Effects of Mixed Saline and Fresh Water Sprinkler Irrigation on the Rhizosphere Soil Microbial Community of Summer Maize
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Site Summary
2.2. Experiment Design
2.3. Measuring Indicators
2.3.1. Measurement of Soil Properties
2.3.2. Measurements of Soil Microbiology
2.3.3. Bacterial 16SrRNA Sequencing
2.4. Data Processing
3. Results
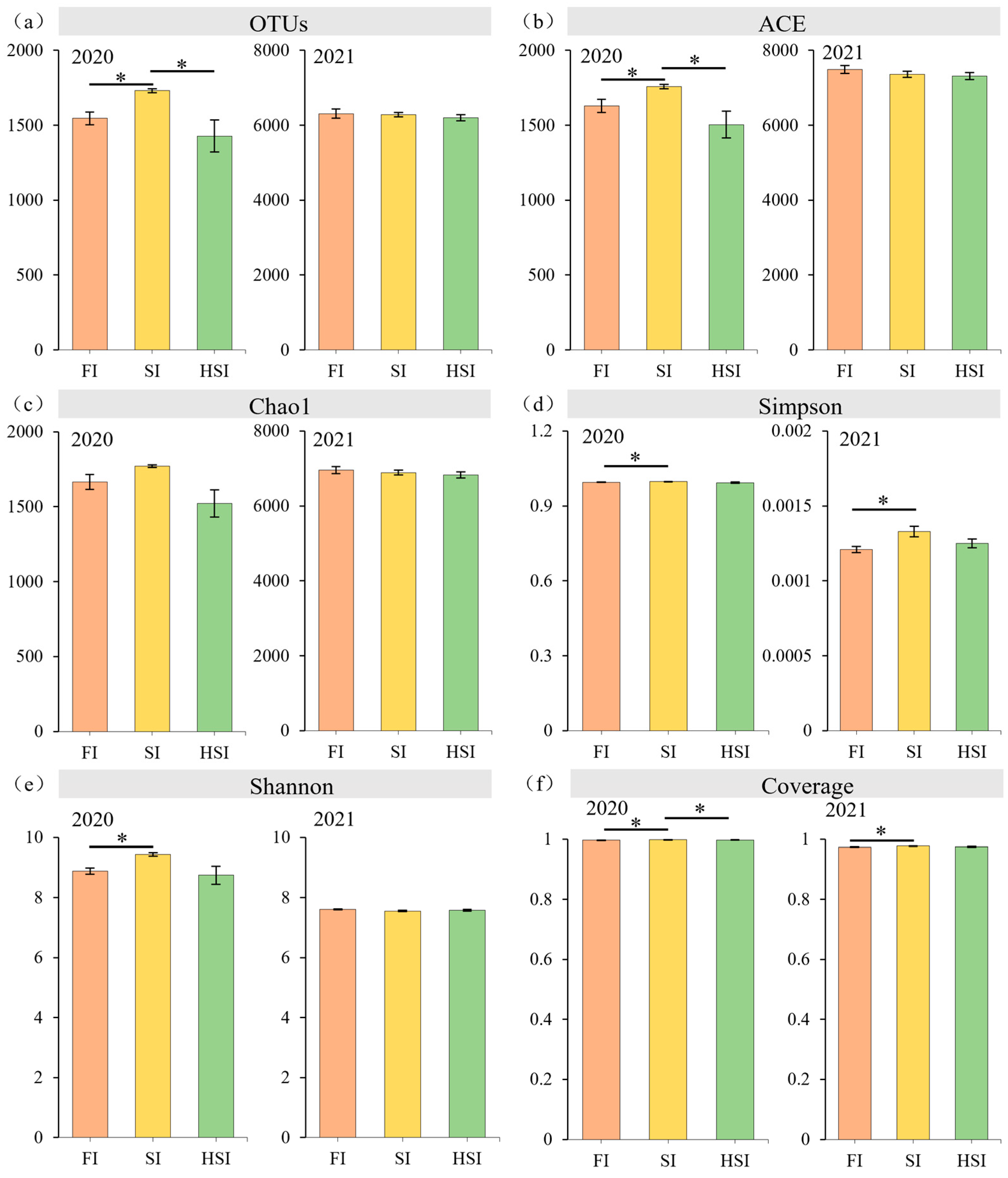
3.1. Analysis of OTUs and Alpha Diversity of Tillage Bacteria
3.2. OTU–Venn Diagram Analysis of Tillage Bacteria
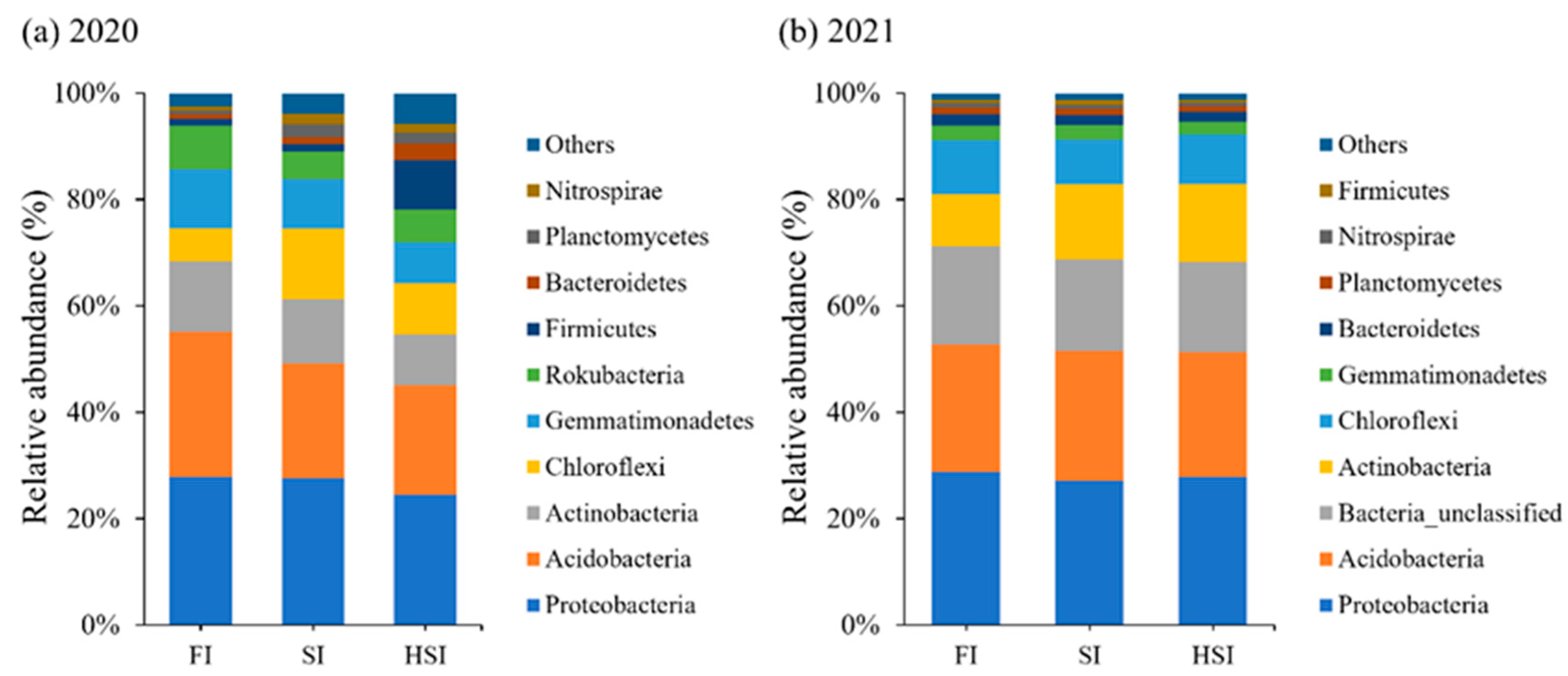
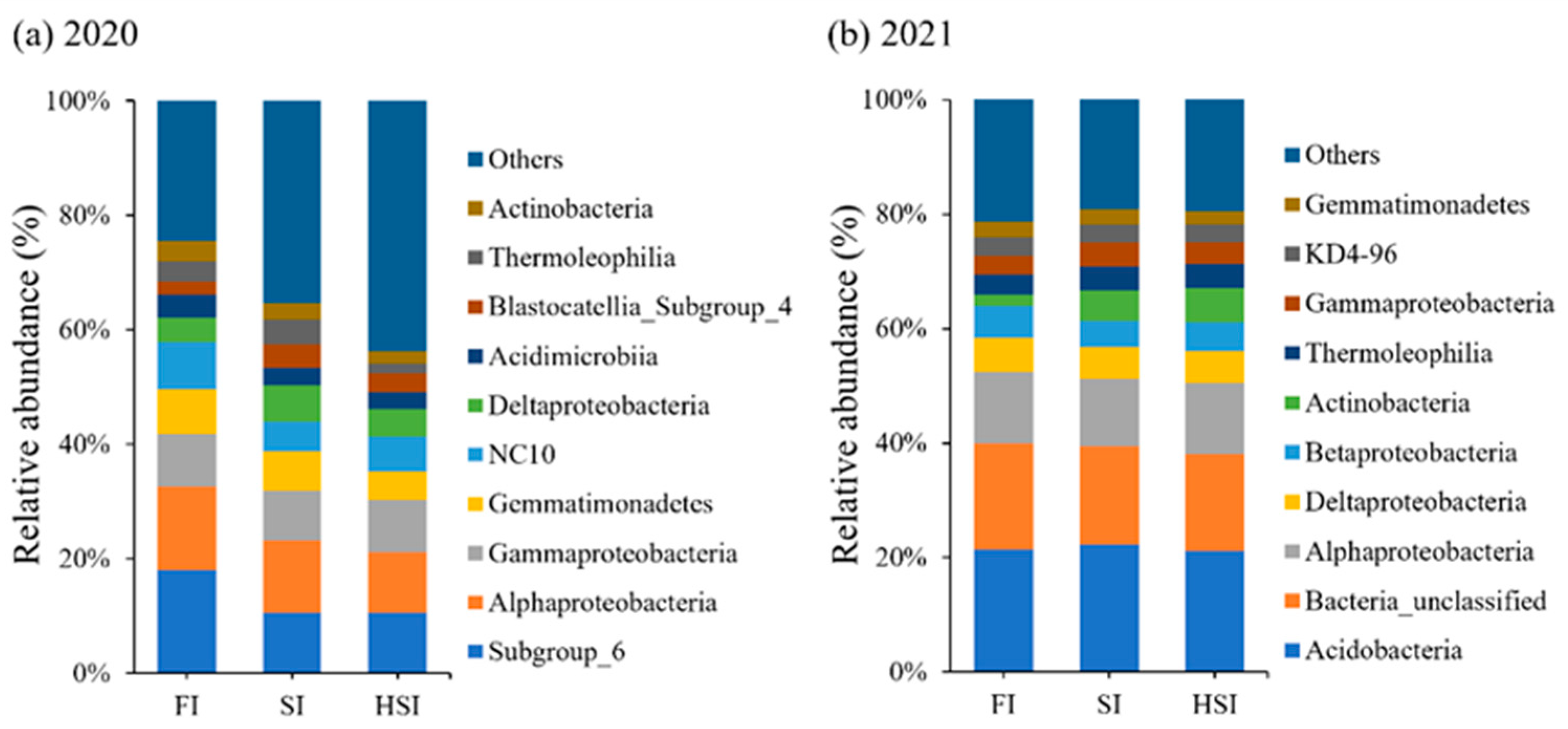
3.3. Population Structure Analysis of Bacteria in Each Sample
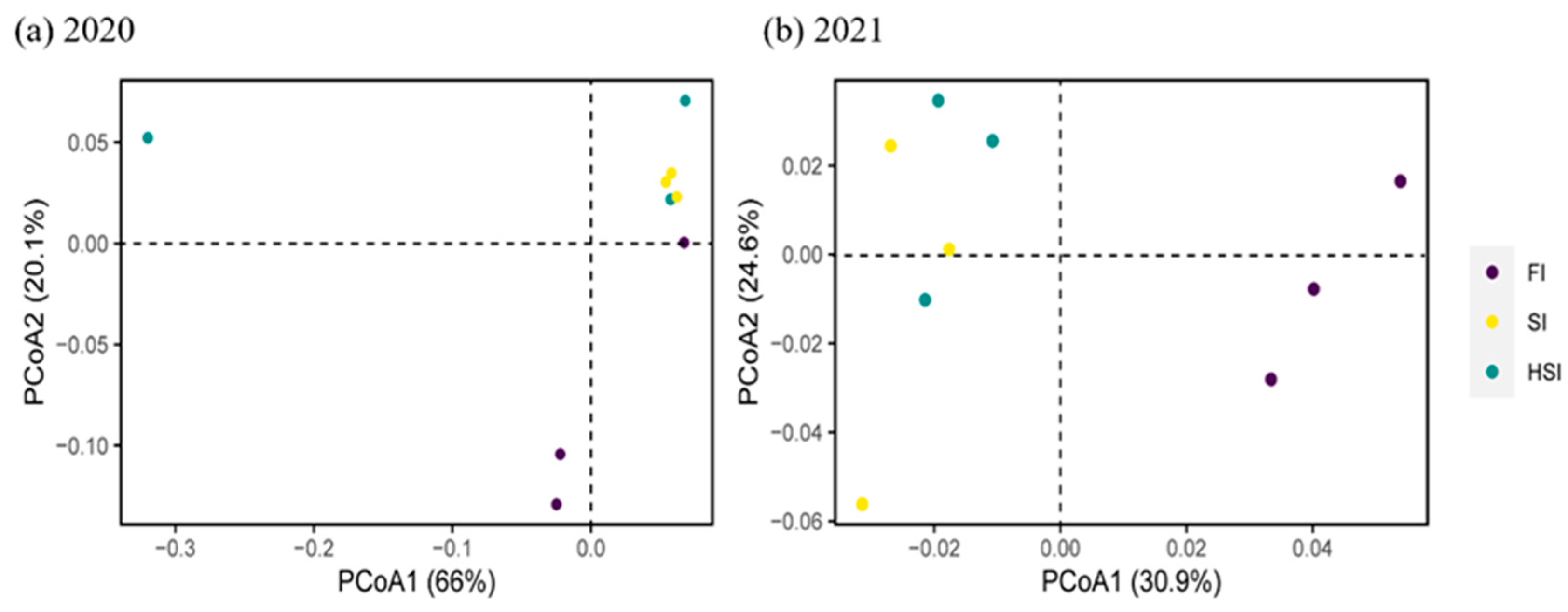
3.4. Beta Diversity Analysis of Bacteria
3.5. Correlation Analysis Between Bacterial Microbial Communities and Soil Physico-Chemical Properties
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, S.; Liu, J.J.; Banerjee, S.; Zhou, N.; Zhao, Z.Y.; Zhang, K.; Tian, C.Y. Soil pH is equally important as salinity in shaping bacterial communities in saline soils under halophytic vegetation. Sci. Rep. 2018, 8, 4550. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Qi, K.B.; Bhusal, D.R.; Huang, J.S.; Chen, W.J.; Wu, Q.S.; Hussain, A.; Pang, X.Y. Soil microbial community and enzymatic activity in soil particle-size fractions of spruce plantation and secondary birch forest. Eur. J. Soil. Biol. 2020, 99, 103196. [Google Scholar] [CrossRef]
