Abstract
Viruses are silent enemies that intrude and take control of the plant cell’s machinery for their own multiplication. Infection by viruses and the resulting damage is still a major challenge in the agriculture sector. Plants have the capability to fight back, but the ability of viruses to mutate at a fast rate helps them to evade the host’s response. Therefore, classical approaches for introgressing resistance genes by breeding have obtained limited success in counteracting the virus menace. Genetic modification (GM)-based strategies have been successful in engineering artificial resistance in plants. Several different approaches based on pathogen-derived resistance, antisense constructs, hairpin RNAs, double-stranded RNA, etc., have been used to enhance plants’ resistance to viruses. Recently, genome editing (GE) strategies mainly involving the CRISPR/Cas-mediated modifications are being used for virus control. In this review, we discuss the developments and advancements in GM- and GE-based methods for tackling viral infection in plants.
1. Introduction
Viruses constitute nearly half of the microbial pathogens that are accountable for the emerging and re-emerging epidemics worldwide [1]. They also pose serious challenges to agriculture by causing major plant diseases like the bunchy top of bananas, tobacco mosaic, wheat streak mosaic, potato tuber necrotic ring spot, cucumber green mottle mosaic, tomato yellow leaf curl, African cassava mosaic, and so on. So far, 131 families containing 803 genera and 4853 species of plant virus are reported by International Committee on Taxonomy of Viruses [2]. Viruses are responsible for a worldwide crop loss of $30 billion annually and is predicted to reach up to $60–80 billion [3,4,5]. Plant viral diseases threaten global food security as they can cause severe food limitations for the world population, which is projected to elevate from 8 billion in 2022 to 10 billion by 2050 [6,7].
For the resolution of any problem, it is important to implement appropriate prevention and control measures. However, viruses cannot be controlled by chemicals like pesticides, although their carriers or vectors can be restrained to some extent [8]. This simply leaves the possibility of finding or generating plant species with inherent resistances to viral infections. A silver lining emerged from McKinney’s observation in 1929 that the mild infection caused by the Tobacco mosaic virus (TMV) green mosaic strain protected the tobacco plants from severe infection by the TMV yellow mosaic strain, which suggested the existence of mechanisms for cross-protection [9]. This phenomenon involved the defense response of the plant to a mild virus and the formation of a memory of the response in the host plant to combat the challenge posed by a related virus [10]. These early observations prompted the search for host-based resistance genes and an understanding of plant–virus interactions [11,12].
Studies performed by different groups over the last century have shown that plants have several physical barriers like cuticles, thorns, trichomes, etc., and chemical barriers like the accumulation of lignin, callose, terpenoids, phenolics, etc., for primary protection against viral invasion [13]. In addition, they have several complex and overlapping natural defense mechanisms to tackle the viruses that manage to invade. Plant immunity can be categorized as a pathogen-triggered immunity (PTI) and an effector-triggered immunity (ETI). A PTI relies on the effective detection of the invading pathogen and limits their growth and spread across host cells [14]. For successful invasion, the pathogens produce different types of avirulence factors (AVRs) for evading or suppressing PTI pathways. The host cells, in turn, produce specific resistance (R) proteins that can recognize the effectors [15]. This R–AVR interaction leads to the activation of an ETI [16,17]. The first line of defense in cases of viral pathogens is the activation of the RNA silencing or RNA interference (RNAi) pathways by viral dsRNAs. The viral triggers for PTI and ETI are not clearly defined, though the viral dsRNAs are generally paralleled to the PAMPs. Some virus-encoded proteins have the ability to suppress the silencing process and they can act as effectors that lead to the generation of an ETI.
Different R genes such as NBS-LRR, RTM1, RTM2, and RTM3 were discovered in Arabidopsis thaliana, which provided resistance against viruses, especially the potyviruses, by preventing the systemic spread through the phloem [18,19]. In addition, the mutations or recessive alleles of certain host-encoded factors which are essential for virus replication provided a unique recessive resistance against plant viruses [20].
The selection of dominant and recessive genes associated with virus resistance played an important role in the breeding of naturally resistant plant varieties as a cheaper and better alternative for controlling viral diseases [5]. Classical approaches such as the introgression of resistance genes by marker-assisted selection (MAS), quantitative-trait loci (QTL) mapping and gene pyramiding were extensively used to tackle the menace of viruses. Another reverse genetics approach known as TILLING (Targeting induced local lesions in genomes) was used to create novel mutations in alleles in order to repress the expression of essential factors that are needed by a virus to complete its infection cycle [21]. However, there were several bottlenecks, such as the non-availability of markers near the R genes, the transfer of undesired traits, and the screening of plants for several generations, which made the process tedious and time-consuming, thereby limiting the success of these techniques.
The era of genetic engineering accelerated the process by allowing the incorporation of genes in the plant genome as per the requirement. In the early 1980s, the concept of pathogen-derived resistance (PDR) laid the foundation of artificial resistance. PDR employed the use of virus proteins like the coat protein (CP) to disrupt virus replication by inactivating the viral components [11]. Subsequent studies unraveled the role of RNA silencing in symptom recovery and cross-protection [22,23]. It also strengthened the possibilities for employing RNA silencing for generating virus resistance in plants. However, the silencing pathways have some limitations due to the presence of silencing suppression activity in the viral-encoded proteins. In recent times, CRISPR/Cas-based tools are being used to edit the plant genome for generating virus-resistant plants.
Both genetic modification (GM) and genome editing (GE) approaches are quite efficient in accelerating the generation of virus-resistant plant varieties without the need of backcrossing. These techniques are very precise and time-saving as compared to traditional breeding. In particular, for virus management, many transgenic plants, such as the squash, papaya, bean, and cassava, have received approval for field trials and commercial cultivation in various countries [24]. The various aspects and examples are described in the following sections.
3. GM Approaches
These approaches involve the over-expression or knock-down of genes using a variety of genetic engineering tools. The methodology usually involves cloning the gene of interest (trans-gene) into the desired vector in either a sense or antisense orientation followed by its transformation in plants [64]. Many old reports are available on engineering virus resistance in plants via the transgenic approach [65,66].
3.1. Pathogen-Derived Resistance
This approach involves the use of the pathogen’s own genetic material to generate a resistance in the host plant. The concept of PDR was first presented by Sanford and Johnston in 1985 [11] and a year later it was proven to be successful in cases of viral infections. The transgenic plants expressing the coat protein (CP) gene of TMV were shown to exhibit a resistance against infection [67]. This ground-breaking discovery initiated the development of various virus-resistant transgenic plants [6]. PDR can be classified into different types depending on the strategy used to generate the resistance towards the virus in the host plant.
3.1.1. Protein-Mediated Resistance
This process entails the expression of viral-encoded proteins like CP, MP (movement protein) or Rep (replicase) in plants to counteract viruses [6]. In 1986, Beachy’s group first demonstrated that the expression of the CP gene of TMV in transgenic tobacco plants could provide considerable protection against viral infection [68]. It was observed that the expression of these viral proteins interfered with the disassembly of the virus by the formation of pseudo-capsids [6]. Several transgenics were made using the CP of the Cucumber mosaic virus (CMV), Papaya ringspot virus (PRSV), PVX, Plum pox virus (PPV), Zucchini yellow mosaic virus (ZYMV), Watermelon mosaic virus (WMV), Tomato yellow leaf curl virus (TYLCV), Tomato mosaic virus (ToMV), and so on [69].
PDR also provided cross-protection against infection by related viruses. This technique was more effective in cases of closely related viruses than those which are distantly related [70]. When TMV CP-transgenic plants were infected by different viruses of the Tobamovirus family, their inhibition was differentially compromised. Viruses like the ToMV, Pepper mild mosaic virus (PMMV) and Tomato mild green mosaic virus (TMGMV) were inhibited completely while no resistance was observed against others like CMV, African mosaic virus (AMV), PVX, and Potato virus Y (PVY), though there was a decrease in symptoms and spread [71]. In field trials also, CP-TMV expressing tomato and CP-PVX/PVY expressing potato transgenics performed better than the wild-type [72].
In cases of PVX infection, it was shown that the interaction of the 5′ end of viral RNA with CP expressed in the host plant suppressed the expression of the RdRP encoded by the virus [73,74]. The region and quantity of protein expression are important determinants for the generation of resistance. For instance, if CP is expressed in mesophyll but the viral RNA is introduced in epidermal cells then resistance did not develop [75]. The expression of mutant CP in transgenics also resulted in a resistance against wild-type CP, as the mutant CP could interact with the wild-type CP subunits or the host factors, which were involved in virion disassembly [76,77]. The extent of the aggregation of the CP correlated with the level of the coat protein-mediated resistance (CPMR) [78]. This observation suggested that homology between the amino acid sequences of the challenging viruses provided cross-protection in the transgenic organism [6].
Likewise, the MPs, which have a role in the cell-to-cell spread of the virus, were also utilized for triggering PDR. This technique worked better for large genera of viruses irrespective of the viral MP expressed in plants. The expression of defective or mutant MPs could also block virus transmission since the mutant MPs competed with the wild-type MPs of the virus for the plasmodesmatal sites [79]. Mutant TMV-MP provided resistance against Potexviruses, Tobamoviruses, and Cucumoviruses [80]. Several groups tried to multiplex this approach by generating transgenics with more than one type of MP by using overlapping regions of the ORFs which are also known as a triple gene block (TGB). However, this approach did not provide broader resistance, possibly due to the insufficient interaction of TGB with the plasmodesmata [81].
In subsequent studies, it was shown that the use of modified non-structural proteins like Rep/RdRP were able to provide better results even at low levels of expression [82]. Mutations in the ori or NTP-binding sites of the Rep gene (AC1) provided resistance against geminiviruses [83,84]. The Rep/RdRP proteins could directly interfere with virus replication or bind with host factors involved in the viral replication process [85]. Detailed investigations into the molecular mechanisms behind the responses demonstrated that these proteins mediated resistance mainly through the RNA-silencing machinery instead of direct protein interactions [86,87].
The commercially produced squash, ZW20, that is resistant to WMV and ZYMV became the first GM crop to receive approvals in the USA [88]. Subsequently, several other virus-resistant crops were produced. Despite several examples of the successful implementation of this technology, it faced some challenges due to evolutionary pressure provided by the expression of the transgene. The genomic content of the virus interacting with the transgenics was altered due to the recombination between the transgenic and viral CP transcripts [89]. This trans-capsidation of the CP in transgenics imparted a new potential to the infecting virus isolate. Mixed infection by two unrelated viruses was another issue, as it led to the antagonism and synergism of defense responses. For instance, different outcomes were produced during mixed infection with PRSV and Papaya mosaic virus (PapMV). In the case when plants were first infected with PRSV followed by PapMV or simultaneously inoculated with both, a synergistic response was observed. However, when the first inoculation of PapMV was followed by PRSV, an antagonistic response was observed, which caused a reduction in PRSV titers due to the activation of defense responses [90].
3.1.2. Defective Interfering Virus-Mediated Resistance
Plant viruses are often associated with sub-genomic/sub-viral RNAs known variably as satellite (sat) or defective interfering (DI) viruses [91]. The DI viruses lack a critical portion of the virus genome, which makes them defective for replication. They require the presence of the complete functional virus, known as the helper virus, for their replication and encapsidation [92]. DI viruses are basically the defective versions of helper viruses formed by mainly deletions, duplication, and inversion processes [93]. There is often a synergistic interaction between the DI virus and its helper as they can produce mild symptoms on one host plant but elicit severe symptoms on another host or in combination with a different strain of a helper virus [94,95]. According to some reports, a DI virus can attenuate symptoms of an unrelated helper virus by inhibiting their multiplication and long-distance transport [96,97]. Hence, they were used in transgenic approaches to decrease virus symptoms and/or titers [92]. As for satellite RNAs, they can compete with the helper virus genome for binding to replicase for replication, which indirectly suppresses virus replication [97]. In 2015, it was shown that CMV sat-Y-derived siRNAs bind to the viral encoded suppressors of RNA silencing (VSR), 2b of CMV and P19 of Tombusvirus, causing the release of the VSR-bound helper viral siRNAs resulting in the down-regulation of the viral target genes [98]. The major limitation of this strategy was a probable risk that the DI virus and satellites could mutate to a virulent form in conjunction with the infecting virus in the transgenic plants.
