Apulian Autochthonous Olive Germplasm: A Promising Resource to Restore Cultivation in Xylella fastidiosa-Infected Areas
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Infection with the Bacterium X.f. subsp. Pauca
2.2. Olive Genotyping
2.3. Analysis of the Data
2.4. Evaluation of the X.f. Symptoms
2.5. Quantification of X.f. in Plant
3. Results
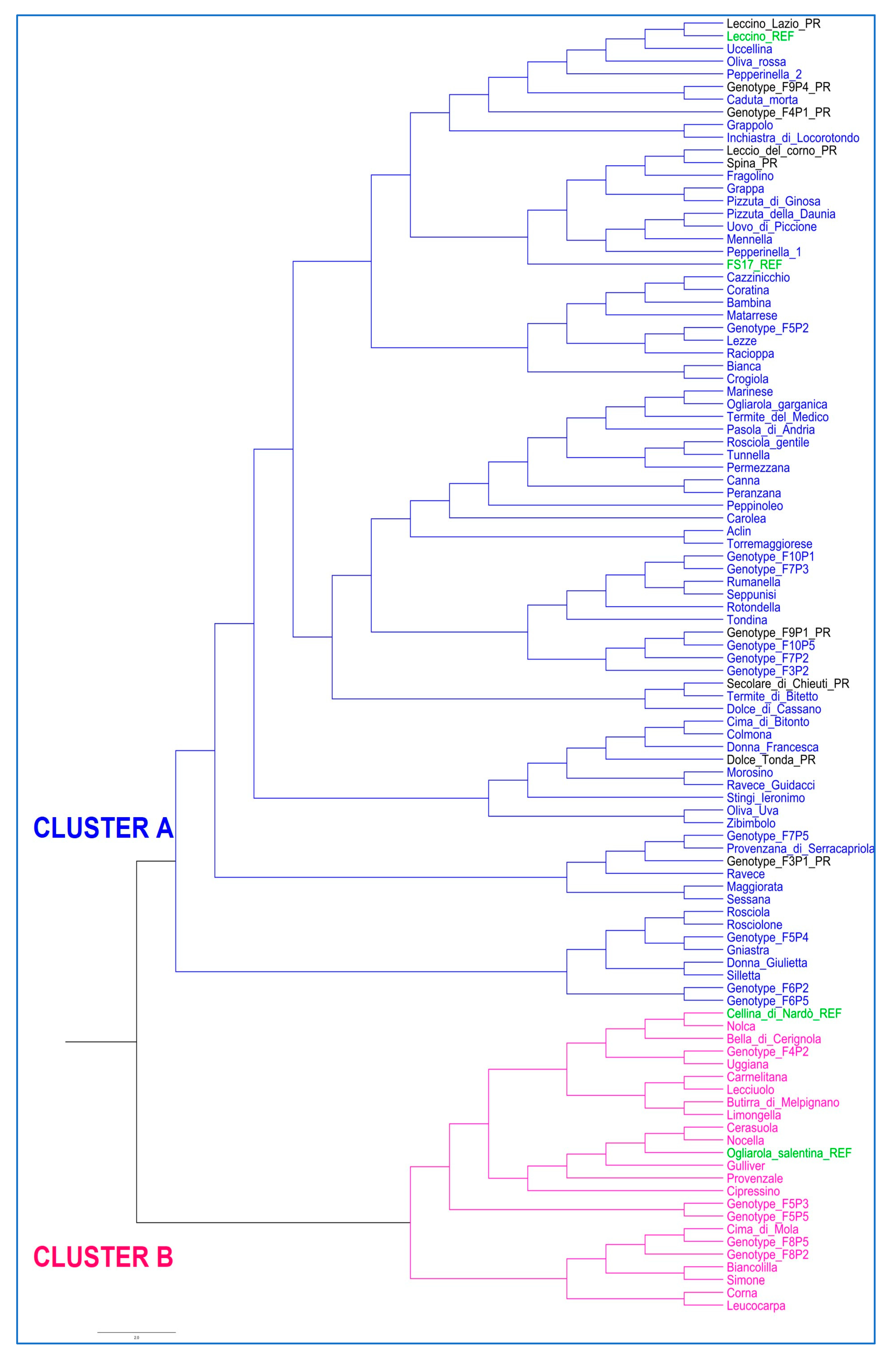
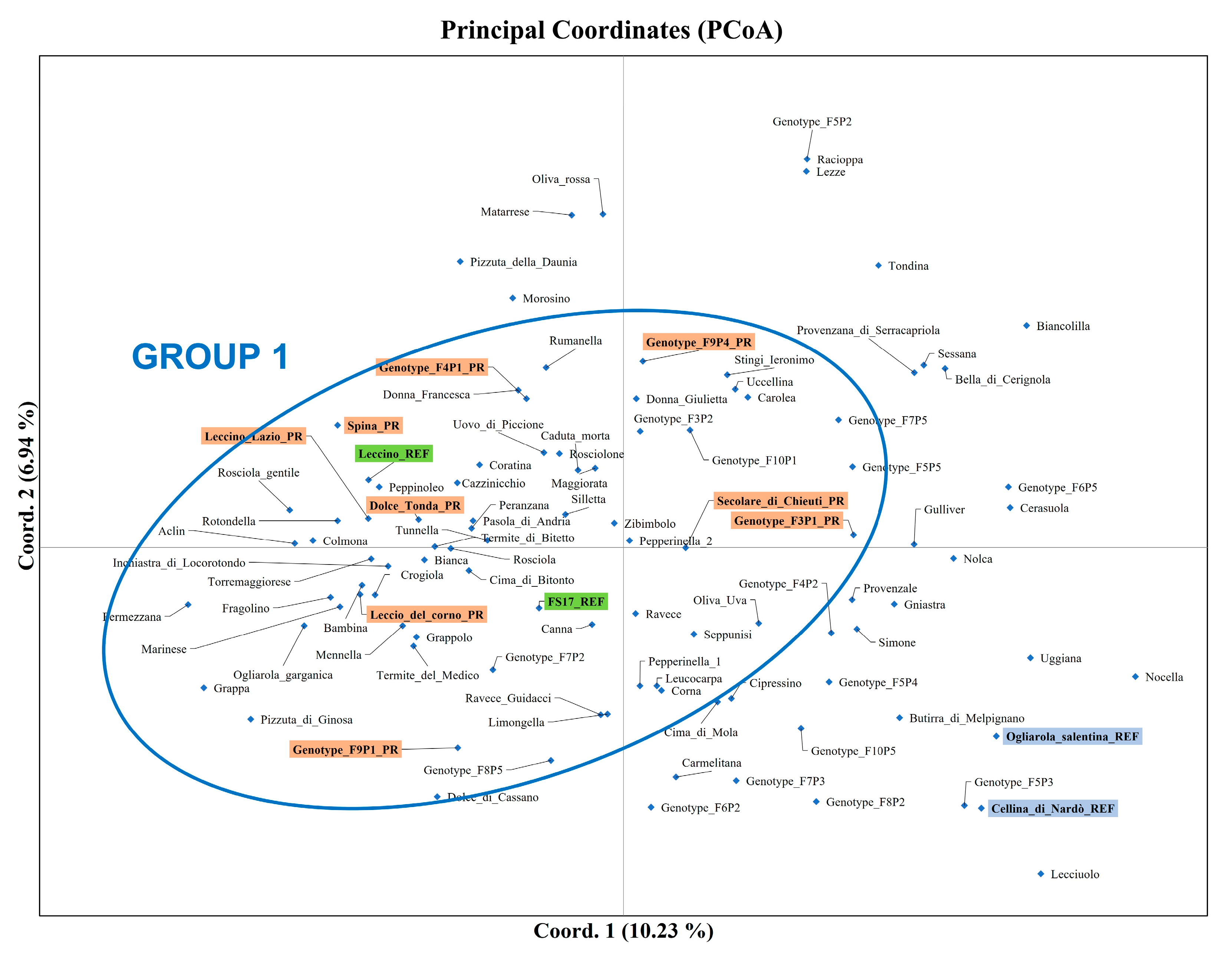
3.1. Olive Genotyping
3.2. In Planta Assessment of Susceptibility to X.f. and the Quantification of the Bacterium
4. Discussion
4.1. Genetic Diversity Assessment
4.2. Evaluation of the Response to X.f. Infection
4.3. Comparison between Genetic Data and the Response to X.f.
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Morelli, M.; García-Madero, J.M.; Jos, Á.; Saldarelli, P.; Dongiovanni, C.; Kovacova, M.; Saponari, M.; Baños, A.; Hackl, E.; Webb, S.; et al. Xylella fastidiosa in Olive: A Review of Control Attempts and Current Management. Microorganisms 2021, 9, 1771. [Google Scholar] [CrossRef]
- Loconsole, G.; Potere, O.; Boscia, D.; Altamura, G.; Djelouah, K.; Elbeaino, T.; Frasheri, D.; Lorusso, D.; Palmisano, F.; Pollastro, S.; et al. Detection of Xylella fastidiosa in olive trees by molecular and serological methods. Eur. J. Plant Pathol. 2014, 96, 7–14. [Google Scholar]
- Purcell, A.H.; Donald, L.H. Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 1996, 34, 131–151. [Google Scholar] [CrossRef]
- Saponari, M.; Boscia, D.; Altamura, G.; Loconsole, G.; Zicca, S.; Datome, G.; Morelli, M.; Palmisano, F.; Saponari, A.; Tavano, D. Isolation and pathogenicity of Xylella fastidiosa associated to the olive quick decline syndrome in southern Italy. Sci. Rep. 2017, 7, 17723. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); Gibin, D.; Pasinato, L.; Delbianco, A. Scientific Report on the update of the Xylella spp. host plant database—Systematic literature search up to 31 December 2022. EFSA J. 2023, 21, 8061. [Google Scholar] [CrossRef]
- Cornara, D.; Sicard, A.; Zeilinger, A.R.; Porcelli, F.; Purcell, A.H.; Almeida, R.P.P. Transmission of Xylella fastidiosa to grapevine by the meadow spittlebug. Phytopathology 2016, 106, 1285–1290. [Google Scholar] [CrossRef]
- Serio, F.D.; Bodino, N.; Cavalieri, V.; Demichelis, S.; Carolo, M.D.; Dongiovanni, C.; Fumarola, G.; Gilioli, G.; Guerrieri, E.; Picciotti, U.; et al. Collection of data and information on biology and control of vectors of Xylella fastidiosa. EFSA Support. Publ. 2019, 16, 1628E. [Google Scholar] [CrossRef]
- Picciotti, U.; Lahbib, N.; Sefa, V.; Porcelli, F.; Garganese, F. Aphrophoridae role in Xylella fastidiosa subsp. pauca ST53 invasion in southern Italy. Pathogens 2021, 10, 1035. [Google Scholar] [CrossRef]
- Lahbib, N.; Picciotti, U.; Boukhris-Bouhachem, S.; Garganese, F.; Porcelli, F. Morphs of Philaenus species; candidate Xylella fastidiosa vectors. Bull. Insectol. 2022, 75, 197–209. [Google Scholar]
- Fierro, A.; Liccardo, A.; Porcelli, F. A lattice model to manage the vector and the infection of the Xylella fastidiosa on olive trees. Sci. Rep. 2019, 9, 8723. [Google Scholar] [CrossRef]
- Cardone, G.; Digiaro, M.; Djelouah, K.; El Bilali, H.; Frem, M.; Fucilli, V.; Ladisa, G.; Rota, C.; Yaseen, T. Potential socio-economic impact of Xylella fastidiosa in the Near East and North Africa (NENA): Risk of introduction and spread; risk perception and socio-economic effects. New Medit. 2021, 20, 27–51. [Google Scholar]
- Bozzo, F.; Frem, M.; Fucilli, V.; Cardone, G.; Garofoli, P.F.; Geronimo, S.; Petrontino, A. Landscape and vegetation patterns zoning is a methodological tool for management costs implications due to Xylella fastidiosa invasion. Land 2022, 11, 1105. [Google Scholar] [CrossRef]
- Liccardo, A.; Fierro, A.; Garganese, F.; Picciotti, U.; Porcelli, F. A biological control model to manage the vector and the infection of Xylella fastidiosa on olive trees. PLoS ONE 2020, 15, e0232363. [Google Scholar] [CrossRef] [PubMed]
- Baù, A.; Delbianco, A.; Stancanelli, G.; Tramontini, S. Susceptibility of Olea europaea L. varieties to Xylella fastidiosa subsp. pauca ST53: Systematic literature search up to 24 March 2017. EFSA J. 2017, 15, e04772. [Google Scholar] [CrossRef]