- Deng, S.; Ke, T.; Li, L.; Cai, S.; Zhou, Y.; Liu, Y.; Guo-, L.; Chen, L.; Zhang, D. Impacts of environmental factors on the whole microbial communities in the rhizosphere of a metal-tolerant plant: Elsholtzia haichowensis Sun. Environ. Pollut. 2018, 237, 1088–1097. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.G.; Wu, X.N.; Qiu, Q.T.; Duan, C.Q.; Jones, D.L. Seasonal variations in soil microbial communities under different land restoration types in a subtropical mountains region, Southwest China. Appl. Soil. Ecol. 2020, 153, 103634. [Google Scholar] [CrossRef]
- Qin, X.; Huang, Q.; Liu, Y.; Zhao, L.; Xu, Y.; Liu, Y. Effects of sepiolite and biochar on microbial diversity in acid red soil from southern China. Chem. Ecol. 2019, 35, 846–860. [Google Scholar] [CrossRef]
- Doran, J.W.; Zeiss, M.R. Soil health and sustainability: Managing the biotic component of soil quality. Appl. Soil. Ecol. 2000, 15, 3–11. [Google Scholar] [CrossRef]
- Yao, H.; He, Z.; Wilson, M.; Campbell, C. Microbial biomass and community structure in a sequence of soils with increasing fertility and changing land use. Microb. Ecol. 2000, 40, 223–237. [Google Scholar] [CrossRef]
- Waldrop, M.; Balser, T.; Firestone, M. Linking microbial community composition to function in a tropical soil. Soil. Biol. Biochem. 2000, 32, 1837–1846. [Google Scholar] [CrossRef]
- Szoboszlay, M.; Näther, A.; Liu, B.; Carrillo, A.; Castellanos, T.; Smalla, K.; Jia, Z.; Tebbe, C.C. Contrasting microbial community responses to salinization and straw amendment in a semiarid bare soil and its wheat rhizosphere. Sci. Rep. 2019, 9, 9795. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Gaurav, A.K.; Srivastava, S.; Verma, J.P. Plant growth-promoting bacteria: Biological tools for the mitigation of salinity stress in plants. Front. Microbiol. 2020, 11, 1216. [Google Scholar] [CrossRef]