3.2. Non-Coding RNA Shields in Virus Resistance
RNA silencing has been effectively used for generating virus resistant plants [99]. A variety of small RNAs have been used through various strategies to trigger the RNA silencing-mediated defense responses against viruses (Table 1). The regions of the viral genome that encode VSRs, e.g., P69, HC-Pro, and AC2, are usually selected as the targets of the small RNAs. The expression of small RNAs can be from various precursors, which may be in cis or trans and single or polycistronic [100,101]. Initial GM approaches involved the use of antisense constructs or hp-RNAs as precursors to generate a pool of siRNAs, which could target the viral transcripts. Subsequently, these were replaced by more specific tools through the use of constructs expressing antiviral artificial microRNAs (amiRNAs) and synthetic trans-acting small interfering RNAs (syn-tasiRNAs).

Table 1.
List of small RNA-based GM approaches used against plant viruses.
3.2.1. microRNA Shield
MicroRNAs (miRNAs) are genome-encoded small RNAs that are processed by DCL1 from imperfectly paired, hairpin precursor RNAs [124,125]. The miRNAs belong to large gene families and each miRNA can modulate a plethora of targets involved in plant immunity and phytohormone pathways [126]. High throughput sequencing tools have provided useful insights for identifying the miRNAs involved in virus resistance. miR156, miR395, miR159, miR166, miR168, miR160, and miR444 have important roles in virus attacks [127,128,129,130]. A recent report has shown that a novel miRNA, Seq119, is down-regulated during Rice stripe virus (RSV) infection. The overexpression of this miRNA reverses the symptoms caused by RSV [131].
In many cases, amiRNAs have been used to over-express a synthetic sequence designed to target a key viral gene. In Arabidopsis plants, amiRNAs were used first to target TYMV and Turnip mosaic virus (TuMV) [132]. This technique has the advantage of enhanced specificity and reduced off-target effects. The amiRNA is composed of two components, namely, a miRNA precursor (pre-miRNA) scaffold and a synthetic RNA insert. Special consideration is placed on the selection of the pre-miRNA backbone to ensure that the correct amiRNA is processed by the cellular machinery [133]. Studies with TuMV population dynamics showed that inadequate levels of amiRNAs build up more mutations in the target site and reduce the efficacy of virus resistance [134].
To synthesize the amiRNA, the miRNA/miRNA* sequence of the pre-miRNA is replaced by a synthetic sequence complementary to the viral target by performing an overlapping PCR [135]. The sequence of the amiRNA is designed to have maximum complementarity with the target and minimum hybridization energy while ensuring sufficient mismatches to retain the secondary structures or bulges [136,137]. Instability at the 5′ end and the avoidance of mismatches at the 10 or 11th nucleotides are also crucial parameters. The design of an effective amiRNA also necessitates the incorporation of Uracil (U) at the first base and Adenine (A) or U at the 10th nucleotide position for making amiRNA biologically active [136,138]. Targeting an exposed region in a viral genome can increase the chances of DCL accessibility and the efficiency of the amiRNA [139]. Tomato transgenics overproducing the amiRNAs to silence the conserved regions of ToLCV- AV2/AC2 genes could tolerate various leaf curl viruses of tomatoes [140]. The amiRNA technique faced the limitations of its stringent design principles and the level of amiRNA expression.
3.2.2. siRNA Shield
The siRNAs are 21–24 nt in length and are derived from perfectly paired dsRNA molecules [141,142]. The siRNA-mediated gene silencing serves as a primary defense mechanism against plant viruses [52,143,144,145,146]. The transitivity of siRNAs is useful in propagating the silencing signal through the production of secondary siRNAs [31,147].
It was observed that better virus resistance was achieved when a dsRNA of the viral sequence was expressed in a plant cell compared to the case of expressing either sense or antisense viral RNA [148]. This resulted in the construction of intron splice-able hairpin RNA (hpRNA) constructs with viral gene or gene fragments. The hpRNA constructs offered the strongest resistance to RNA and DNA viruses [149,150,151]. The GM approaches involve the expression of short-hairpin RNA (shRNA) constructs as precursors of the siRNAs in the plants to tackle the virus. The constructs are mobilized into the Agrobacterium and stably integrated into plants [152]. In the majority of cases, the VSR genes have been used as a silencing target to generate virus resistance [150]. In Brazil, a transgenic bean resistant to a Bean golden mosaic virus (BGMV) was created by expressing a hpRNA of Rep gene, and it is being commercially cultivated [149].
Tomato plants expressing hpRNA corresponding to the TYLCV-Rep coding sequence produce 21 and 22 nt siRNAs and are resistant to TYLCV [153]. Similarly, Cassava transgenics containing hpRNA homologous to the overlapping region of South African cassava mosaic virus (SACMV)-Rep and the AC1/AC4 were tolerant to cassava mosaic disease [154]. In another study, three distinct intron hpRNA constructs comprising sequences of AC2, AC4, and a fusion of AC2 and AC4 (AC2 + AC4) of seven begomoviruses were used. All transgenic lines showed a resistance against MYMIV infection when compared with untransformed controls [155].
3.2.3. Synthetic tasiRNA Shield
The tasiRNAs are derived from non-protein-coding TAS transcripts that are capped as well as poly-adenylated and contain a binding site mostly for 22 nt miRNAs [156]. The miRNA cleavage products are stabilized by the Suppressor of Gene Silencing 3 (SGS3) and converted into a dsRNA form by RDR6 [157,158]. The dsRNA intermediate is then processed by DCL4 and dedicated dsRNA-Binding Protein 4 (DRB4) to phased 21 nt siRNAs in a ‘head-to-tail’ phased pattern. The transitive siRNAs are incorporated into AGO-RISC for targeting complementary sequences [157,158,159,160]. In Arabidopsis thaliana, eight tasiRNA-producing loci have been identified that fall into four TAS groups (TAS1–TAS4).
Since TAS precursors are able to generate multiple secondary siRNAs once they are cleaved by (with) the miRNA, they work more efficiently against the rapidly evolving viruses that can evade the single amiRNA approach. The Arabidopsis TAS DNA sequences can be engineered to silence targeted viral sequences by replacing some of the phased tasiRNA producing sequences by single or multiple siRNAs of different sequences but of an equivalent length of base-pairs. When the replacing siRNAs are processed in the tasiRNA pathway from the engineered vectors as desired, these are called synthetic-tasiRNAs (syn-tasiRNAs), which consequently silence their targets in a usual manner using the RNA-silencing machinery of the host plants. It was observed that most plants expressing higher levels of syn-tasiRNA were resistant to TSWV (Tomato Spotted Wilt Virus), while plants with particularly low levels of syn-tasiRNAs were infected. Several automated design tools have been used for designing syn-tasiRNA constructs, which are able to target distinct locations within a single RNA or multiple viral RNAs. A more efficient, long-lasting, and widespread resistance could be produced by the simultaneous co-expression of multiple syn-tasiRNAs [161,162].
3.2.4. Long Non-Coding RNA Shield
Long non-coding RNAs (lncRNAs) are 200 nt or longer and are mainly produced by the action of polymerase II [163]. The lncRNAs resemble the mRNAs in having a 5′m7G cap and 3′poly(A) tail [164,165], though their sequences show less conservation [166]. On the basis of their origin, they can be categorized as intergenic, intronic, or exonic. They can act either in cis or trans and may have sense or antisense orientations [167,168]. NGS analysis has played an important role in the identification of differentially expressed lncRNAs and their roles in host interaction studies [169]. An RNA-seq of TYLCV-resistant tomato cultivar showed that some lncRNAs such as slylnc0048, slylnc00449, slylnc0483, slylnc0531, and slylnc0934 were up-regulated while slylnc0475, slylnc0476, slylnc0673, and slylnc1052 were down-regulated after virus infection [170].
LncRNAs can act as sources for the generation of small RNAs and regulate DNA or histone methylation by guiding the DNA methyltransferase. LMT1 encoded by the Citrus tristeza virus (CTV) plays an important role in the susceptibility of host plants. The mutation in LMT1 caused an increase in the SA levels and enhanced virus resistance [171]. Recent reports have shown a relation between RNA silencing and viroid replication. Viroids are free RNA molecules which do not code for any protein and are similar to circular lncRNAs [172]. Several horticulture crops such as the apple, avocado, grapevine, peach, tomato, etc., are facing viroid infections. The Potato spindle tuber viroid (PSTVd) is a good model for studying viroid–host interactions. PSTVd-induced small RNAs cause the cleavage of several host transcripts coding for pyrophosphatase, callose, etc. [173,174].
The lncRNAs can also quench the function of small RNAs and act as target mimics of the miRNAs [167,175]. The SlLNR1 (lncRNA of tomato), involved in regulating leaf growth and development, contained sequences complementary to the vsiRNA derived from the intergenic region (IR) of the virus. In susceptible lines, viral intergenic siRNAs targeted the SlLNR1 resulting in stunted growth and the curling of leaves [176].
4. Genome Editing (GE) Approaches
Genome editing (GE) offers the most precise tool to modify a target gene by using the endonucleases to create breaks in the DNA strands that lead to both target-specific mutagenesis (addition, removal, or alteration) and gene replacement [177,178]. It offers the advantage of being less expensive, simple to design and execute, and more acceptable. Several GE techniques have been implemented against different viruses and have produced better outcomes (Table 2). By complementing it with NGS, it will become easier to identify the fast-mutating viral sequences and redesign the targets which influence the susceptibility response in host–virus interactions [179]. Genome editing is now emerging and expanding as a modern tool in biotechnology. From basic techniques such as Meganucleases (MNs), ZFNs (zinc finger nucleases), TALENs (transcription activator-like effector nucleases) to advanced techniques and the CRISPR/Cas system. GE methods rely on the introduction of specific mutations to inactivate certain target genes. Modifications like CRISPR ON and OFF, and the use of base editors and prime editors restrict the possibilities of accidental host targets.
4.1. Different Genome Editing (GE) Tools
The onset of GE lies in the use of specialized restriction endonucleases known as Meganucleases (MNs), which could target broad recognition sites of around 12–40 bp [180,181]. These nucleases create the double-stranded breaks (DSBs) that were repaired by non-homologous end joining (NHEJ), resulting in gene mutations [182]. MNs have been studied in model plant systems such as Arabidopsis, Gossypium, and maize. They exhibited a high efficiency in target recognition and a moderate mutation rate. The length of the target sequence recognized by the MN was around 68–88 bp, which was quite long as compared to others, and it did not require another endonuclease [183]. However, the major limitation was that the production of these enzymes was costlier and time-consuming. It was not easy to manipulate the MNs as their catalytic and DNA-binding domains were intermingled and became difficult to detach [184].
Zinc-finger nucleases (ZFN) emerged as specialized cutters that could cleave dsDNA sequences [185,186,187]. ZFN-induced double-strand breaks were subject to cellular DNA repair processes. ZFNs were engineered by fusing the non-specific DNA cleavage domain of Fok1 (Flavobacterium okeanokoites 1) with the Cys2-His2 ZF DNA-binding domain [188,189]. Fok1 is a type II endonuclease that can bind to DNA regions in a non-specific manner, but it can cleave the double strand only when two monomers bind to the target sequence (overall 9–18 bp). Each monomer binds to 5–6 bp-long sequences within the target region, so 4 monomers were required for generating 2 cleavage sites [190]. The breaks are repaired by homology directed repair (HDR) or NHEJ (when homologous sequences are not found) resulting in errors [191]. NHEJ creates deletions or insertions at target sites that further leads to a frame shift and results in gene knockout, while HDJ can insert new sequences at the break site that result in knock-in [192].