- Luvisi, A.; Aprile, A.; Sabella, E.; Vergine, M.; Nicolì, F.; Nutricati, E.; Miceli, A.; Negro, C.; De Bellis, L. Xylella fastidiosa subsp. pauca (CoDiRO strain) infection in four olive (Olea europaea L.) cultivars: Profile of phenolic compounds in leaves and progression of leaf scorch symptoms. Phytopathol. Mediterr. 2017, 56, 259–273. [Google Scholar]
- Walker, N.C.; White, S.M.; Fletcher, D.M.; Ruiz, S.A.; Rankin, K.E.; De Stradis, A.; Saponari, M.; Williams, K.A.; Petroselli, C.; Roose, T. The impact of xylem geometry on olive cultivar resistance to Xylella fastidiosa: An image-based study. Plant Pathol. 2023, 72, 521–535. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Morelli, M.; Saponari, M.; Loconsole, G.; Chiumenti, M.; Boscia, D.; Savino, V.N.; Martelli, G.P.; Saldarelli, P. Transcriptome profiling of two olive cultivars in response to infection by the CoDiRO strain of Xylella fastidiosa subsp. pauca. BMC Genom. 2016, 17, 475. [Google Scholar] [CrossRef]
- Sabella, E.; Luvisi, A.; Aprile, A.; Negro, C.; Vergine, M.; Nicolì, F.; Miceli, A.; De Bellis, L. Xylella fastidiosa induces differential expression of lignification related-genes and lignin accumulation in tolerant olive trees cv. Leccino. J. Plant Physiol. 2018, 220, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Vergine, M.; Nicolì, F.; Sabella, E.; Aprile, A.; Negro, C.; Fanelli, V.; Savoia, M.A.; Montilon, V.; Susca, L.; et al. Screening of Olive Biodiversity Defines Genotypes Potentially Resistant to Xylella fastidiosa. Front. Plant Sci. 2021, 12, 723879. [Google Scholar] [CrossRef] [PubMed]
- Di Rienzo, V.; Miazzi, M.M.; Fanelli, V.; Sabetta, W.; Montemurro, C. The preservation and characterization of Apulian olive germplasm biodiversity. Acta Hortic. 2016, 1199, 1–6. [Google Scholar] [CrossRef]
- Sion, S.; Taranto, F.; Montemurro, C.; Mangini, G.; Camposeo, S.; Falco, V.; Gallo, A.; Mita, G.; Saddoud Debbabi, O.; Ben Amar, F.; et al. Genetic characterization of Apulian olive germplasm as potential source in new breeding programs. Plants 2019, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Miazzi, M.M.; di Rienzo, V.; Mascio, I.; Montemurro, C.; Sion, S.; Sabetta, W.; Vivaldi, G.A.; Camposeo, S.; Caponio, F.; Squeo, G.; et al. Re. Ger. OP: An Integrated Project for the Recovery of Ancient and Rare Olive Germplasm. Front. Plant Sci. 2020, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Muzzalupo, I.; Stefanizzi, F.; Perri, E. Evaluation of olives cultivated in southern Italy by simple sequence repeat markers. HortScience 2009, 44, 582–588. [Google Scholar] [CrossRef]
- Boucheffa, S.; Tamendjari, A.; Sanchez-Gimeno, A.C.; Rovellini, P.; Venturini, S.; di Rienzo, V.; Miazzi, M.M.; Montemurro, C. Diversity assessment of Algerian wild and cultivated olives (Olea europaea L.) by molecular; morphological; and chemical traits. Eur. J. Lipid Sci. Technol. 2019, 121, 1800302. [Google Scholar] [CrossRef]
- Islam, A.F.; Sanders, D.; Mishra, A.K.; Joshi, V. Genetic Diversity and Population Structure Analysis of the USDA Olive Germplasm Using Genotyping-By-Sequencing (GBS). Genes 2021, 12, 2007. [Google Scholar] [CrossRef]
- Taranto, F.; D’Agostino, N.; Pavan, S.; Fanelli, V.; Di Rienzo, V.; Sabetta, W.; Miazzi, M.M.; Zelasco, S.; Perri, E.; Montemurro, C. Single nucleotide polymorphism (SNP) diversity in an olive germplasm collection. Acta Hort. 2018, 1199, 27–31. [Google Scholar] [CrossRef]
- Bazakos, C.; Alexiou, K.G.; Ramos-Onsins, S.; Koubouris, G.; Tourvas, N.; Xanthopoulou, A.; Mellidou, I.; Moysiadis, T.; Vourlaki, I.T.; Metzidakis, I.; et al. Whole genome scanning of a Mediterranean basin hotspot collection provides new insights into olive tree biodiversity and biology. Plant J. 2023, 2, 1–17. [Google Scholar] [CrossRef]
- Pasqualone, A.; Di Rienzo, V.; Nasti, R.; Blanco, A.; Gomes, T.; Montemurro, C. Traceability of Italian protected designation of origin (PDO) table olives by means of microsatellite molecular markers. J. Agric. Food Chem. 2013, 61, 3068–3073. [Google Scholar] [CrossRef]
- Sion, S.; Savoia, M.A.; Gadaleta, S.; Piarulli, L.; Mascio, I.; Fanelli, V.; Montemurro, C.; Miazzi, M.M. How to choose a good marker to analyze the olive germplasm (Olea europaea L.) and derived products. Genes 2021, 12, 1474. [Google Scholar] [CrossRef]