- Amini, S.; Ghadiri, H.; Chen, C.; Marschner, P. Salt-affected soils, reclamation, carbon dynamics, and biochar: A review. J. Soils Sediments 2016, 16, 939–953. [Google Scholar] [CrossRef]
- Ke, C.R.; Li, Z.Y.; Liang, Y.M.; Tao, W.Q.; Du, M.C. Impacts of chloride, de-icing salt on bulk soils, fungi, and bacterial populations surrounding the plant rhizosphere. Appl. Soil. Ecol. 2013, 72, 69–78. [Google Scholar] [CrossRef]
- Chowdhury, N.; Marschner, P.; Burns, R. Response of microbial activity and community structure to decreasing soil osmotic and matric potential. Plant Soil. 2011, 344, 241–254. [Google Scholar] [CrossRef]
- Wichern, J.; Wichern, F.; Joergensen, R.G. Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils. Geoderma 2006, 137, 100–108. [Google Scholar] [CrossRef]
- Kamble, P.N.; Gaikwad, V.B.; Kuchekar, S.R.; Bååth, E. Microbial growth, biomass, community structure and nutrient limitation in high pH and salinity soils from Pravaranagar (India). Eur. J. Soil. Biol. 2014, 65, 87–95. [Google Scholar] [CrossRef]
- Rath, K.M.; Maheshwari, A.; Bengtson, P.; Rousk, J. Comparative toxicities of salts on microbial processes in soil. Appl. Microbiol. Biotechnol. 2016, 82, 2012–2020. [Google Scholar] [CrossRef]