ZFNs were used to mutate the AC1 gene, which encodes the Rep protein, in TYLCV [193]. Artificial ZFN proteins (AZPs) have been designed using conserved sequences for blocking the DNA binding sites that are used by viruses [194,195]. For instance, AZPs bind to the IR region of the Beet severe curly top virus (BSCTV) and interfere with the binding of Rep, thus suppressing the replication of the virus in the host [196]. The ZFN-based technique allowed for easy manipulation and could target any site, but the efficiency was limited by chances of off-target cleavage. ZFN-mediated gene editing has been used in many crops such as Arabidopsis, tobacco, maize, and soybean, etc. [182,188,197,198].
TALENs (Transcription Activator-like effectors Nucleases) were produced by fusing the DNA binding domains of transcription activator-like (TAL) effectors to the DNA cleavage domains of Fok1 to induce dsDNA breaks that could be repaired by NHEJ or HDR repair mechanisms [199]. TAL effectors are produced by bacteria Xanthomonas in plants through the type III secretion system [200]. These effectors alter the transcripts of the host by binding with the promoter region via repetitive amino acid residues in the central domain and activating gene expression [201,202]. TALENs were used to generate disease-resistant plants by mutagenizing the susceptible genes in host plants [203]. TALENs were used to develop the resistance of tobacco plants against the Tobacco curly shoot virus (TbCSV) and Tomato leaf curl China virus (TYLCCNV) [204].
Recently, CRISPR-Cas has become popular in the scientific community as it has tremendous potential to improve crop traits (Section 3.2). The attractive features of this technology include its ease, efficiency, and rapidity of genetic manipulations. The CRISPR system is rapidly becoming diverse and now includes different types of base editors, prime editors, and an endogenous tRNA structure. These modifications have given it greater specificity and multiplex targeting abilities, which ultimately make GE more precise (Table 2).
4.2. Manipulation of the Genome through the CRISPR/Cas System
The CRISPR-Cas system was first identified as a unique strategy used by bacteria and archaea for adaptive immune response against invading phages or plasmids [205,206,207]. In this system, the prokaryotes were able to acquire small pieces of the infecting virus and then use it as memory for protection against further attacks. CRISPR stands for cluster regularly interspaced short palindromic repeats, and the Cas protein is the RNA-dependent DNA endonuclease. The Cas protein interacts with two special non-coding RNAs namely CRISPR RNA (crRNA) and transactivating crRNA (tracrRNA). The crRNA functions as a short guide as it consists of a unique 20-base sequence that enables target recognition. The tracrRNA is a trans-encoded small RNA, which provides the stem loop structure [208]. The fusion of the crRNA and tracrRNA generates a synthetic RNA of around 100 nucleotides (nt), called the single guide RNA (gRNA).
In the initial step, Cas9 protein binds to gRNA to form a catalytically active Cas9-gRNA duplex [209,210]. The Cas 9 protein contains two REC domains, an arginine-rich Bridge Helix, a proto-spacer adjacent motif (PAM) interacting domain, and two DNA nuclease domains (HNH and RuvC). The REC I domain is responsible for binding to the single gRNA, while the bridge helix is crucial for initiating cleavage activity upon the binding of target DNA [211]. The PAM domain helps in recognizing the consensus NGG (N, any nucleotide; G, guanine) at the 3′ end of the target DNA strand for initiating the binding of the Cas9 to the target DNA [208,212,213,214]. After binding, the Cas9 unzips the DNA to allow the crRNA (within gRNA) to anneal to the target DNA. After annealing, both the strands are cleaved three bases upstream to the PAM, thereby creating blunt ends at the target site. The DSB can be repaired by the error prone NHEJ or HDJ to generate the mutations.
4.2.1. Cas9 Editing
This technique has been beneficial for targeting DNA and RNA viruses with precision and high efficiency (Table 2) [215,216]. Studies using Arabidopsis thaliana or Nicotiana benthamiana showed that single gRNAs targeting the stem-loop sequence within the geminiviral genome or the non-coding intergenic region (IR) showed a stronger interference as compared with those targeting the viral genes like CP or Rep. When gRNAs were used to target non-coding IRs in the Cotton leaf curl Kokhran virus (CLCuKoV) and Tomato yellow leaf curl Sardinia virus (TYLCSV), their systemic movement in the host was blocked [217]. The gRNAs also rendered a broad-spectrum resistance to different mono- and bipartite geminiviruses [218]. The combination of two single gRNAs against the IR and Rep of Cotton leaf curl Multan virus (CLCuMuV) in a single plasmid enhanced the overall resistance of the tobacco plants [219]. CRISPR/Cas9 based approaches have also been used to target the host susceptibility factors such as the eukaryotic translation initiation protein (eIF4E) to improve resistance against many potyviruses [220,221].
Several extensions or modifications of the CRISPR/Cas system have been developed either by mutating the Cas 9 enzyme or by using other variants of Cas. One of the class two effectors, Cas12a (also called Cpf1), has only a RuvC domain and no HNH domain, hence, it generates staggered DSBs instead of blunt [222,223]. The staggered DSB is close to the 3′ end of the target sequence, which creates a 5′ overhang. This could be beneficial for knock-in experiments and also helpful in the insertion of DNA fragments precisely and with perfect orientation through complementary ends by HDR [223]. Also, it requires shorter crRNA than Cas9 and there is no requirement for tracrRNA as per the current reports [224].
4.2.2. Epigenetic Editors
An engineered Cas variant known as dead Cas9 (dCas9) contains inactivated HNH and RuvC nuclease domains [191]. It targets specific genomic regions by recruiting chromatin modifiers and effector proteins, which leads to programmable epigenetic editing [225,226]. dCas9 can be used as a transcriptional activator (CRISPRa) or transcriptional repressor (CRISPRi) [227]. There is enormous potential for dCas9 to gain insights into plant–virus interactions and for performing transcriptional engineering and chromatin alterations [228].
4.2.3. RNA Editors
The FnCas9 (class 2, Type II) derived from Francisella novicida, has RNA-targeting and editing properties [229,230]. The FnCas9 with RNA-targeting gRNA has been used to cleave the ssRNA viruses for resistance against CMV and TMV in N. benthamiana and A. thaliana [231].
Cas13 is a new type of protein isolated from Ruminococcus flavefaciens that comes in class 2 type VI effectors [232]. It has programmable RNA-targeting properties, which broaden the application of the CRISPR/Cas system [215]. Recent studies have demonstrated the effectiveness of using various Cas13 systems successfully for targeting multiple RNA viruses [233]. Transgenic rice and tobacco harboring LshCas13a (Leptotrichia shahii Cas13) with a specific protospacer showed resistance to the Rice stripe mosaic virus (RSMV), TMV, and the Southern rice black-streaked dwarf virus (SRBSDV) [234]. The effectiveness of using LshCas13a to target the helper component proteinase (HC-Pro) and coat protein (CP) segments of the Turnip Mosaic Virus (TuMV) RNA genome in N. benthamiana and A. thaliana, respectively, has been demonstrated. Cas13 has many variants, of which Cas13d is more advantageous than Cas13a, Cas13b, or others [235]. CRISPR techniques, especially CRISPR-Cas12a and CRISPR-Cas13a/d, are also useful in the detection of DNA and RNA virus amplicons generated through isothermal amplification [236].
4.2.4. Base Editors
These are formed by the fusion of the enzyme deaminase with the Cas nuclease domain. Base editors are used to create point mutations without the requirement of a donor template and DSBs [237,238]. This approach is useful for targeting genes responsible for host susceptibility [239]. Cytosine base editors (CBE) and Adenosine base editors (ABE) are two commonly used editors in base editing. The former converts C:G to T:A and the latter converts A:T to G:C [240]. In this process, the availability of the PAM is important to the editing of the target DNA. Advancements in the base editing technology such as variants of CBEs and ABEs are being used to create randomized mutations in targets [241,242]. In Arabidopsis, resistance to Clover yellow vein virus (ClYVV) was obtained by using a novel CBE to convert a susceptible allele of eIF4E1 into a resistance allele through a point mutation of N176K [239].
4.2.5. Prime Editors
Prime editing was achieved by using an engineered nCas9 fused with a reverse transcriptase and prime-editing guide RNA (pegRNA). The pegRNA has a protospacer sequence that directs a Cas nuclease to cleave a target region and add extra sequences, which are responsible for specific changes [243,244]. Like base editing, this technique does not require double-stranded breaks (DSBs). This technique was used to edit PBS1, a decoy host protein, which is responsible for disease resistance. The cleavage of PBS1 by pathogen proteases such as AvrRpt2 from Pseudomonas syringae and Nla from Turnip mosaic virus (TuMV) generates host immune responses. This protein was edited in such a way that it became amenable to cleavage by other pathogen proteases [245]. Hence, precision editing could be used as a promising tool to edit several immune receptors for improving pathogen resistance.
4.2.6. Multiplexing CRISPR/Cas Editing
Targeting multiple sites on a virus in a single go is being developed as an effective strategy to minimize the chances of viral escape. In cotton, three gRNAs were designed to target three viral regions for modifying six overlapping genes (AV2/AV1, AC2/AC3, and AC1/AC4). The analysis showed relatively low virus titers in the plants transformed with all three gRNAs as compared to plants expressing a single gRNA and the wild-type plants [246].
This includes the simultaneous delivery of Cas9 mRNA with multiple gRNAs expression cassettes in one plasmid, artificial gRNAs with Cas9 flanked by Csy4/HH-HDV ribozymes for the cleavage and release of gRNAs, multiple gRNA precursors with Cas9 flanked by tRNAs, etc. [247]. The coupling of CRISPR/Cas13d with an endogenous tRNA system for the expression of four gRNAs was found to be effective against PVY, PVX, PVS, or PLRV viruses in potatoes [248]. The combination of dCas9 with cytosine/adenosine deaminase and an gRNA complex was used for generating point mutations in targets [237,249].
The multiple gRNA strategy is also helpful in the case of mixed viral infections [250]. In potatoes, CRISPR/Cas13a was coupled with an endogenous tRNA system and four different gRNAs to produce a polycistronic tRNA-gRNA (PTG) against PVY. However, the transgenic lines exhibited similar levels of tolerance as observed in the case of the single gRNA lines, indicating that multiple target sites did not affect viral interference [248].
The major hurdles to the GE approaches are the generation of off-targets due to the constitutive expression of the Cas nuclease or sequence mismatches in gRNA [179]. The error-prone repairing process creates selection pressure on viruses which results in a generation of viral variants having a resistance towards editing. The replacement of a constitutive promoter by a virus inducible promoter for the expression of the Cas nuclease is quite useful for bringing down the off-targets [251].

Table 2.
List of GE approaches available against Plant DNA and RNA viruses.
Table 2.