- Saddoud Debbabi, O.; Miazzi, M.M.; Elloumi, O.; Fendri, M.; Ben Amar, F.; Savoia, M.; Sion, S.; Souabni, H.; Mnasri, S.R.; Ben Abdelaali, S.; et al. Recovery, Assessment, and Molecular Characterization of Minor Olive Genotypes in Tunisia. Plants 2020, 9, 382. [Google Scholar] [CrossRef]
- Marchese, A.; Bonanno, F.; Marra, F.P.; Trippa, D.A.; Zelasco, S.; Rizzo, S.; Giovino, A.; Imperiale, V.; Ioppolo, A.; Sala, G.; et al. Recovery and genotyping ancient Sicilian monumental olive trees. Front. Conserv. Sci. 2023, 4, 1206832. [Google Scholar] [CrossRef]
- Spadoni, A.; Sion, S.; Gadaleta, S.; Savoia, M.; Piarulli, L.; Fanelli, V.; di Rienzo, V.; Taranto, F.; Miazzi, M.M.; Montemurro, C.; et al. A simple and rapid method for genomic DNA extraction and microsatellite analysis in tree plants. J. Agric. Sci. Technol. 2019, 21, 1215–1226. [Google Scholar]
- Sefc, K.M.; Lopes, M.S.; Lefort, F.; Botta, R.; Roubelakis-Angelakis, K.A.; Ibanez, J.; Pejić, I.; Wagner, H.W.; Glössl, J.; Steinkellner, H. Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theor. Appl. Genet. 2000, 100, 498–505. [Google Scholar] [CrossRef]
- Carriero, F.; Fontanazza, G.; Cellini, F.; Giorio, G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor. Appl. Genet. 2002, 104, 301–307. [Google Scholar] [CrossRef] [PubMed]
- De La Rosa, R.; James, C.M.; Tobutt, K.R. Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Mol. Ecol. Notes 2002, 2, 265–267. [Google Scholar] [CrossRef]
- Lynch, M.; Ritland, K. Estimation of pairwise relatedness with molecular markers. Genetics 1999, 152, 1753–1766. [Google Scholar] [CrossRef]
- Peakall, R. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Gower, J.C. Wiley StatsRef: Statistics Reference Online; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; Volume 70. [Google Scholar]
- Nutter, F.W., Jr.; Teng, P.S.; Shokes, F.M. Disease assessment terms and concepts. Plant Dis. 1991, 75, 1187–1188. [Google Scholar]
- Bock, C.H.; Chiang, K.S.; Del Ponte, E.M. Plant Disease Severity Estimated Visually: A Century of Research, Best Practices, and Opportunities for Improving Methods and Practices to Maximize Accuracy. Trop. Plant Pathol. 2022, 47, 25–42. [Google Scholar] [CrossRef]
- James, W.C. Assessment of Plant Diseases and Losses. Annu. Rev. Phytopathol. 1974, 12, 27–48. [Google Scholar] [CrossRef]
- Harper, S.J.; Ward, L.I.; Clover, G.R.G. Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosafor quarantine and field applications. Phytopathology 2010, 100, 1282–1288. [Google Scholar] [CrossRef]
- Gaut, B.S.; Díez, C.M.; Morrell, P.L. Genomics and the contrasting dynamics of annual and perennial domestication. Trends Genet. 2015, 31, 709–719. [Google Scholar] [CrossRef]
- Besnard, G.; Terral, J.F.; Cornille, A. On the origins and domestication of the olive: A review and perspectives. Ann. Bot. 2018, 121, 385–403. [Google Scholar] [CrossRef]
- Barazani, O.; Dag, A.; Kerem, Z.; Lavee, S.; Kadereit, J.W. Local old olive landrace varieties in Israel—Valuable plant genetic resources in olive cultivation. Isr. J. Plant Sci. 2008, 56, 265–271. [Google Scholar] [CrossRef]
- Ben Ayed, R.; Ercişli, S.; Hanana, M.; Rebai, A.; Moreau, F. Assessment of population structure, genetic diversity and relationship of Mediterranean olive accessions using SSR markers and computational tools. Biotechnol. Lett. 2022, 44, 113–127. [Google Scholar] [CrossRef]
- Francis, M.; Lin, H.; Cabrera-La Rosa, J.; Doddapaneni, H.; Civerolo, E.L. Genome-based PCR primers for specific and sensitive detection and quantification of Xylella fastidiosa. Eur. J. Plant Pathol. 2006, 115, 203–213. [Google Scholar] [CrossRef]
- White, S.M.; Navas-Cortés, J.A.; Bullock, J.M.; Boscia, D.; Chapman, D.S. Estimating the epidemiology of emerging Xylella fastidiosa outbreaks in olives. Plant Pathol. 2020, 69, 1403–1413. [Google Scholar] [CrossRef]
- Frisullo, S.; Camele, I.; Agosteo, G.E.; Boscia, D.; Martelli, G.P. Brief historical account of olive leaf scorch (brusca) in the Salento peninsula of Italy and state-of-the- art of the olive quick decline syndrome. J. Plant. Pathol. 2014, 96, 441–449. [Google Scholar]
- Martelli, G.P. The current status of the quick decline syndrome of olive in Southern Italy. Phytoparasitica 2016, 44, 1–10. [Google Scholar] [CrossRef]