- Hollister, E.B.; Engledow, A.S.; Hammett, A.J.; Provin, T.L.; Wilkinson, H.H.; Gentry, T.J. Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J. 2010, 4, 829–838. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Knight, R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef]
- Kuczynski, J.; Lauber, C.L.; Walters, W.A.; Parfrey, L.W.; Clemente, J.C.; Gevers, D.; Knight, R. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 2012, 13, 47–58. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Boutros, P.C. Venn Diagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Sakaki, T.; Takeshima, T.; Tominaga, M.; Hashimoto, H.; Kawaguchi, S. Recurrence of ICA-PCoA aneurysms after neck clipping. J. Neurosurg. 1994, 80, 58–63. [Google Scholar] [CrossRef]
- Castro, S.P.; Cleland, E.E.; Wagner, R.; Sawad, R.A.; Lipson, D.A. Soil microbial responses to drought and exotic plants shift carbon metabolism. ISME J. 2019, 13, 1776–1787. [Google Scholar] [CrossRef]
- Hu, L.F.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.N.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, X.Y.; Wang, L.M.; Cao, Y.B.; Song, W.B.; Shi, J.P.; Lai, J.; Jiang, C. A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat. Plants 2019, 5, 1297–1308. [Google Scholar] [CrossRef]
- Dang, Q.L.; Tan, W.B.; Zhao, X.Y.; Li, D.; Li, Y.P.; Yang, T.X.; Li, R.; Zu, G.; Xi, B. Linking the response of soil microbial community structure in soils to long-term wastewater irrigation and soil depth. Sci. Total Environ. 2019, 688, 26–36. [Google Scholar] [CrossRef]
- Jha, Y.; Subramanian, R.B.; Patel, S. Combination of endophytic and rhizospheric plant growth promoting rhizobacteria in Oryza sativa shows higher accumulation of osmoprotectant against saline stress. Acta Physiol. Plant 2011, 33, 797–802. [Google Scholar] [CrossRef]