List of GE approaches available against Plant DNA and RNA viruses.
| Strategy | Virus | Target | Plant | References |
|---|---|---|---|---|
| For DNA Viruses | ||||
| ZFN | Tomato yellow leaf curl China virus, Tobacco curly shoot virus | AC1 gene | Tobacco | [193] |
| Bean yellow dwarf virus | Cis-acting long and short intergenic region | [252] | ||
| TALENs | Tomato yellow leaf curl China virus, Tobacco curly shoot virus, Tomato leaf curl yellow virus | Motif 1 and 2 of AC1 ORF | Tobacco | [204] |
| AZP | Beet severe curly top virus | Rep protein | Arabidopsis | [194,253,254,255] |
| Tomato yellow leaf curl virus | ||||
| Rice tungro bacilliform virus | RTBV promoter | [256] | ||
| CRISPR/Cas9 | Cotton leaf curl Kokhran virus | CP and Rep | Tobacco | [217] |
| Merremia mosaic virus Tomato yellow leaf curl virus | Intergenic region | |||
| Bean severe curly top virus | Viral genome | Tobacco | [257] | |
| Bean yellow dwarf virus | Rep, short and long intergenic region | Tobacco | [258] | |
| Chilli leaf curl virus | Intergenic region and overlapping (V2/V1) and C1/C4) | Tobacco | [259] | |
| Cauliflower mosaic virus | CP gene | Arabidopsis | [260] | |
| Wheat dwarf virus | MP, CP, Rep/RepA, long intergenic | Barley | [261] | |
| Banana streak virus strain Obino I’Evai African cassava mosaic virus | ORF1, ORF2 and ORF3 AC2 and AC3 | Banana Cassava | [262] [263] | |
| For RNA Viruses | ||||
| MNs | Tobacco rattle virus | Dihydroflavonol 4-reductase (DFR) | Tobacco | [264] |
| FnCas9 | Tobacco mosaic virus, Cucumber mosaic virus | ORF1a/2a/2b/3a/CP | Tobacco | [231] |
| Cucumber mosaic virus | Arabidopsis | |||
| LshCas13a | Turnip mosaic virus | HC-Pro | Arabidopsis | [216] |
| Southern rice black-streaked dwarf virus | Viral dsRNA | Rice | [234] | |
| Rice stripe mosaic virus | Viral ssRNA | |||
| Cas13d/PTG | Potato virus X, Potato leafroll virus, Potato virus Y, Potato virus S | CP | Potato | [248] |
| Cas13a/PTG | Potato virus Y | P3, CI, NIb and CP | Potato | [265] |
| Cas13a with multiplex gRNAs | Potato virus Y | PI, HC-Pro, P3, Cl, Cl2, VPg | Potato | [266] |
| LbCas12a | Cotton leaf curl Multan virus | C1, C2/C3, V1 | Cotton | [267] |
| Cas12f | Cotton leaf curl Kokhran virus and Cotton leaf curl Multan virus | CP | Cotton | [268] |
5. Summary and Future Perspectives
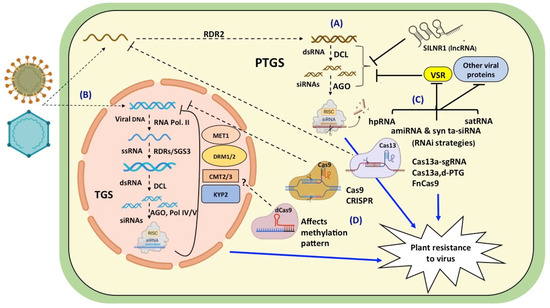
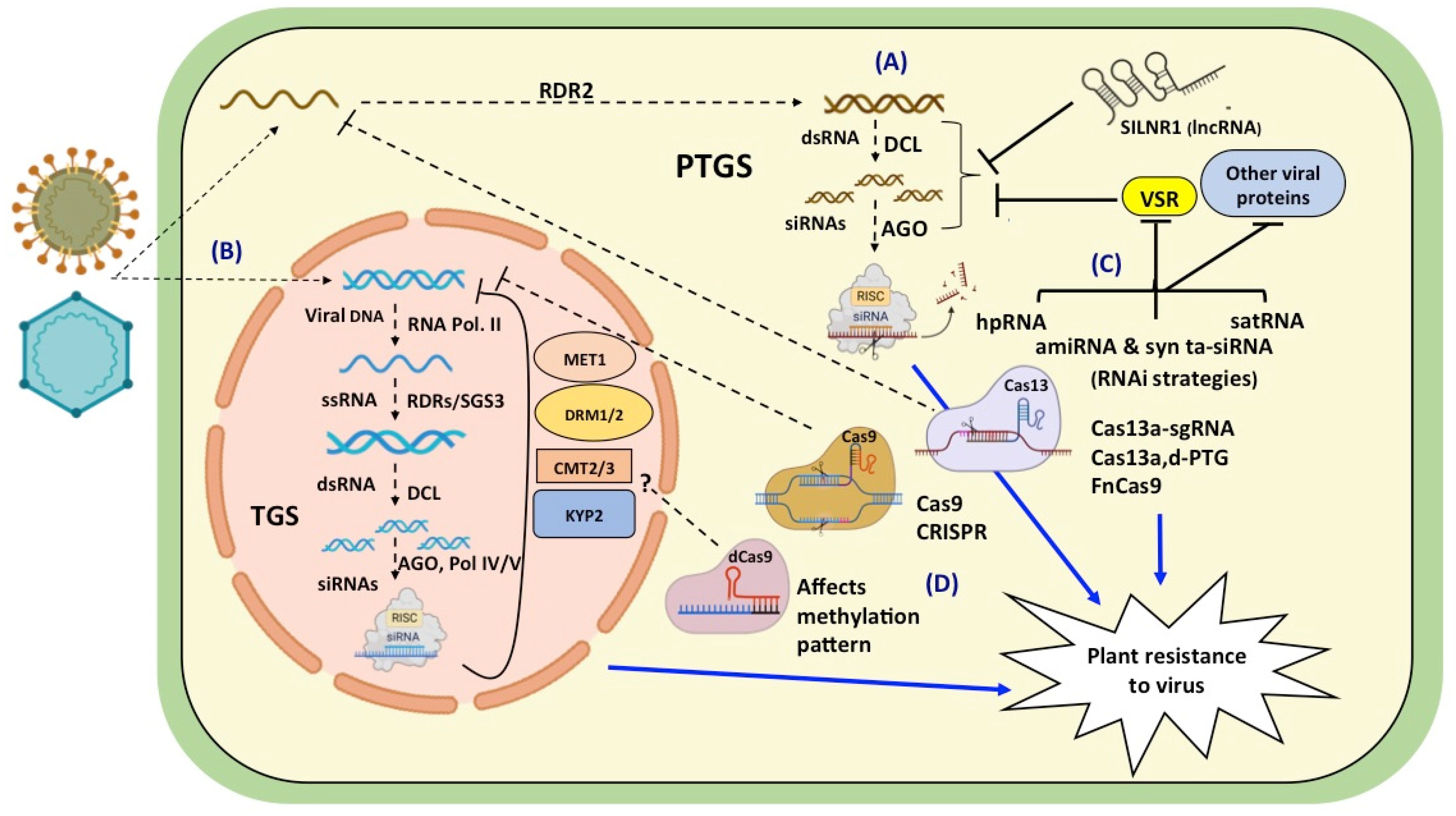
Viruses invade plant cells and hijack the host’s cellular machinery for their own multiplication. The plants fight back by activating cellular defense responses resulting in genetic and epigenetic events to block viral gene expression. Several attempts were made to introgress resistance to a virus in cultivated plants by breeding with its wild relatives, but were unsuccessful. Subsequently, strategies were changed to directly target the virus for conferring resistance. The GM and GE approaches for targeting the virus offered great promise in saving crops from viral infection (Figure 1).
The early GM approaches were based on engineering PDR by over-expressing CP or other viral proteins, and it proved to be effective against many viruses. However, with the discovery and understanding of the underlying principles of RNA silencing and its role in providing resistance to plant viruses, there was a paradigm shift in these strategies. The first silencing tools employed the dsRNA or hpRNA precursors directed against specific viral genes. The transgenic papayas expressing siRNAs against the Papaya ringspot virus (PRSV) proved to be a boon for the Hawaiian papaya industry. The EMBRAPA 5.1 bean was the first RNA silencing-based GM crop approved and cultivated in Brazil [269]. It was developed by transforming plants with the AC1 gene of the Bean golden mosaic virus. The RNAi-based cassava showing resistance to the Cassava brown streak virus was also approved for release by the Kenya national Biosafety authority (NBA). GM technology has helped to save the papaya grown in Hawaii and reduce potato crop losses in the USA and Canada. These case studies pointed out the potential of genetic engineering for generating strong resistances against plant viruses.
The siRNAs had the limitations of being less specific and generating undesirable off targets [270]. This resulted in the development of amiRNA tools by engineering the natural pre-miRNA backbones. These second sets of tools were more specific but had a limited targeting capacity, so syn-tasiRNA vectors were favored and developed as the third set of tools. However, the skepticism regarding the acceptance of GM crops by the public has hindered the active implementation of this technology. Regulations of GM crops vary from country to country, e.g., the USA focusses on the risk assessment of the ultimate food product rather than the process by which it was made [271]. With the help of deep sequencing, a wide range of endogenization of viral genomes was observed in plants which clearly suggests the incorporation of foreign genes is not that unnatural [272]. Also, there is still a lack of evidence in support of claims of the adverse effects of GM crops on humans and animals [273].
As an alternative strategy, the search for an exogenous application of dsRNA precursors or small RNAs to activate the host-silencing pathways was started (Table 3). Several reports have highlighted the use of high pressure (4–6 bar) for the efficient transfer of dsRNA in the nucleus for spray-induced epigenetic modifications (SPIEM). When high pressure spraying was used to transfer 333 bp of dsRNA in N. benthamiana, there was no induction of methylation at the target region at three days post spraying (dps), while 28% of samples were methylated at six dps and 40% of samples were methylated at ten dps [274]. However, spraying with low pressure did not result in any methylation. The RNA-based bioformulations are in various stages of development and are anticipated to prove useful in tackling viral infections.

Table 3.
List of exogenously delivered dsRNAs for the induction of RNA silencing against plant viruses.
The emergence of GE tools has added another arsenal in the fight against viruses infecting crop plants. CRISPR/Cas9 provides a robust system for generating more effective and prolonged resistances against viruses. The recent additions of Cas13 and Cas12 variants have proved very effective in targeting RNA viruses. The efficacy of CRISPR-based approaches primarily depends on the method of delivery and the design of the gRNA sequence. A number of delivery methods including Agrobacterium-mediated T-DNA transformation, protoplast transfection, microprojectile bombardment, and virus-based methods have been deployed to introduce CRISPR/Cas components into plants. As viruses have the tendency to mutate at a faster rate, multiplexing technologies can contribute to precise editing and broadening the range of the targeted viruses. Although the use of CRISPR/Cas for the development of viral-resistant plants has been demonstrated to be effective, further improvements in the technology are awaited to attain efficient, durable, and broad-spectrum resistance.