- Newman, K.L.; Almeida, R.P.; Purcell, A.H.; Lindow, S.E. Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl. Environ. Microbiol. 2003, 69, 7319–7327. [Google Scholar] [CrossRef]
- Alves, E.; Marucci, C.R.; Lopes, J.R.S.; Leite, B. Leaf symptoms on plum; coffee and citrus and the relationship with the extent of xylem vessels colonized by Xylella fastidiosa. J. Phytopathol. 2004, 152, 291–297. [Google Scholar] [CrossRef]
- Krell, R.K.; Perring, T.M.; Farrar, C.A.; Park, Y.L.; Gispert, C. Intraplant sampling of grapevines for Pierce’s disease diagnosis. Plant Dis. 2006, 90, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Gambetta, G.A.; Fei, J.; Rost, T.L.; Matthews, M.A. Leaf scorch symptoms are not correlated with bacterial populations during Pierce’s disease. J. Exp. Bot. 2007, 58, 4037–4046. [Google Scholar] [CrossRef]
- Hopkins, D.L. Effects of plant-growth regulators on development of Pierces disease symptoms in grapevine. Plant Dis. 1985, 69, 944–946. [Google Scholar]
- Perez-Donoso, A.G.; Greve, L.C.; Walton, J.H.; Shackel, K.A.; Labavitch, J.M. Xylella fastidiosa infection and ethylene exposure result in xylem and water movement disruption in grapevine shoots. Plant Physiol. 2007, 143, 1024–1036. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Baptista, P.; Morelli, M.; Cameirão, C.; Lino Neto, T.; Costa, D.; D’Attoma, G.; Kubaa, R.A.; Altamura, G.; Saponari, M.; et al. Differences in the endophytic microbiome of olive cultivars infected by Xylella fastidiosa across seasons. Pathogens 2020, 9, 723. [Google Scholar] [CrossRef]
- Surano, A.; Abou Kubaa, R.; Nigro, F.; Altamura, G.; Losciale, P.; Saponari, M.; Saldarelli, P. Susceptible and resistant olive cultivars show differential physiological response to Xylella fastidiosa infections. Front. Plant Sci. 2022, 13, 968934. [Google Scholar] [CrossRef]
- Chatelet, D.S.; Matthews, M.A.; Rost, T.L. Xylem structure and connectivity in grapevine (Vitis vinifera) shoots provides a passive mechanism for the spread of bacteria in grape plants. Ann. Bot. 2006, 98, 483–494. [Google Scholar] [CrossRef]
- Fontanazza, G.; Baldoni, L.; Corona, C. Osservazioni sull’impiego di portainnesti clonali negli olivi “Ascolana tenera” e “Giarraffa”. Riv. Frutticol. 1992, 11, 65–69. [Google Scholar]
- Ranalli, A.; Lucera, L.; Contento, S.; Fontanazza, G.; Patumi, M. Assessment of physico-chemical, sensory and nutritional parameters in virgin olive oil from the new genotype Favolosa (FS17). Acta Hortic. 2008, 791, 697–704. [Google Scholar] [CrossRef]


| Genotypes | Origin | Provinces | Purpose | |
|---|---|---|---|---|
| Table | Oil | |||
| Ac’lin | Castellana Grotte | Bari | √ | √ |
| Bambina | Gravina In Puglia | Bari | √ | √ |
| Bella Di Cerignola | Ascoli Satriano | Foggia | √ | |
| Bianca | Ceglie Messapica | Brindisi | √ | |
| Biancolilla | Valenzano (screen house) | Bari | √ | |
| Butirra Di Melpignano | Melpignano | Lecce | √ | √ |
| Caduta Morta | Terlizzi | Bari | √ | √ |
| Canna | Polignano A Mare | Bari | √ | √ |
| Carmelitana | San Severo | Foggia | √ | √ |
| Carolea | Valenzano (screen house) | Bari | √ | √ |
| Cazzinicchio | Bari | Bari | √ | |
| Cellina Di Nardo’ | Carpignano Salentino | Lecce | √ | |
| Cerasuola | Valenzano (screen house) | Bari | √ | √ |
| Cima Di Bitonto | Bitonto | Bari | √ | |
| Cima Di Mola | Monopoli | Bari | √ | |
| Cipressino | Castellana Grotte | Bari | √ | √ |
| Colmona | Ginosa | Taranto | √ | √ |
| Coratina | Andria | Bari | √ | |
| Corna | Ceglie Messapica | Brindisi | √ | √ |
| Crogiola | Ceglie Messapica | Brindisi | √ | √ |
| Dolce Di Cassano | Cassano Delle Murge | Bari | √ | √ |
| Dolce Tonda | Sannicandro | Bari | √ | √ |
| Donna Francesca | Modugno | Bari | √ | |
| Donna Giulietta | Modugno | Bari | √ | |
| Fragolino | Chieuti | Foggia | √ | √ |
| Genotype_F10P1 | Valenzano (screen house) | Bari | √ | |
| Genotype_F10P5 | Valenzano (screen house) | Bari | √ | |
| Genotype_F3P1 | Valenzano (screen house) | Bari | √ | |
| Genotype_F3P2 | Valenzano (screen house) | Bari | √ | |
| Genotype_F4P1 | Valenzano (screen house) | Bari | √ | |
| Genotype_F4P2 | Valenzano (screen house) | Bari | √ | |
| Genotype_F5P2 | Valenzano (screen house) | Bari | √ | |
| Genotype_F5P3 | Valenzano (screen house) | Bari | √ | |
| Genotype_F5P4 | Valenzano (screen house) | Bari | √ | |