- Sun, M.Y.; Dafforn, K.A.; Johnston, E.L.; Brown, M.V. Core sediment bacteria drive community response to anthropogenic contamination over multiple environmental gradients. Environ. Microbiol. 2013, 15, 2517–2531. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Encinas, C.; Neria-Gonzalez, I.; Alcantara-Hernandez, R.J.; Estrada-Alvarado, I.; Zavala-Diaz de la Serna, F.J.; Dendooven, L.; Marsch, R. Changes in the bacterial populations of the highly alkaline saline soil of the former lake Texcoco (Mexico) following flooding. Extremophiles 2009, 13, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Samaddar, S.; Chatterjee, P.; Krishnamoorthy, R.; Jeon, S.; Sa, T. Structural and functional responses of microbial community with respect to salinity levels in a coastal reclamation land. Appl. Soil. Ecol. 2019, 137, 96–105. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, G.; Tang, X.; Shao, K.; Gong, Y. Pyrosequencing analysis of bacterial communities in Lake Bosten, a large brackish inland lake in the arid northwest of China. Can. J. Microbiol. 2016, 62, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Rath, K.M.; Fierer, N.; Murphy, D.V.; Rousk, J. Linking bacterial community composition to soil salinity along environmental gradients. ISME J. 2019, 13, 836–846. [Google Scholar] [CrossRef]