Author Contributions
A.R. wrote the manuscript and drew the figures. N.S.-M. edited and improved the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by grants from ICGEB. A.R. acknowledges Fellowship support from CSIR.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef]
- Chandra, P.; Awasthi, L.P. Plant virus taxonomy. In Applied Plant Virology; Academic Press: Cambridge, MA, USA, 2020; pp. 421–434. [Google Scholar]
- Jones, R.A. Global plant virus disease pandemics and epidemics. Plants 2021, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Sastry, K.S.; AZitter, T.; Sastry, K.S.; Zitter, T.A. Management of virus and viroid diseases of crops in the tropics. In Plant Virus and Viroid Diseases in the Tropics: Volume 2: Epidemiology and Management; Springer: Berlin/Heidelberg, Germany, 2014; pp. 149–480. [Google Scholar]
- Zhao, Y.; Yang, X.; Zhou, G.; Zhang, T. Engineering plant virus resistance: From RNA silencing to genome editing strategies. Plant Biotechnol. J. 2020, 18, 328–336. [Google Scholar] [CrossRef]
- Johnson, A.A.; Gopal, D.S.; Sudhakar, C. GM Crops for Plant Virus Resistance: A Review. Genet. Modif. Crops Curr. Status Prospect. Chall. 2021, 2, 257–337. [Google Scholar]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef]
- Legg, J.P.; Shirima, R.; Tajebe, L.S.; Guastella, D.; Boniface, S.; Jeremiah, S.; Nsami, E.; Chikoti, P.; Rapisarda, C. Biology and management of Bemisia whitefly vectors of cassava virus pandemics in Africa. Pest Manag. Sci. 2014, 70, 1446–1453. [Google Scholar] [CrossRef]
- Lu, B.; Stubbs, G.; Culver, J.N. Coat protein interactions involved in tobacco mosaic tobamovirus cross-protection. Virology 1998, 248, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Muller, G.W.; Rezende, J.A. Preimmunization: Applications and perspectives in virus disease control. In Diseases of Fruits and Vegetables Volume I: Diagnosis and Management; Springer: Dordrecht, The Netherlands, 2004; pp. 361–395. [Google Scholar]
- Sanford, J.C.; Johnston, S.A. The concept of parasite-derived resistance—Deriving resistance genes from the parasite’s own genome. J. Theor. Biol. 1985, 113, 395–405. [Google Scholar] [CrossRef]
- Nicaise, V. Crop immunity against viruses: Outcomes and future challenges. Front. Plant Sci. 2014, 5, 660. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.C.; Beattie, G.A. An Overview of Plant Defenses against Pathogens and Herbivores. In Plant Pathology and Microbiology; Iowa State University: Ames, IA, USA, 2008. [Google Scholar]
- Thomma, B.P.; Nürnberger, T.; Joosten, M.H. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.H. The complementary genic systems in flax and flax rust. Adv. Genet. 1956, 8, 29–54. [Google Scholar]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Sinha, K.V.; Sopory, S.K.; Sanan-Mishra, N. Influence of virus–host interactions on plant response to abiotic stress. Plant Cell Rep. 2021, 40, 2225–2245. [Google Scholar] [CrossRef] [PubMed]
- Cosson, P.; Schurdi-Levraud, V.; Le, Q.H.; Sicard, O.; Caballero, M.; Roux, F.; Le Gall, O.; Candresse, T.; Revers, F. The RTM resistance to potyviruses in Arabidopsis thaliana: Natural variation of the RTM genes and evidence for the implication of additional genes. PLoS ONE 2012, 7, e39169. [Google Scholar] [CrossRef] [PubMed]
- de Ronde, D.; Butterbach, P.; Kormelink, R. Dominant resistance against plant viruses. Front. Plant Sci. 2014, 5, 307. [Google Scholar] [CrossRef]
- Diaz-Pendon, J.A.; Truniger, V.; Nieto, C.; Garcia-Mas, J.O.R.D.I.; Bendahmane, A.; Aranda, M.A. Advances in understanding recessive resistance to plant viruses. Mol. Plant Pathol. 2004, 5, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Colbert, T.; Till, B.J.; Tompa, R.; Reynolds, S.; Steine, M.N.; Yeung, A.T.; McCallum, C.M.; Comai, L.; Henikoff, S. High-throughput screening for induced point mutations. Plant Physiol. 2001, 126, 480–484. [Google Scholar] [CrossRef]
- Galvez, L.C.; Banerjee, J.; Pinar, H.; Mitra, A. Engineered plant virus resistance. Plant Sci. 2014, 228, 11–25. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Pixley, K.V.; Falck-Zepeda, J.B.; Paarlberg, R.L.; Phillips, P.W.; Slamet-Loedin, I.H.; Dhugga, K.S.; Campos, H.; Gutterson, N. Genome-edited crops for improved food security of smallholder farmers. Nat. Genet. 2022, 54, 364–367. [Google Scholar] [CrossRef]
- Ibrahim, A.B.; Aragão, F.J. RNAi-mediated resistance to viruses in genetically engineered plants. In Plant Gene Silencing: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2015; pp. 81–92. [Google Scholar]
- Mueller, E.; Gilbert, J.; Davenport, G.; Brigneti, G.; Baulcombe, D.C. Homology-dependent resistance: Transgenic virus resistance in plants related to homology-dependent gene silencing. Plant J. 1995, 7, 1001–1013. [Google Scholar] [CrossRef]
- Schwind, N.; Zwiebel, M.; Itaya, A.; Ding, B.; Wang, M.B.; Krczal, G.; Wassenegger, M. RNAi-mediated resistance to Potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol. Plant Pathol. 2009, 10, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Prins, M.; Goldbach, R. The emerging problem of tospovirus infection and nonconventional methods of control. Trends Microbiol. 1998, 6, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, H.; Rajasubramaniam, S.; Rajam, M.V.; Dasgupta, I. RNA-interference in rice against Rice tungro bacilliform virus results in its decreased accumulation in inoculated rice plants. Transgenic Res. 2008, 17, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, D.N.; Martin, D.P.; Thomson, J.A. Transgenic strategies for developing crops resistant to geminiviruses. Plant Sci. 2009, 176, 1–11. [Google Scholar] [CrossRef]
- Sanan-Mishra, N.; Abdul Kader Jailani, A.; Mandal, B.; Mukherjee, S.K. Secondary siRNAs in plants: Biosynthesis, various functions, and applications in virology. Front. Plant Sci. 2021, 12, 610283. [Google Scholar] [CrossRef]
- Tabassum, B.; Nasir, I.A.; Aslam, U.; Husnain, T. How RNA interference combat viruses in plants. In Functional Genomics; InTech: Rijeka, Croatia, 2012; pp. 113–130. [Google Scholar]
- Mette, M.F.; Aufsatz, W.; Van der Winden, J.; Matzke, M.A.; Matzke, A.J.M. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000, 19, 5194–5201. [Google Scholar] [CrossRef] [PubMed]
- Kooter, J.M.; Matzke, M.A.; Meyer, P. Listening to the silent genes: Transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 1999, 4, 340–347. [Google Scholar] [CrossRef]
- Brodersen, P.; Voinnet, O. The diversity of RNA silencing pathways in plants. TRENDS Genet. 2006, 22, 268–280. [Google Scholar] [CrossRef]
- Palauqui, J.C.; Elmayan, T.; Pollien, J.M.; Vaucheret, H. Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997, 16, 4738–4745. [Google Scholar] [CrossRef]
- Voinnet, O.; Vain, P.; Angell, S.; Baulcombe, D.C. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 1998, 95, 177–187. [Google Scholar] [CrossRef]
- Kumakura, N.; Takeda, A.; Fujioka, Y.; Motose, H.; Takano, R.; Watanabe, Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009, 583, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Pélissier, T.; Thalmeir, S.; Kempe, D.; Sänger, H.L.; Wassenegger, M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 1999, 27, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Wassenegger, M. RNA-directed DNA methylation. In Plant Gene Silencing; Springer: Berlin/Heidelberg, Germany, 2000; pp. 83–100. [Google Scholar]
- Melnyk, C.W.; Molnar, A.; Bassett, A.; Baulcombe, D.C. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr. Biol. 2011, 21, 1678–1683. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, C.A.; Mitter, N.; Christie, M.; Smith, N.A.; Waterhouse, P.M.; Carroll, B.J. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 14741–14746. [Google Scholar] [CrossRef]
- Pikaard, C.S.; Haag, J.R.; Ream, T.; Wierzbicki, A.T. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 2008, 13, 390–397. [Google Scholar] [CrossRef]
- Sigman, M.J.; Panda, K.; Kirchner, R.; McLain, L.L.; Payne, H.; Peasari, J.R.; Husbands, A.Y.; Slotkin, R.K.; McCue, A.D. An siRNA-guided ARGONAUTE protein directs RNA polymerase V to initiate DNA methylation. Nat. Plants 2021, 7, 1461–1474. [Google Scholar] [CrossRef]
- Raja, P.; Sanville, B.C.; Buchmann, R.C.; Bisaro, D.M. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 2008, 82, 8997–9007. [Google Scholar] [CrossRef]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011, 14, 267–274. [Google Scholar] [CrossRef]
- Wassenegger, M. The role of the RNAi machinery in heterochromatin formation. Cell 2005, 122, 13–16. [Google Scholar] [CrossRef]
- Aufsatz, W.; Mette, M.; Matzke, A.; Matzke, M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol. Biol. 2004, 54, 793–804. [Google Scholar] [CrossRef]
- Cao, X.; Aufsatz, W.; Zilberman, D.; Mette, M.F.; Huang, M.S.; Matzke, M.; Jacobsen, S.E. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 2003, 13, 2212–2217. [Google Scholar] [CrossRef] [PubMed]
- Pooggin, M.M. How can plant DNA viruses evade siRNA-directed DNA methylation and silencing? Int. J. Mol. Sci. 2013, 14, 15233–15259. [Google Scholar] [CrossRef] [PubMed]
- Raja, P.; Wolf, J.N.; Bisaro, D.M. RNA silencing directed against geminiviruses: Post-transcriptional and epigenetic components. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2010, 1799, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Sanfaçon, H. Symptom recovery in virus-infected plants: Revisiting the role of RNA silencing mechanisms. Virology 2015, 479, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Bischof, S.; Wang, H.; Feng, S.; Lee, T.F.; Teng, C.; Chen, X.; Park, S.Y.; Liu, L.; Gallego-Bartolome, J.; et al. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 2015, 163, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.I.; Eskelin, K.; Bašić, M.; De, S.; Lõhmus, A.; Varjosalo, M.; Mäkinen, K. Molecular insights into the function of the viral RNA silencing suppressor HC-Pro. Plant J. 2016, 85, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Mäkinen, K.; De, S. The significance of methionine cycle enzymes in plant virus infections. Curr. Opin. Plant Biol. 2019, 50, 67–75. [Google Scholar] [CrossRef]
- Vaucheret, H.; Béclin, C.; Fagard, M. Post-transcriptional gene silencing in plants. J. Cell Sci. 2001, 114, 3083–3091. [Google Scholar] [CrossRef]
- Scholthof, K.B.G. Taking Some of the Mystery out of Host∶ Virus Interactions. PLoS Pathog. 2011, 7, e1002033. [Google Scholar] [CrossRef]
- Jaubert, M.; Bhattacharjee, S.; Mello, A.F.; Perry, K.L.; Moffett, P. ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 2011, 156, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Singh, J.; Li, D.; Qu, F. Temperature-dependent survival of Turnip crinkle virus-infected Arabidopsis plants relies on an RNA silencing-based defense that requires dcl2, AGO2, and HEN1. J. Virol. 2012, 86, 6847–6854. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, P.; Obrępalska-Stęplowska, A. A single amino acid substitution in movement protein of tomato torrado virus influences ToTV infectivity in Solanum lycopersicum. Virus Res. 2016, 213, 32–36. [Google Scholar] [CrossRef]
- Chen, P.Y.; Weinmann, L.; Gaidatzis, D.; Pei, Y.; Zavolan, M.; Tuschl, T.; Meister, G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA Soc. 2008, 14, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, R.; Doudna, J.A. dsRNA with 5′ overhangs contribute to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009, 28, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Glick, E.; Zrachya, A.; Levy, Y.; Mett, A.; Gidoni, D.; Belausov, E.; Citovsky, V.; Gafni, Y. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc. Natl. Acad. Sci. USA 2008, 105, 157–161. [Google Scholar] [CrossRef]