| Genotype_F5P5 | Valenzano (screen house) | Bari | √ | |
| Genotype_F6P2 | Valenzano (screen house) | Bari | √ | |
| Genotype_F6P5 | Valenzano (screen house) | Bari | √ | |
| Genotype_F7P2 | Valenzano (screen house) | Bari | √ | |
| Genotype_F7P3 | Valenzano (screen house) | Bari | √ | |
| Genotype_F7P5 | Valenzano (screen house) | Bari | √ | |
| Genotype_F8P2 | Valenzano (screen house) | Bari | √ | |
| Genotype_F8P5 | Valenzano (screen house) | Bari | √ | |
| Genotype_F9P1 | Valenzano (screen house) | Bari | √ | |
| Genotype_F9P4 | Valenzano (screen house) | Bari | √ | |
| Gniastra | Valenzano (screen house) | Bari | √ | |
| Grappa | Ostuni | Brindisi | √ | √ |
| Grappolo | Fasano | Brindisi | √ | |
| Gulliver | Chieuti | Foggia | √ | |
| Inchiastra Di Locorotondo | Locorotondo | Bari | √ | |
| Leccino Lazio | Valenzano (screen house) | Bari | √ | |
| Leccino_Ref | Valenzano (screen house) | Bari | √ | |
| Leccio Del Corno | Valenzano (screen house) | Bari | √ | |
| Lecciuolo | Valenzano (screen house) | Bari | √ | |
| Leucocarpa | Ascoli Satriano | Foggia | √ | |
| Lezze | Ceglie Messapica | Brindisi | √ | |
| Limongella | Polignano A Mare | Bari | √ | √ |
| Maggiorata | Bitetto | Bari | √ | √ |
| Marinese | Cerignola | Foggia | √ | |
| Matarrese | Turi | Bari | √ | |
| Mennella | Ceglie Messapica | Brindisi | √ | |
| Morosino | Torremaggiore | Foggia | √ | √ |
| Nocella | Santa Cesarea Terme | Lecce | √ | √ |
| Nolca | Bitonto | Bari | √ | √ |
| Ogliarola Garganica | Biccari | Foggia | √ | |
| Oliva Rossa | Locorotondo | Bari | √ | |
| Oliva Uva | Turi | Bari | √ | |
| Pasola Di Andria | Andria | Bari | √ | √ |
| Pepperinella 1 | Chieuti | Foggia | √ | √ |
| Pepperinella 2 | Chieuti | Foggia | √ | √ |
| Peppino Leo | Cassano Delle Murge | Bari | √ | |
| Peranzana | San Severo | Foggia | √ | √ |
| Permezzana | San Giovanni Rotondo | Foggia | √ | √ |
| Pizzuta Della Daunia | Volturino | Foggia | √ | |
| Pizzuta Di Ginosa | Ginosa | Taranto | √ | |
| Provenzale | Chieuti | Foggia | √ | √ |
| Provenzale Di Serracapriola | Serracapriola | Foggia | √ | √ |
| Racioppa | Adelfia | Bari | √ | |
| Ravece | Orsara Di Puglia | Foggia | √ | |
| Ravece Guidacci | Orsara Di Puglia | Foggia | √ | |
| Rosciola | Chieuti | Foggia | √ | |
| Rosciola Gentile | Serracapriola | Foggia | √ | |
| Rosciolone | Serracapriola | Foggia | √ | √ |
| Rotondella | Cerignola | Foggia | √ | √ |
| Rumanella | San Marco La Catola | Foggia | √ | √ |
| Secolare Di Chieuti | Chieuti | Foggia | √ | √ |
| Seppunisi | Ceglie Messapica | Brindisi | √ | √ |
| Sessana | Ostuni | Brindisi | √ | |
| Silletta | Rutigliano | Bari | √ | |
| Simone | Castellana Grotte | Bari | √ | √ |
| Spina | Ceglie Messapica | Brindisi | √ | √ |
| Stingi Ieronimo | Volturino | Foggia | √ | |
| Termite Del Medico | Modugno | Bari | √ | √ |
| Termite Di Bitetto | Bitetto | Bari | √ | √ |
| Tondina | San Severo | Foggia | √ | √ |
| Torremaggiorese | Torremaggiore | Foggia | √ | |
| Tunnella | Chieuti | Foggia | √ | |
| Uccellina | San Paolo Di Civitate | Foggia | √ | √ |
| Uggiana | Carpignano Salentino | Lecce | √ | |
| Uovo Di Piccione | Massafra | Taranto | √ | √ |
| Zibimbolo | San Severo | Foggia | √ | |
| Genotypes with 0.40 < LRM < 0.50 | ||
|---|---|---|
| Pepperinella1 | Ravece Guidacci | 0.50 |
| Morosino | Pizzuta della Daunia | 0.50 |
| Lezze | Racioppa | 0.45 |
| Grappa | Pizzuta di Ginosa | 0.42 |
| Rosciola | Rotondella | 0.41 |
| F1 Genotype | First Candidate | Pair Loci Mismatching | Second Candidate | Pair Loci Mismatching |
|---|---|---|---|---|
| F10P1 | Dolce di Sannicandro | 1 | - | - |
| F4P1 | Leccino REF | 0 | - | - |
| F5P2 | Lezze | 0 | Racioppa | 0 |
| F6P5 | Framichele | 1 | - | - |
| F8P5 | Sivigliana | 0 | - | - |
| F9P1 | Caduta morta | 1 | - | - |
| Genotypes | Spring 2021 | Spring 2023 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SYM (0–5) | Cq | SD | CFU/mL | SD | SYM (0–5) | Cq | SD | CFU/mL | SD | |
| Bella Di Cerignola | 0 | 24.17 | 4.48 | 7.22 × 105 | 3.73 × 105 | 3 | 22.32 | 0.55 | 6.72 × 105 | 2.36 × 105 |
| Bianca | 0 | 22.44 | 2.41 | 1.05 × 106 | 8.72 × 105 | 4 | 21.77 | 2.78 | 2.32 × 106 | 2.54 × 106 |
| Butirra Di Melpignano | 0 | 24.19 | 0.05 | 1.74 × 105 | 4.76 × 103 | 1 | 24.45 | 1.87 | 2.36 × 105 | 2.04 × 105 |