- Rousk, J.; Baath, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Liu, B.; Hu, Y.; Wang, Y.; Xue, H.; Li, Z.; Li, M. Effects of saline-alkali stress on bacterial and fungal community diversity in Leymus chinensis rhizosphere soil. Environ. Sci. Pollut. Res. 2022, 29, 70000–70013. [Google Scholar] [CrossRef]
- Yang, J.; Ma, L.; Jiang, H.C.; Wu, G.; Dong, H.L. Salinity shapes microbial diversity and community structure in surface sediments of the Qinghai-Tibetan Lakes. Sci. Rep. 2016, 6, 25078. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Guo, Z.; Wang, H.; Li, W.; Sheng, W.; Zhang, S.; Zhang, D. Effects of Mixed Saline and Fresh Water Sprinkler Irrigation on the Rhizosphere Soil Microbial Community of Summer Maize. Agriculture 2024, 14, 2237. https://doi.org/10.3390/agriculture14122237
Wang T, Guo Z, Wang H, Li W, Sheng W, Zhang S, Zhang D. Effects of Mixed Saline and Fresh Water Sprinkler Irrigation on the Rhizosphere Soil Microbial Community of Summer Maize. Agriculture. 2024; 14(12):2237. https://doi.org/10.3390/agriculture14122237
Chicago/Turabian StyleWang, Tieqiang, Zikang Guo, Hanbo Wang, Weidong Li, Wenxu Sheng, Shuantang Zhang, and Dasheng Zhang. 2024. "Effects of Mixed Saline and Fresh Water Sprinkler Irrigation on the Rhizosphere Soil Microbial Community of Summer Maize" Agriculture 14, no. 12: 2237. https://doi.org/10.3390/agriculture14122237
APA StyleWang, T., Guo, Z., Wang, H., Li, W., Sheng, W., Zhang, S., & Zhang, D. (2024). Effects of Mixed Saline and Fresh Water Sprinkler Irrigation on the Rhizosphere Soil Microbial Community of Summer Maize. Agriculture, 14(12), 2237. https://doi.org/10.3390/agriculture14122237






