 | |
| 811-Locus 6 | Total Identified Organosulfur Compounds (OS) | 0.015 | 0.039 |
| 826-Locus 6 | 0.009 | 0.039 | |
| 826-Locus 9 | Shape of bulb base (SBB) | 0.037 | 0.016 |
| 846-Locus 6 | 0.007 | 0.03 | |
| 826-Locus 9 | Shape of Mature Bulb (SMB) | 0.018 | 0.07 |
| 846-Locus 5 | 0.006 | 0.019 | |
| 812-Locus 5 | Total soluble solids (TSS) content (oBrix) | 0.008 | 0.027 |
| 818-Locus 11 | Anthocyanin stripes on dry external scales of bulb (ASBDES) | 0.042 | 0.036 |
| 846-Locus 6 | 0.050 | 0.012 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papaioannou, C.; Fassou, G.; Petropoulos, S.A.; Lamari, F.N.; Bebeli, P.J.; Papasotiropoulos, V. Evaluation of the Genetic Diversity of Greek Garlic (Allium sativum L.) Accessions Using DNA Markers and Association with Phenotypic and Chemical Variation. Agriculture 2023, 13, 1408. https://doi.org/10.3390/agriculture13071408
Papaioannou C, Fassou G, Petropoulos SA, Lamari FN, Bebeli PJ, Papasotiropoulos V. Evaluation of the Genetic Diversity of Greek Garlic (Allium sativum L.) Accessions Using DNA Markers and Association with Phenotypic and Chemical Variation. Agriculture. 2023; 13(7):1408. https://doi.org/10.3390/agriculture13071408
Chicago/Turabian StylePapaioannou, Charikleia, Georgia Fassou, Spyridon A. Petropoulos, Fotini N. Lamari, Penelope J. Bebeli, and Vasileios Papasotiropoulos. 2023. "Evaluation of the Genetic Diversity of Greek Garlic (Allium sativum L.) Accessions Using DNA Markers and Association with Phenotypic and Chemical Variation" Agriculture 13, no. 7: 1408. https://doi.org/10.3390/agriculture13071408
APA StylePapaioannou, C., Fassou, G., Petropoulos, S. A., Lamari, F. N., Bebeli, P. J., & Papasotiropoulos, V. (2023). Evaluation of the Genetic Diversity of Greek Garlic (Allium sativum L.) Accessions Using DNA Markers and Association with Phenotypic and Chemical Variation. Agriculture, 13(7), 1408. https://doi.org/10.3390/agriculture13071408