| Carmelitana | 0 | 25.24 | 2.44 | 2.28 × 105 | 1.74 × 105 | 2 | 22.73 | 2.7 | 4.01 × 105 | 3.47 × 105 |
| Cerasuola | 0 | NA | NA | NA | NA | 1 | 26.95 | 7.14 | 2.14 × 105 | 4.28 × 105 |
| Cima Di Mola | 0 | 21.22 | 1.44 | 1.72 × 106 | 1.05 × 106 | 1 | 22.63 | 1.40 | 9.98 × 105 | 7.40 × 105 |
| Cipressino | 0 | 26.11 | 5.33 | 3.18 × 105 | 3.14 × 105 | 2 | 26.55 | 2.78 | 7.95 × 104 | 7.64 × 104 |
| Corna | 0 | 26.90 | 1.84 | 3.82 × 104 | 2.74 × 104 | 1 | 23.33 | 2.24 | 5.24 × 105 | 3.30 × 105 |
| Genotype_F4P2 | 0 | 28.85 | 0.69 | 7.50 × 103 | 3.95 × 103 | 2 | 23.64 | 0.51 | 2.67 × 105 | 9.68 × 104 |
| Genotype_F5P3 | 0 | 22.65 | 0.37 | 5.17 × 105 | 7.18 × 104 | 2 | 22.52 | 1.91 | 6.47 × 105 | 7.54 × 105 |
| Genotype_F5P5 | 0 | 23.06 | 1.01 | 4.30 × 105 | 1.97 × 105 | 2 | 21.93 | 1.22 | 6.62 × 105 | 7.82 × 105 |
| Genotype_F8P2 | 0 | 29.08 | 4.33 | 2.48 × 104 | 3.41 × 104 | 1 | 23.43 | 1.46 | 3.00 × 105 | 3.38 × 105 |
| Genotype_F8P5 | 0 | 27.77 | 5.43 | 2.75 × 105 | 2.71 × 105 | 3 | 20.75 | 0.85 | 2.12 × 106 | 1.16 × 106 |
| Gulliver | 1 | 28.67 | 4.47 | 3.54 × 104 | 3.45 × 104 | 1 | 25.54 | 3.57 | 2.26 × 105 | 2.60 × 105 |
| Lecciuolo | 0 | NA | NA | NA | NA | 1 | 23.70 | 6.15 | 1.25 × 106 | 2.50 × 106 |
| Leucocarpa | 0 | 21.94 | 0.67 | 8.95 × 105 | 2.58 × 105 | 4 | 24.07 | 2.64 | 4.53 × 105 | 4.93 × 105 |
| Limongella | 0 | 26.94 | 4.18 | 1.96 × 105 | 1.87 × 105 | 2 | 23.03 | 1.93 | 6.82 × 105 | 6.59 × 105 |
| Nocella | 0 | 23.78 | 3.22 | 5.87 × 105 | 3.24 × 105 | 1 | 25.57 | 3.51 | 1.46 × 105 | 1.87 × 105 |
| Nolca | 0 | 27.19 | 6.23 | 2.32 × 105 | 2.31 × 105 | 1 | 22.39 | 1.73 | 8.99 × 105 | 8.01 × 105 |
| Provenzale | 0 | 27.38 | 0.00 | 19075.58 | 0.00 | 1 | 23.99 | 2.61 | 3.83 × 105 | 2.77 × 105 |
| Simone | 0 | 25.08 | 6.06 | 9.25 × 105 | 1.30 × 106 | 3 | 22.08 | 0.41 | 7.77 × 105 | 2.25 × 105 |
| Cellina Di Nardo’ | 0 | 23.43 | 2.77 | 6.16 × 105 | 3.03 × 105 | 2 | 21.45 | 1.84 | 1.79 × 106 | 1.41 × 106 |
| Uggiana | 0 | 22.83 | 0.00 | 4.48 × 105 | 0.00 | 2 | 25.58 | 2.39 | 8.92 × 104 | 9.80 × 104 |
| Ac’lin | 0 | 30.30 | 2.23 | 4.21 × 103 | 3.36 × 103 | 2 | 22.95 | 1.44 | 6.12 × 105 | 6.58 × 105 |
| Bambina | 0 | 27.70 | 0.12 | 1.53 × 104 | 1.24 × 103 | 1 | 21.45 | 3.63 | 1.62 × 106 | 1.81 × 106 |
| Biancolilla | 0 | 28.17 | 0.92 | 1.32 × 104 | 5.20 × 103 | 2 | 23.06 | 1.15 | 4.73 × 105 | 3.13 × 105 |
| Caduta Morta | 0 | 30.80 | 0.00 | 1.78 × 103 | 0.00 | 1 | 23.48 | 2.94 | 5.14 × 105 | 6.95 × 105 |
| Canna | 0 | 28.89 | 0.00 | 6.71 × 103 | 0.00 | 1 | 22.67 | 2.00 | 9.22 × 105 | 1.08 × 106 |
| Carolea | 0 | 23.24 | 0.00 | 3.36 × 105 | 0.00 | 1 | 22.97 | 2.97 | 5.20 × 105 | 4.26 × 105 |
| Cazzinicchio | 0 | 28.27 | 4.47 | 9.95 × 104 | 9.63 × 104 | 3 | 23.52 | 0.28 | 2.81 × 105 | 5.72 × 104 |
| Cima Di Bitonto | 0 | 22.10 | 1.13 | 9.17 × 105 | 4.21 × 105 | 2 | 22.05 | 0.06 | 3.83 × 105 | 4.43 × 105 |
| Colmona | 0 | 31.11 | 0.00 | 1.44 × 103 | 0.00 | 1 | 25.15 | 2.00 | 1.05 × 105 | 1.14 × 105 |
| Coratina | 0 | 23.33 | 2.53 | 5.95 × 105 | 5.04 × 105 | 1 | 23.67 | 2.09 | 5.07 × 105 | 6.36 × 105 |
| Crogiola | 0 | 21.88 | 0.96 | 9.65 × 105 | 4.24 × 105 | 2 | 21.66 | 1.05 | 1.22 × 106 | 9.74 × 105 |
| Dolce Di Cassano | 0 | 20.64 | 0.00 | 2.05 × 106 | 0.00 | 1 | 25.39 | 3.70 | 5.18 × 105 | 9.40 × 105 |
| Dolce Tonda_PR | 0 | 36.30 | 0.00 | 6.44 × 104 | 0.00 | 0 | 30.72 | 0.00 | 6.28 × 102 | 1.09 × 103 |
| Donna Francesca | 0 | 30.77 | 1.02 | 2.06 × 103 | 9.59 × 102 | 1 | 26.23 | 2.80 | 6.61 × 104 | 7.39 × 104 |
| Donna Giulietta | 0 | NA | NA | NA | NA | 2 | 22.77 | 3.74 | 1.79 × 106 | 1.92 × 106 |
| Fragolino | 0 | 27.71 | 0.61 | 1.21 × 104 | 6.85 × 103 | 1 | 25.25 | 3.38 | 1.14 × 105 | 2.18 × 105 |
| Genotype_F10P1 | 0 | 23.49 | 1.87 | 4.11 × 105 | 2.98 × 105 | 2 | 22.57 | 2.39 | 1.49 × 106 | 2.33 × 106 |
| Genotype_F10P5 | 0 | 25.32 | 4.42 | 3.54 × 105 | 3.45 × 105 | 2 | 22.87 | 1.54 | 5.65 × 105 | 5.12 × 105 |
| Genotype_F3P1_PR | 0 | 29.51 | 0.00 | 4.37 × 103 | 0.00 | 2 | 30.68 | 4.56 | 4.29 × 104 | 8.44 × 104 |
| Genotype_F3P2 | 0 | 26.38 | 0.77 | 4.10 × 104 | 1.49 × 104 | 1 | 24.23 | 0.73 | 1.39 × 105 | 1.26 × 105 |
| Genotype_F4P1_PR | 0 | 23.65 | 0.00 | 2.53 × 105 | 0.00 | 0 | 28.04 | 0.84 | 6.59 × 103 | 8.70 × 103 |
| Genotype_F5P2 | 0 | 27.01 | 3.52 | 1.08 × 105 | 9.59 × 104 | 1 | 23.34 | 2.81 | 7.91 × 105 | 9.74 × 105 |