- Sharma, K.K.; Bhatnagar-Mathur, P.; Thorpe, T.A. Genetic transformation technology: Status and problems. Vitr. Cell. Dev. Biol. Plant 2005, 41, 102–112. [Google Scholar] [CrossRef]
- Fitchen, J.H.; Beachy, R.N. Genetically engineered protection against viruses in transgenic plants. Annu. Rev. Microbiol. 1993, 47, 739–763. [Google Scholar] [CrossRef]
- Baulcombe, D. Novel strategies for engineering virus resistance in plants. Curr. Opin. Biotechnol. 1994, 5, 117–124. [Google Scholar] [CrossRef]
- Abel, P.P.; Nelson, R.S.; De, B.; Hoffmann, N.; Rogers, S.G.; Fraley, R.T.; Beachy, R.N. Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 1986, 232, 738–743. [Google Scholar] [CrossRef]
- Beachy, R.N. Coat–protein–mediated resistance to tobacco mosaic virus: Discovery mechanisms and exploitation. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 1999, 354, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, I.; Malathi, V.G.; Mukherjee, S.K. Genetic engineering for virus resistance. Curr. Sci. 2003, 84, 341–354. [Google Scholar]
- Nejidat, A.; Beachy, R.N. Transgenic tobacco plants expressing a coat protein gene. Mol. Plant-Microbe Interact. 1990, 3, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Stark, D.M.; Nelson, R.S.; Powell, P.A.; Tumer, N.E.; Beachy, R.N. Transgenic plants that express the coat protein genes of tobacco mosaic virus or alfalfa mosaic virus interfere with disease development of some nonrelated viruses. Phytopathology 1989, 79, 1284–1290. [Google Scholar] [CrossRef]
- Lawson, C.; Kaniewski, W.; Haley, L.; Rozman, R.; Newell, C.; Sanders, P.; Tumer, N.E. Engineering resistance to mixed virus infection in a commercial potato cultivar: Resistance to potato virus X and potato virus Y in transgenic Russet Burbank. Bio/Technology 1990, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.; Kavanagh, T.; Baulcombe, D. Potato virus X as a vector for gene expression in plants. Plant J. 1992, 2, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Taschner, P.E.; Van Marle, G.; Brederode, F.T.; Tumer, N.E.; Bol, J.F. Plants transformed with a mutant alfalfa mosaic virus coat protein gene are resistant to the mutant but not to wild-type virus. Virology 1994, 203, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Reimann-Philipp, U.; Beachy, R.N. December. The mechanism (s) of coat protein-mediated resistance against tobacco mosaic virus. In Seminars in Virology; Academic Press: Cambridge, MA, USA, 1993; Volume 4, pp. 349–356. [Google Scholar]
- Yusibov, V.M.; Loesch-Fries, L.S. N-terminal basic amino acids of alfalfa mosaic virus coat protein involved in the initiation of infection. Virology 1995, 208, 405–407. [Google Scholar] [CrossRef]
- Clark, W.G.; Fitchen, J.; Nejidat, A.; Deom, C.M.; Beachy, R.N. Studies of coat protein-mediated resistance to tobacco mosaic virus (TMV), I.I. Challenge by a mutant with altered virion surface does not overcome resistance conferred by TMV coat protein. J. Gen. Virol. 1995, 76, 2613–2617. [Google Scholar] [CrossRef]
- Asurmendi, S.; Berg, R.H.; Smith, T.J.; Bendahmane, M.; Beachy, R.N. Aggregation of TMV CP plays a role in CP functions and in coat-protein-mediated resistance. Virology 2007, 366, 98–106. [Google Scholar] [CrossRef]
- Lapidot, M.; Gafny, R.; Ding, B.; Wolf, S.; Lucas, W.J.; Beachy, R.N. A dysfunctional movement protein of tobacco mosaic virus that partially modifies the plasmodesmata and limits virus spread in transgenic plants. Plant J. 1993, 4, 959–970. [Google Scholar] [CrossRef]
- Cooper, B.; Lapidot, M.; Heick, J.A.; Dodds, J.A.; Beachy, R.N. A defective movement protein of TMV in transgenic plants confers resistance to multipleviruses whereas the functional analog increases susceptibility. Virology 1995, 206, 307–313. [Google Scholar] [CrossRef]
- Beck, D.L.; Van Dolleweerd, C.J.; Lough, T.J.; Balmori, E.; Voot, D.M.; Andersen, M.T.; O’Brien, I.E.; Forster, R.L. Disruption of virus movement confers broad-spectrum resistance against systemic infection by plant viruses with a triple gene block. Proc. Natl. Acad. Sci. USA 1994, 91, 10310–10314. [Google Scholar] [CrossRef]
- Golemboski, D.B.; Lomonossoff, G.P.; Zaitlin, M. Plants transformed with a tobacco mosaic virus nonstructural gene sequence are resistant to the virus. Proc. Natl. Acad. Sci. USA 1990, 87, 6311–6315. [Google Scholar] [CrossRef]
- Hanson, S.F.; Maxwell, D.P. trans-Dominant inhibition of geminiviral DNA replication by bean golden mosaic geminivirus rep gene mutants. Phytopathology 1999, 89, 480–486. [Google Scholar] [CrossRef]
- Sangaré, A.; Deng, D.; Fauquet, C.M.; Beachy, R.N. Resistance to African cassava mosaic virus conferred by a mutant of the putative NTP-binding domain of the Rep gene (AC1) in Nicotiana benthamiana. Mol. Breed. 1999, 5, 95–102. [Google Scholar] [CrossRef]
- Donson, J.; Kearney, C.M.; Turpen, T.H.; Khan, I.A.; Kurath, G.; Turpen, A.M.; Jones, G.E.; Dawson, W.O.; Lewandowski, D.J. Broad resistance to tobamoviruses is mediated by a modified tobacco mosaic virus replicase transgene. Mol. Plant-Microbe Interact. 1993, 6, 635–642. [Google Scholar] [CrossRef]
- Tenllado, F.; García-Luque, I.; Serra, M.T.; Díaz-Ruíz, J.R. Resistance to pepper mild mottle tobamovirus conferred by the 54-kDa gene sequence in transgenic plants does not require expression of the wild-type 54-kDa protein. Virology 1996, 219, 330–335. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marano, M.R.; Baulcombe, D. Pathogen-derived resistance targeted against the negative-strand RNA of tobacco mosaic virus: RNA strand-specific gene silencing? Plant J. 1998, 13, 537–546. [Google Scholar] [CrossRef]
- Tricoll, D.M.; Carney, K.J.; Russell, P.F.; McMaster, J.R.; Groff, D.W.; Hadden, K.C.; Himmel, P.T.; Hubbard, J.P.; Boeshore, M.L.; Quemada, H.D. Field evaluation of transgenic squash containing single or multiple virus coat protein gene constructs for resistance to cucumber mosaic virus, watermelon mosaic virus 2, and zucchini yellow mosaic virus. Bio/Technology 1995, 13, 1458–1465. [Google Scholar] [CrossRef]
- Greene, A.E.; Allison, R.F. Recombination between viral RNA and transgenic plant transcripts. Science 1994, 263, 1423–1425. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Calvillo, G.; Contreras-Paredes, C.A.; Mora-Macias, J.; Noa-Carrazana, J.C.; Serrano-Rubio, A.A.; Dinkova, T.D.; Carrillo-Tripp, M.; Silva-Rosales, L. Antagonism or synergism between papaya ringspot virus and papaya mosaic virus in Carica papaya is determined by their order of infection. Virology 2016, 489, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.C.; Hsu, Y.H.; Lin, N.S. Satellite RNAs and satellite viruses of plants. Viruses 2009, 1, 1325–1350. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P.; Roossinck, M.J.; Dietzgen, R.G.; Francki, R.I. Cucumber mosaic virus. Adv. Virus Res. 1992, 41, 281–348. [Google Scholar] [PubMed]
- Patil, B.L.; Dasgupta, I. Defective interfering DNAs of plant viruses. Crit. Rev. Plant Sci. 2006, 25, 47–64. [Google Scholar] [CrossRef]
- Burgyan, J.; Grieco, F.; Russo, M. A defective interfering RNA molecule in cymbidium ringspot virus infections. J. Gen. Virol. 1989, 70, 235–239. [Google Scholar] [CrossRef]
- Stanley, J.; Frischmuth, T.; Ellwood, S. Defective viral DNA ameliorates symptoms of geminivirus infection in transgenic plants. Proc. Natl. Acad. Sci. USA 1990, 87, 6291–6295. [Google Scholar] [CrossRef]
- Kong, Q.; Wang, J.; Simon, A.E. Satellite RNA-mediated resistance to turnip crinkle virus in Arabidopsis involves a reduction in virus movement. Plant Cell 1997, 9, 2051–2063. [Google Scholar]
- Budzyńska, D.; Zwart, M.P.; Hasiów-Jaroszewska, B. Defective RNA Particles of Plant Viruses—Origin, Structure and Role in Pathogenesis. Viruses 2022, 14, 2814. [Google Scholar] [CrossRef]
- Shen, W.X.; Au, P.C.K.; Shi, B.J.; Smith, N.A.; Dennis, E.S.; Guo, H.S.; Zhou, C.Y.; Wang, M.B. Satellite RNAs interfere with the function of viral RNA silencing suppressors. Front. Plant Sci. 2015, 6, 281. [Google Scholar] [CrossRef]
- Voinnet, O. Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 2005, 6, 206–220. [Google Scholar] [CrossRef]
- Fahim, M.; Millar, A.A.; Wood, C.C.; Larkin, P.J. Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol. J. 2012, 10, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Kung, Y.J.; Lin, S.S.; Huang, Y.L.; Chen, T.C.; Harish, S.S.; Chua, N.H.; Yeh, S.D. Multiple artificial microRNAs targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded RNA plant virus. Mol. Plant Pathol. 2012, 13, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Niu, Y.; Zhang, K.; Liu, Y.; Zhou, X. Virus-derived transgenes expressing hairpin RNA give immunity to Tobacco mosaic virus and Cucumber mosaic virus. Virol. J. 2011, 8, 41. [Google Scholar] [CrossRef]
- Nicola-Negri, E.D.; Brunetti, A.; Tavazza, M.; Ilardi, V. Hairpin RNA-mediated silencing of Plum pox virus P1 and HC-Pro genes for efficient and predictable resistance to the virus. Transgenic Res. 2005, 14, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, S.; Mochizuki, A.; Nishiguchi, M.; Tabei, Y. Transgenic Nicotiana benthamiana plants resistant to cucumber green mottle mosaic virus based on RNA silencing. Plant Cell Rep. 2007, 26, 1283–1288. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, A.; Ramos, P.L.; Fiallo, E.; Callard, D.; Sánchez, Y.; Peral, R.; Rodríguez, R.; Pujol, M. Intron–hairpin RNA derived from replication associated protein C1 gene confers immunity to Tomato yellow leaf curl virus infection in transgenic tomato plants. Transgenic Res. 2006, 15, 291–304. [Google Scholar] [CrossRef]
- Zrachya, A.; Kumar, P.P.; Ramakrishnan, U.; Levy, Y.; Loyter, A.; Arazi, T.; Lapidot, M.; Gafni, Y. Production of siRNA targeted against TYLCV coat protein transcripts leads to silencing of its expression and resistance to the virus. Transgenic Res. 2007, 16, 385–398. [Google Scholar] [CrossRef]
- Arif, M.; Azhar, U.; Arshad, M.; Zafar, Y.; Mansoor, S.; Asad, S. Engineering broad-spectrum resistance against RNA viruses in potato. Transgenic Res. 2012, 21, 303–311. [Google Scholar] [CrossRef]
- Krubphachaya, P.; Juricek, M.; Kertbundit, S. Induction of RNA-mediated resistance to papaya ringspot virus type W. BMB Rep. 2007, 40, 404–411. [Google Scholar] [CrossRef]
- Tougou, M.; Furutani, N.; Yamagishi, N.; Shizukawa, Y.; Takahata, Y.; Hidaka, S. Development of resistant transgenic soybeans with inverted repeat-coat protein genes of soybean dwarf virus. Plant Cell Rep. 2006, 25, 1213–1218. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, M.J.; Pak, J.H.; Im, H.H.; Lee, D.H.; Kim, K.H.; Lee, J.H.; Kim, D.H.; Choi, H.K.; Jung, H.W.; et al. RNAi-mediated Soybean mosaic virus (SMV) resistance of a Korean Soybean cultivar. Plant Biotechnol. Rep. 2016, 10, 257–267. [Google Scholar] [CrossRef]
- Ludlow, E.J.; Mouradov, A.; Spangenberg, G.C. Post-transcriptional gene silencing as an efficient tool for engineering resistance to white clover mosaic virus in white clover (Trifolium repens). J. Plant Physiol. 2009, 166, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- Vanderschuren, H.; Alder, A.; Zhang, P.; Gruissem, W. Dose-dependent RNAi-mediated geminivirus resistance in the tropical root crop cassava. Plant Mol. Biol. 2009, 70, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, U.K.; Ganapathi, T.R.; Hadapad, A.B. Transgenic banana plants expressing small interfering RNAs targeted against viral replication initiation gene display high-level resistance to banana bunchy top virus infection. J. Gen. Virol. 2012, 93, 1804–1813. [Google Scholar] [CrossRef]
- Soler, N.; Plomer, M.; Fagoaga, C.; Moreno, P.; Navarro, L.; Flores, R.; Pena, L. Transformation of Mexican lime with an intron-hairpin construct expressing untranslatable versions of the genes coding for the three silencing suppressors of Citrus tristeza virus confers complete resistance to the virus. Plant Biotechnol. J. 2012, 10, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Winterhagen, P.; Dubois, C.; Sinn, M.; Wetzel, T.; Reustle, G.M. Gene silencing and virus resistance based on defective interfering constructs in transgenic Nicotiana benthamiana is not linked to accumulation of siRNA. Plant Physiol. Biochem. 2009, 47, 739–742. [Google Scholar] [CrossRef] [PubMed]
- Patil, B.L.; Bagewadi, B.; Yadav, J.S.; Fauquet, C.M. Mapping and identification of cassava mosaic geminivirus DNA-A and DNA-B genome sequences for efficient siRNA expression and RNAi based virus resistance by transient agro-infiltration studies. Virus Res. 2016, 213, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Ogamino, T.; Hiraguri, A.; Nakazono-Nagaoka, E.; Uehara-Ichiki, T.; Nakajima, M.; Akutsu, K.; Omura, T.; Sasaya, T. Strong resistance against Rice grassy stunt virus is induced in transgenic rice plants expressing double-stranded RNA of the viral genes for nucleocapsid or movement proteins as targets for RNA interference. Phytopathology 2013, 103, 513–519. [Google Scholar] [CrossRef]
- Liang, C.; Liu, H.; Hao, J.; Li, J.; Luo, L. Expression profiling and regulatory network of cucumber microRNAs and their putative target genes in response to cucumber green mottle mosaic virus infection. Arch. Virol. 2019, 164, 1121–1134. [Google Scholar] [CrossRef]
- Ai, T.; Zhang, L.; Gao, Z.; Zhu, C.X.; Guo, X. Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol. 2011, 13, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Lafforgue, G.; Martínez, F.; Niu, Q.W.; Chua, N.H.; Daròs, J.A.; Elena, S.F. Improving the effectiveness of artificial microRNA (amiR)-mediated resistance against Turnip mosaic virus by combining two amiRs or by targeting highly conserved viral genomic regions. J. Virol. 2013, 87, 8254–8256. [Google Scholar] [CrossRef] [PubMed]
- Gago-Zachert, S.; Schuck, J.; Weinholdt, C.; Knoblich, M.; Pantaleo, V.; Grosse, I.; Gursinsky, T.; Behrens, S.E. Highly efficacious antiviral protection of plants by small interfering RNAs identified in vitro. Nucleic Acids Res. 2019, 47, 9343–9357. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cheng, X.; Cai, J.; Zhan, L.; Wu, X.; Liu, Q.; Wu, X. Multiple virus resistance using artificial trans-acting siRNAs. J. Virol. Methods 2016, 228, 16–20. [Google Scholar] [CrossRef] [PubMed]
- López-Dolz, L.; Spada, M.; Daròs, J.A.; Carbonell, A. Fine-tune control of targeted RNAi efficacy by plant artificial small RNAs. Nucleic Acids Res. 2020, 48, 6234–6250. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Bazzini, A.A.; Hopp, H.E.; Beachy, R.N.; Asurmendi, S. Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc. Natl. Acad. Sci. USA 2007, 104, 12157–12162. [Google Scholar] [CrossRef]
- Tagami, Y.; Inaba, N.; Kutsuna, N.; Kurihara, Y.; Watanabe, Y. Specific enrichment of miRNAs in Arabidopsis thaliana infected with Tobacco mosaic virus. DNA Res. 2007, 14, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 2015, 4, e05733. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiao, X.; Kong, X.; Hamera, S.; Wu, Y.; Chen, X.; Fang, R.; Yan, Y. A signaling cascade from miR444 to RDR1 in rice antiviral RNA silencing pathway. Plant Physiol. 2016, 170, 2365–2377. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Zhai, Y.; Zhou, L.; Ai, X.; Chen, J.; Yan, F. A Novel miRNA in Rice Associated with the Low Seed Setting Rate Symptom of Rice Stripe Virus. Int. J. Mol. Sci. 2023, 24, 3675. [Google Scholar] [CrossRef]
- Niu, Q.W.; Lin, S.S.; Reyes, J.L.; Chen, K.C.; Wu, H.W.; Yeh, S.D.; Chua, N.H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006, 24, 1420–1428. [Google Scholar] [CrossRef]
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Lafforgue, G.; Martínez, F.; Sardanyés, J.; De La Iglesia, F.; Niu, Q.W.; Lin, S.S.; Solé, R.V.; Chua, N.H.; Daròs, J.A.; Elena, S.F. Tempo and mode of plant RNA virus escape from RNA interference-mediated resistance. J. Virol. 2011, 85, 9686–9695. [Google Scholar] [CrossRef]
- Cisneros, A.E.; Carbonell, A. Artificial small RNA-based silencing tools for antiviral resistance in plants. Plants 2020, 9, 669. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 2006, 18, 1121–1133. [Google Scholar] [CrossRef]
- Ramesh, S.V.; Ratnaparkhe, M.B.; Kumawat, G.; Gupta, G.K.; Husain, S.M. Plant miRNAome and antiviral resistance: A retrospective view and prospective challenges. Virus Genes 2014, 48, 1–14. [Google Scholar] [CrossRef]
- Reynolds, A.; Leake, D.; Boese, Q.; Scaringe, S.; Marshall, W.S.; Khvorova, A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004, 22, 326–330. [Google Scholar] [CrossRef]
- Duan, C.G.; Wang, C.H.; Fang, R.X.; Guo, H.S. Artificial microRNAs highly accessible to targets confer efficient virus resistance in plants. J. Virol. 2008, 82, 11084–11095. [Google Scholar] [CrossRef]
- Van Vu, T.; Choudhury, N.R.; Mukherjee, S.K. Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res. 2013, 172, 35–45. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bartel, B. MicroRNAs directing siRNA biogenesis. Nat. Struct. Mol. Biol. 2005, 12, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.B.; Masuta, C.; Smith, N.A.; Shimura, H. RNA Silencing and Plant Viral Diseases. Mol. Plant-Microbe Interact. 2012, 25, 1275–1285. [Google Scholar] [CrossRef]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Revers, F.; Nicaise, V. Plant Resistance to Infection by Viruses; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014. [Google Scholar]
- Khalid, A.; Zhang, Q.; Yasir, M.; Li, F. Small RNA based genetic engineering for plant viral resistance: Application in crop protection. Front. Microbiol. 2017, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.D.; Gan, Q.H.; Chi, X.Y.; Qin, S. Roles of microRNA in plant defense and virus offense interaction. Plant Cell Rep. 2008, 27, 1571–1579. [Google Scholar] [CrossRef]
- Waterhouse, P.M.; Graham, M.W.; Wang, M.B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 1998, 95, 13959–13964. [Google Scholar] [CrossRef] [PubMed]
- Rosa, C.; Kuo, Y.W.; Wuriyanghan, H.; Falk, B.W. RNA interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610. [Google Scholar] [CrossRef]
- Gaffar, F.Y.; Koch, A. Catch me if you can! RNA silencing-based improvement of antiviral plant immunity. Viruses 2019, 11, 673. [Google Scholar] [CrossRef]
- Voloudakis, A.E.; Kaldis, A.; Patil, B.L. RNA-based vaccination of plants for control of viruses. Annu. Rev. Virol. 2022, 9, 521–548. [Google Scholar] [CrossRef]
- Tenllado, F.; Dıaz-Ruız, J.R. Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 2001, 75, 12288–12297. [Google Scholar] [CrossRef]
- Fuentes, A.; Carlos, N.; Ruiz, Y.; Callard, D.; Sánchez, Y.; Ochagavía, M.E.; Seguin, J.; Malpica-López, N.; Hohn, T.; Lecca, M.R.; et al. Field trial and molecular characterization of RNAi-transgenic tomato plants that exhibit resistance to tomato yellow leaf curl geminivirus. Mol. Plant-Microbe Interact. 2016, 29, 197–209. [Google Scholar] [CrossRef]
- Walsh, H.A.; Vanderschuren, H.; Taylor, S.; Rey, M.E.C. RNA silencing of South African cassava mosaic virus in transgenic cassava expressing AC1/AC4 hp-RNA induces tolerance. Biotechnol. Rep. 2019, 24, e00383. [Google Scholar] [CrossRef]
- Kumar, S.; Tanti, B.; Patil, B.L.; Mukherjee, S.K.; Sahoo, L. RNAi-derived transgenic resistance to Mungbean yellow mosaic India virus in cowpea. PLoS ONE 2017, 12, e0186786. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Mao, L.; Qi, Y. Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 2012, 160, 990–999. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Peragine, A.; Park, M.Y.; Poethig, R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005, 19, 2164–2175. [Google Scholar] [CrossRef]
- Vazquez, F.; Gasciolli, V.; Crété, P.; Vaucheret, H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004, 14, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Gasciolli, V.; Mallory, A.C.; Bartel, D.P.; Vaucheret, H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005, 15, 1494–1500. [Google Scholar] [CrossRef]
- Carbonell, A.; Takeda, A.; Fahlgren, N.; Johnson, S.C.; Cuperus, J.T.; Carrington, J.C. New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 2014, 165, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A. Secondary small interfering RNA-based silencing tools in plants: An update. Front. Plant Sci. 2019, 10, 687. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicki, A.T.; Blevins, T.; Swiezewski, S. Long noncoding RNAs in plants. Annu. Rev. Plant Biol. 2021, 72, 245–271. [Google Scholar] [CrossRef]
- Rai, M.I.; Alam, M.; Lightfoot, D.A.; Gurha, P.; Afzal, A.J. Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Kaya, S.B.; Cagirici, H.B. Long non-coding RNA in plants in the era of reference sequences. Front. Plant Sci. 2020, 11, 276. [Google Scholar] [CrossRef] [PubMed]
- Taliansky, M.; Samarskaya, V.; Zavriev, S.K.; Fesenko, I.; Kalinina, N.O.; Love, A.J. RNA-based technologies for engineering plant virus resistance. Plants 2021, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Gelaw, T.A.; Sanan-Mishra, N. Non-coding RNAs in response to drought stress. Int. J. Mol. Sci. 2021, 22, 12519. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. The state of long non-coding RNA biology. Non-Coding RNA 2018, 4, 17. [Google Scholar] [CrossRef]
- Prasad, A.; Prasad, M. Host-virus interactions mediated by long non-coding RNAs. Virus Res. 2021, 298, 198402. [Google Scholar] [CrossRef]
- Wang, J.; Yu, W.; Yang, Y.; Li, X.; Chen, T.; Liu, T.; Ma, N.; Yang, X.; Liu, R.; Zhang, B. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 2015, 5, 16946. [Google Scholar] [CrossRef]
- Kang, S.H.; Sun, Y.D.; Atallah, O.O.; Huguet-Tapia, J.C.; Noble, J.D.; Folimonova, S.Y. A long non-coding RNA of Citrus tristeza virus: Role in the virus interplay with the host immunity. Viruses 2019, 11, 436. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Bujarski, J.J. Long noncoding RNAs in plant viroids and viruses: A review. Pathogens 2020, 9, 765. [Google Scholar] [CrossRef]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Di Serio, F. Viroids: How to infect a host and cause disease without encoding proteins. Biochimie 2012, 94, 1474–1480. [Google Scholar] [CrossRef] [PubMed]
- Dadami, E.; Boutla, A.; Vrettos, N.; Tzortzakaki, S.; Karakasilioti, I.; Kalantidis, K. DICER-LIKE 4 but not DICER-LIKE 2 may have a positive effect on potato spindle tuber viroid accumulation in Nicotiana benthamiana. Mol. Plant 2013, 6, 232–234. [Google Scholar] [CrossRef]