| Genotype_F5P4 | 0 | 26.64 | 5.07 | 1.94 × 105 | 2.70 × 105 | 3 | 23.89 | 1.49 | 2.88 × 105 | 1.95 × 105 |
| Genotype_F6P2 | 0 | 25.67 | 1.70 | 9.71 × 104 | 5.99 × 104 | 2 | 22.60 | 0.24 | 5.29 × 105 | 9.15 × 104 |
| Genotype_F6P5 | 0 | 26.26 | 0.00 | 4.16 × 104 | 0.00 | 2 | 25.36 | 4.10 | 2.53 × 105 | 2.03 × 105 |
| Genotype_F7P2 | 0 | 27.01 | 3.52 | 1.08 × 105 | 9.59 × 104 | 1 | 23.34 | 2.81 | 7.91 × 105 | 9.74 × 105 |
| Genotype_F7P3 | 0 | 30.47 | 0.00 | 2.24 × 103 | 0.00 | 2 | 26.56 | 4.35 | 2.36 × 105 | 4.60 × 105 |
| Genotype_F7P5 | 0 | 29.72 | 2.41 | 6.74 × 103 | 5.59 × 103 | 2 | 21.37 | 1.37 | 1.25 X 106 | 1.51 X 106 |
| Genotype_F9P1_PR | 0 | 28.80 | 0.00 | 7.12 × 103 | 0.00 | 0 | 31.67 | 0.00 | 9.75 × 102 | 0.00 |
| Genotype_F9P4_PR | 0 | 30.40 | 0.00 | 2.92 × 105 | 0.00 | 2 | 28.09 | 2.29 | 2.48 × 104 | 3.24 × 104 |
| Gniastra | 0 | 25.27 | 2.16 | 1.59 × 105 | 1.11 × 105 | 1 | 26.72 | 4.42 | 6.67 × 104 | 1.31 × 105 |
| Grappa | 0 | 25.80 | 0.87 | 2.26 × 105 | 0.00 | 1 | 22.14 | 0.07 | 3.60 × 105 | 4.17 × 105 |
| Grappolo | 0 | 23.06 | 1.45 | 4.82 × 105 | 2.96 × 105 | 2 | 21.77 | 2.24 | 1.92 × 106 | 2.08 × 106 |
| Inchiastra Di Locorotondo | 0 | 26.20 | 0.00 | 4.34 × 104 | 0.00 | 3 | 22.56 | 1.10 | 6.58 × 105 | 4.28 × 105 |
| Leccino Lazio_PR | 0 | 33.49 | 0.72 | 3.02 × 102 | 1.62 × 102 | 1 | 32.33 | 0.00 | 0.00 | 0.00 |
| Leccino_Ref | 0 | - | - | - | - | 0 | 32.93 | 0.00 | 9.67 × 103 | 1.32 × 104 |
| Leccio Del Corno_PR | 0 | 33.66 | 5.10 | 1.51 × 103 | 2.10 × 103 | 1 | 28.66 | 5.32 | 2.67 × 104 | 5.34 × 104 |
| Lezze | 0 | 21.27 | 1.65 | 1.78 × 106 | 1.19 × 106 | 1 | 20.57 | 1.27 | 2.81 × 106 | 2.11 × 106 |
| Maggiorata | 0 | 27.04 | 0.00 | 3.41 × 103 | 0.00 | 3 | 20.97 | 1.06 | 2.02 × 106 | 1.64 × 106 |
| Marinese | 0 | 25.40 | 1.73 | 1.05 × 105 | 7.23 × 104 | 2 | 23.77 | 0.41 | 1.80 × 105 | 1.34 × 105 |
| Matarrese | 0 | 24.66 | 0.00 | 1.25 × 105 | 0.00 | 2 | 21.44 | 1.44 | 1.25 × 106 | 1.63 × 106 |
| Mennella | 0 | 23.50 | 0.00 | 2.82 × 105 | 0.00 | 2 | 23.02 | 1.24 | 5.44 × 105 | 5.60 × 105 |
| Morosino | 0 | 22.55 | 0.66 | 5.81 × 105 | 1.35 x105 | 1 | 22.72 | 1.64 | 5.18 × 105 | 5.96 × 105 |
| Ogliarola Garganica | 0 | 25.27 | 0.00 | 8.23 × 104 | 0.00 | 1 | 24.04 | 0.98 | 1.08 × 105 | 1.48 × 105 |
| Oliva Rossa | 0 | 23.33 | 1.49 | 4.57 × 105 | 2.73 × 105 | 2 | 21.70 | 1.17 | 1.22 × 106 | 8.56 × 105 |
| Oliva Uva | 0 | 23.80 | 0.00 | 2.28 × 105 | 0.00 | 3 | 22.20 | 1.03 | 3.94 × 105 | 5.39 × 105 |
| Pasola Di Andria | 0 | 21.43 | 0.83 | 1.28 × 106 | 4.94 × 105 | 2 | 21.48 | 1.05 | 1.40 × 106 | 1.05 × 106 |
| Pepperinella 1 | 0 | 27.30 | 8.02 | 5.17 × 105 | 5.16 × 105 | 2 | 23.49 | 2.84 | 8.34 × 105 | 1.21 × 106 |
| Pepperinella 2 | 0 | 26.93 | 4.55 | 1.23 × 105 | 1.20 × 105 | 1 | 26.22 | 0.90 | 3.67 × 104 | 3.61 × 104 |
| Peppino Leo | 0 | 23.98 | 0.00 | 2.02 × 105 | 0.00 | 0 | 26.69 | 3.81 | 7.78 × 104 | 1.05 × 105 |
| Peranzana | 0 | 21.90 | 2.40 | 1.51 × 106 | 1.25 × 106 | 2 | 20.80 | 0.33 | 1.85 × 106 | 4.28 × 105 |
| Permezzana | 0 | 26.27 | 0.00 | 4.12 × 104 | 0.00 | 0 | 25.62 | 2.28 | 5.48 × 104 | 9.60 × 104 |
| Pizzuta Della Daunia | 0 | 26.90 | 0.00 | 2.67 × 104 | 0.00 | 3 | 24.60 | 2.68 | 3.65 × 105 | 4.67 × 105 |
| Pizzuta Di Ginosa | 0 | 25.35 | 3.51 | 2.14 × 105 | 1.06 × 105 | 2 | 22.58 | 1.44 | 7.49 × 105 | 6.10 × 105 |
| Provenzale Di Serracapriola | 0 | 28.38 | 0.00 | 9.57 × 103 | 0.00 | 1 | 24.71 | 0.00 | 3.04 × 104 | 6.07 × 104 |
| Racioppa | 0 | 22.20 | 0.05 | 6.93 × 105 | 1.86 × 104 | 2 | 22.64 | 3.65 | 1.45 × 106 | 1.26 × 106 |
| Ravece | 0 | 29.92 | 0.00 | 3.29 × 103 | 0.00 | 1 | 22.74 | 0.15 | 4.77 × 105 | 5.18 × 104 |
| Ravece Guidacci | 0 | 23.98 | 0.00 | 2.02 × 105 | 0.00 | 2 | 22.47 | 1.31 | 7.29 × 105 | 4.61 × 105 |
| Rosciola | 0 | 22.43 | 0.00 | 5.92 × 105 | 0.00 | 3 | 21.25 | 1.06 | 1.64 × 106 | 1.22 × 106 |
| Rosciola Gentile | 3 | 20.70 | 0.00 | 1.96 × 106 | 0.00 | 2 | 24.01 | 0.00 | 1.97 × 105 | 0.00 |
| Rosciolone | 0 | 22.69 | 2.20 | 8.36 × 105 | 3.98 × 105 | 2 | 21.27 | 1.39 | 1.91 × 106 | 1.97 × 106 |
| Rotondella | 0 | 25.08 | 4.94 | 5.29 × 105 | 5.57 × 105 | 3 | 20.93 | 0.53 | 1.76 × 106 | 6.55 × 105 |
| Rumanella | 0 | 23.20 | 1.98 | 6.32 × 105 | 7.40 × 105 | 2 | 21.89 | 0.86 | 9.96 × 105 | 6.90 × 105 |
| Secolare Di Chieuti_PR | 0 | 30.72 | 6.93 | 1.30 × 105 | 2.24 × 105 | 1 | 27.11 | 2.98 | 1.33 × 105 | 2.49 × 105 |
| Seppunisi | 0 | 25.59 | 0.00 | 6.62 × 104 | 0.00 | 0 | 24.08 | 0.10 | 1.25 × 105 | 1.09 × 105 |
| Sessana | 0 | 30.40 | 0.00 | 2.35 × 103 | 0.00 | 0 | 26.70 | 2.79 | 1.23 × 105 | 2.16 × 105 |
| Silletta | 0 | 24.66 | 0.00 | 1.26 × 105 | 0.00 | 1 | 21.47 | 2.23 | 2.38 × 106 | 2.87 × 106 |
| Spina_PR | 0 | 25.55 | 0.00 | 6.78 × 104 | 0.00 | 2 | 27.37 | 5.70 | 1.05 × 105 | 1.82 × 105 |
| Stingi Ieronimo | 0 | 24.31 | 1.83 | 2.30 × 105 | 2.33 × 105 | 3 | 21.18 | 0.79 | 1.59 × 106 | 9.53 × 105 |
| Termite Del Medico | 0 | 27.62 | 0.00 | 1.62 × 104 | 0.00 | 2 | 23.79 | 2.64 | 5.17 × 105 | 9.28 × 105 |
| Termite Di Bitetto | 0 | 24.40 | 0.00 | 1.51 × 105 | 0.00 | 1 | 25.03 | 1.66 | 1.52 × 105 | 1.47 × 105 |
| Tondina | 0 | 25.49 | 1.43 | 1.00 × 105 | 6.03 × 104 | 1 | 24.21 | 1.86 | 2.90 × 105 | 2.68 × 105 |
| Torremaggiorese | 0 | 24.96 | 1.13 | 1.76 × 106 | 8.14 × 105 | 5 | 21.25 | 1.14 | 1.20 × 106 | 1.17 × 106 |
| Tunnella | 0 | 25.25 | 0.00 | 8.36 × 104 | 0.00 | 1 | 23.63 | 1.57 | 3.52 × 105 | 2.28 × 105 |
| Uccellina | 0 | 26.66 | 4.96 | 1.80 × 105 | 2.51 × 105 | 2 | 22.23 | 0.73 | 7.34 × 105 | 3.32 × 105 |
| Uovo Di Piccione | 0 | 22.20 | 0.00 | 6.94 × 105 | 0.00 | 1 | 23.37 | 0.21 | 1.54 × 105 | 1.80 × 105 |
| Zibimbolo | 0 | 26.25 | 0.00 | 4.18 × 104 | 0.00 | 3 | 25.84 | 0.00 | 5.57 × 104 | 0.00 |
| Varieties | ||||
|---|---|---|---|---|
| Constant Cq | Spring 2021 | Spring 2023 | ||
| Cq | SD | Cq | SD | |
| Pasola Di Andria | 21.43 | ±0.83 | 21.48 | ±1.05 |
| Crogiola | 21.88 | ±0.96 | 21.66 | ±1.05 |
| Cima Di Bitonto | 22.10 | ±1.13 | 22.05 | ±0.06 |
| Genotype_F5P3 | 22.65 | ±0.37 | 22.52 | ±1.91 |
| Racioppa | 22.20 | ±0.05 | 22.64 | ±3.65 |
| Morosino | 22.55 | ±0.66 | 22.72 | ±1.64 |
| Mennella | 23.50 | 0.0 | 23.02 | ±1.24 |
| Coratina | 23.33 | ±2.53 | 23.67 | ±2.09 |
| Butirra Di Melpignano | 24.19 | ±0.05 | 24.45 | ±1.87 |
| Pepperinella 2 | 26.93 | ±4.55 | 26.22 | ±0.90 |
| Cipressino | 26.11 | ±5.33 | 26.55 | ±2.78 |
| Genotype_F3P1_PR | 29.51 | 0.0 | 30.68 | ±4.56 |
| Increasing Cq | ||||
| Rosciola Gentile | 20.70 | 0.0 | 24.01 | 0.00 |
| Leucocarpa | 21.94 | ±0.67 | 24.07 | ±2.64 |
| Dolce Di Cassano | 20.64 | 0.0 | 25.39 | ±3.70 |
| Nocella | 23.78 | ±3.22 | 25.57 | ±3.51 |
| Uggiana | 22.83 | 0.0 | 25.58 | ±2.39 |
| Peppino Leo | 23.98 | 0.0 | 26.69 | ±3.81 |
| Spina _PR | 25.55 | 0.0 | 27.37 | ±5.70 |
| Genotype_F4P1 _PR | 23.65 | 0.0 | 28.04 | ±0.84 |
| Genotype_F9P1_PR | 28.80 | 0.0 | 31.67 | 0.00 |
| PR Genotypes | First Candidate | Pair Loci Mismatching | Second Candidate | Pair Loci Mismatching |
|---|---|---|---|---|
| Dolce_Tonda_PR | Coratina | 0 | Nocellara_del_Belice | 0 |
| Leccino_Lazio_PR | Leccino_REF | 0 | Frantoiana | 0 |
| Secolare_di_Chieuti_PR | Gulliver | 1 | Ciddina_FG | 0 |
| Spina_PR | - | - | Grappa | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savoia, M.A.; Fanelli, V.; Miazzi, M.M.; Taranto, F.; Procino, S.; Susca, L.; Montilon, V.; Potere, O.; Nigro, F.; Montemurro, C. Apulian Autochthonous Olive Germplasm: A Promising Resource to Restore Cultivation in Xylella fastidiosa-Infected Areas. Agriculture 2023, 13, 1746. https://doi.org/10.3390/agriculture13091746
Savoia MA, Fanelli V, Miazzi MM, Taranto F, Procino S, Susca L, Montilon V, Potere O, Nigro F, Montemurro C. Apulian Autochthonous Olive Germplasm: A Promising Resource to Restore Cultivation in Xylella fastidiosa-Infected Areas. Agriculture. 2023; 13(9):1746. https://doi.org/10.3390/agriculture13091746
Chicago/Turabian StyleSavoia, Michele Antonio, Valentina Fanelli, Monica Marilena Miazzi, Francesca Taranto, Silvia Procino, Leonardo Susca, Vito Montilon, Oriana Potere, Franco Nigro, and Cinzia Montemurro. 2023. "Apulian Autochthonous Olive Germplasm: A Promising Resource to Restore Cultivation in Xylella fastidiosa-Infected Areas" Agriculture 13, no. 9: 1746. https://doi.org/10.3390/agriculture13091746
APA StyleSavoia, M. A., Fanelli, V., Miazzi, M. M., Taranto, F., Procino, S., Susca, L., Montilon, V., Potere, O., Nigro, F., & Montemurro, C. (2023). Apulian Autochthonous Olive Germplasm: A Promising Resource to Restore Cultivation in Xylella fastidiosa-Infected Areas. Agriculture, 13(9), 1746. https://doi.org/10.3390/agriculture13091746