- Karlik, E.; Ari, S.; Gozukirmizi, N. LncRNAs: Genetic and epigenetic effects in plants. Biotechnol. Biotechnol. Equip. 2019, 33, 429–439. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, T.; Shen, D.; Wang, J.; Ling, X.; Hu, Z.; Chen, T.; Hu, J.; Huang, J.; Yu, W.; et al. Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLoS Pathog. 2019, 15, e1007534. [Google Scholar] [CrossRef] [PubMed]
- Bak, R.O.; Gomez-Ospina, N.; Porteus, M.H. Gene editing on center stage. Trends Genet. 2018, 34, 600–611. [Google Scholar] [CrossRef]
- Saurabh, S. Genome editing: Revolutionizing the crop improvement. Plant Mol. Biol. Rep. 2021, 39, 752–772. [Google Scholar] [CrossRef]
- Mushtaq, M.; Mukhtar, S.; Sakina, A.; Dar, A.A.; Bhat, R.; Deshmukh, R.; Molla, K.; Kundoo, A.A.; Dar, M.S. Tweaking genome-editing approaches for virus interference in crop plants. Plant Physiol. Biochem. 2020, 147, 242–250. [Google Scholar] [CrossRef]
- Jurica, M.S.; Stoddard, B.L. Homing endonucleases: Structure, function and evolution. Cell. Mol. Life Sci. CMLS 1999, 55, 1304–1326. [Google Scholar] [CrossRef]
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome editing in rice: Recent advances, challenges, and future implications. Front. Plant Sci. 2018, 9, 1361. [Google Scholar] [CrossRef]
- Townsend, J.A.; Wright, D.A.; Winfrey, R.J.; Fu, F.; Maeder, M.L.; Joung, J.K.; Voytas, D.F. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 2009, 459, 442–445. [Google Scholar] [CrossRef]
- Ahmar, S.; Saeed, S.; Khan, M.H.U.; Ullah Khan, S.; Mora-Poblete, F.; Kamran, M.; Faheem, A.; Maqsood, A.; Rauf, M.; Saleem, S.; et al. A revolution toward gene-editing technology and its application to crop improvement. Int. J. Mol. Sci. 2020, 21, 5665. [Google Scholar] [CrossRef]
- Puchta, H. The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 2005, 56, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, K.; Osakabe, Y.; Toki, S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 12034–12039. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Maeder, M.L.; Unger-Wallace, E.; Hoshaw, J.P.; Reyon, D.; Christian, M.; Li, X.; Pierick, C.J.; Dobbs, D.; Peterson, T.; et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 12028–12033. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Curtin, S.J.; Zhang, F.; Sander, J.D.; Haun, W.J.; Starker, C.; Baltes, N.J.; Reyon, D.; Dahlborg, E.J.; Goodwin, M.J.; Coffman, A.P.; et al. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 2011, 156, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Curtin, S.J.; Voytas, D.F.; Stupar, R.M. Genome engineering of crops with designer nucleases. Plant Genome 2012, 5. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ren, S.; Yu, S.; Pan, H.; Li, T.; Ge, S.; Zhang, J.; Xia, N. Methods favoring homology-directed repair choice in response to CRISPR/Cas9 induced-double strand breaks. Int. J. Mol. Sci. 2020, 21, 6461. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Qian, Y.; Wu, X.; Sun, Y.; Wu, X.; Cheng, X. Inhibiting replication of begomoviruses using artificial zinc finger nucleases that target viral-conserved nucleotide motif. Virus Genes 2014, 48, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Sera, T. Inhibition of virus DNA replication by artificial zinc finger proteins. J. Virol. 2005, 79, 2614–2619. [Google Scholar] [CrossRef]
- Khan, Z.; Khan, S.H.; Ahmed, A.; Iqbal, M.U.; Mubarik, M.S.; Ghouri, M.Z.; Ahmad, F.; Yaseen, S.; Ali, Z.; Khan, A.A.; et al. Genome editing in cotton: Challenges and opportunities. J. Cotton Res. 2023, 6, 3. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Lloyd, A.; Plaisier, C.L.; Carroll, D.; Drews, G.N. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 2232–2237. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef]
- Kay, S.; Bonas, U. How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 2009, 12, 37–43. [Google Scholar] [CrossRef]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef]
- Yang, B.; Sugio, A.; White, F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 2006, 103, 10503–10508. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, F.; Cai, J.; Chen, W.; Zhao, N.; Sun, Y.; Guo, Y.; Yang, X.; Wu, X. Artificial TALE as a convenient protein platform for engineering broad-spectrum resistance to begomoviruses. Viruses 2015, 7, 4772–4782. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; Embden, J.D.V.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.V.; Nuñez, J.K.; Doudna, J.A. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S. The heroes of CRISPR. Cell 2016, 164, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Konermann, S.; Shehata, S.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O.; Nishimasu, H.; Ran, F.; Hsu, P. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Biophys. J. 2014, 106, 695a. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1. [Google Scholar] [CrossRef]
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.E.A.; Mahfouz, M.M. CRISPR/Cas9-mediated immunity to geminiviruses: Differential interference and evasion. Sci. Rep. 2016, 6, 26912. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Abul-Faraj, A.; Li, L.; Ghosh, N.; Piatek, M.; Mahjoub, A.; Aouida, M.; Piatek, A.; Baltes, N.J.; Voytas, D.F.; et al. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant 2015, 8, 1288–1291. [Google Scholar] [CrossRef]
- Khan, Z.A.; Kumar, R.; Dasgupta, I. CRISPR/Cas-mediated resistance against viruses in plants. Int. J. Mol. Sci. 2022, 23, 2303. [Google Scholar] [CrossRef]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef]
- Lucioli, A.; Tavazza, R.; Baima, S.; Fatyol, K.; Burgyan, J.; Tavazza, M. CRISPR-Cas9 Targeting of the eIF4E1 gene extends the potato virus Y resistance spectrum of the Solanum tuberosum L. cv. Desirée. Front. Microbiol. 2022, 13, 873930. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Fonfara, I.; Richter, H.; Bratovič, M.; Le Rhun, A.; Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016, 532, 517–521. [Google Scholar] [CrossRef]
- Selma, S.; Orzáez, D. Perspectives for epigenetic editing in crops. Transgenic Res. 2021, 30, 381–400. [Google Scholar] [CrossRef] [PubMed]
- Uranga, M.; Daròs, J.A. Tools and targets: The dual role of plant viruses in CRISPR–Cas genome editing. Plant Genome 2023, 16, e20220. [Google Scholar] [CrossRef]
- Khatodia, S.; Bhatotia, K.; Passricha, N.; Khurana, S.M.P.; Tuteja, N. The CRISPR/Cas genome-editing tool: Application in improvement of crops. Front. Plant Sci. 2016, 7, 506. [Google Scholar] [CrossRef] [PubMed]
- Dubois, A.; Roudier, F. Deciphering plant chromatin regulation via CRISPR/dCas9-based epigenome engineering. Epigenomes 2021, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Price, A.A.; Sampson, T.R.; Ratner, H.K.; Grakoui, A.; Weiss, D.S. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc. Natl. Acad. Sci. USA 2015, 112, 6164–6169. [Google Scholar] [CrossRef]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, Q.; Yi, X.; An, H.; Zhao, Y.; Ma, S.; Zhou, G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 2018, 16, 1415–1423. [Google Scholar] [CrossRef]
- Marqués, M.C.; Sánchez-Vicente, J.; Ruiz, R.; Montagud-Martínez, R.; Márquez-Costa, R.; Gómez, G.; Carbonell, A.; Daròs, J.A.; Rodrigo, G. Diagnostics of infections produced by the plant viruses TMV, TEV, and PVX with CRISPR-Cas12 and CRISPR-Cas13. ACS Synth. Biol. 2022, 11, 2384–2393. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Kavuri, N.R.; Ramasamy, M.; Qi, Y.; Mandadi, K. Applications of CRISPR/Cas13-based RNA editing in plants. Cells 2022, 11, 2665. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.; Ye, J.; Cao, X.; Xu, C.; Chen, B.; An, H.; Jiao, Y.; Zhang, F.; Yang, X.; et al. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019, 17, 1185. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Ghosh, A.; Chakravarti, R.; Singh, R.; Ravichandiran, V.; Swarnakar, S.; Ghosh, D. Cas13d: A new molecular scissor for transcriptome engineering. Front. Cell Dev. Biol. 2022, 10, 866800. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Molla, K.A.; Yang, Y. CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef] [PubMed]
- Bastet, A.; Zafirov, D.; Giovinazzo, N.; Guyon-Debast, A.; Nogué, F.; Robaglia, C.; Gallois, J.L. Mimicking natural polymorphism in eIF 4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 2019, 17, 1736–1750. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Li, C.; Zhang, R.; Meng, X.; Chen, S.; Zong, Y.; Lu, C.; Qiu, J.L.; Chen, Y.H.; Li, J.; Gao, C. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 2020, 38, 875–882. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Jiang, Z.; Hong, X.; Zhang, S.; Yao, R.; Xiao, Y.I. CRISPR base editing and prime editing: DSB and template-free editing systems for bacteria and plants. Synth. Syst. Biotechnol. 2020, 5, 277–292. [Google Scholar]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.M.; Yuan, G.; Chen, J.G.; Tuskan, G.A.; Yang, X. Prime editing technology and its prospects for future applications in plant biology research. BioDesign Res. 2020, 2020, 9350905. [Google Scholar] [CrossRef] [PubMed]
- Binyameen, B.; Khan, Z.; Khan, S.H.; Ahmad, A.; Munawar, N.; Mubarik, M.S.; Riaz, H.; Ali, Z.; Khan, A.A.; Qusmani, A.T. Using multiplexed CRISPR/Cas9 for suppression of cotton leaf curl virus. Int. J. Mol. Sci. 2021, 22, 12543. [Google Scholar] [CrossRef]
- Cao, J.; Xiao, Q.; Yan, Q. The multiplexed CRISPR targeting platforms. Drug Discov. Today Technol. 2018, 28, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Liu, W.; Nie, B.; Zhang, F.; Zhang, J. Cas13d-mediated multiplex RNA targeting confers a broad-spectrum resistance against RNA viruses in potato. Commun. Biol. 2023, 6, 855. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.; Alshareef, S.; Mahfouz, M.M. CRISPR base editors: Genome editing without double-stranded breaks. Biochem. J. 2018, 475, 1955–1964. [Google Scholar] [CrossRef]
- Mubarik, M.S.; Wang, X.; Khan, S.H.; Ahmad, A.; Khan, Z.; Amjid, M.W.; Razzaq, M.K.; Ali, Z.; Azhar, M.T. Engineering broad-spectrum resistance to cotton leaf curl disease by CRISPR-Cas9 based multiplex editing in plants. GM Crops Food 2021, 12, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Si, X.; Zhang, Y.; Zhang, H.; Zhang, F.; Gao, C. Conferring DNA virus resistance with high specificity in plants using virus-inducible genome-editing system. Genome Biol. 2018, 19, 197. [Google Scholar] [CrossRef] [PubMed]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. Plant Cell 2014, 26, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, K.; Koshino-Kimura, Y.; Aoyama, Y.; Sera, T. November. Inhibition of tomato yellow leaf curl virus replication by artificial zinc-finger proteins. Nucleic Acids Symp. Ser. 2007, 51, 429–430. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koshino-Kimura, Y.; Takenaka, K.; Domoto, F.; Ohashi, M.; Miyazaki, T.; Aoyama, Y.; Sera, T. September. Construction of plants resistant to TYLCV by using artificial zinc-finger proteins. Nucleic Acids Symp. Ser. 2009, 53, 281–282. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Takenaka, K.; Domoto, F.; Aoyama, Y.; Sera, T. Inhibition of binding of tomato yellow leaf curl virus rep to its replication origin by artificial zinc-finger protein. Mol. Biotechnol. 2013, 54, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Ordiz, M.I.; Magnenat, L.; Barbas, C.F.; Beachy, R.N. Negative regulation of the RTBV promoter by designed zinc finger proteins. Plant Mol. Biol. 2010, 72, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 15144. [Google Scholar] [CrossRef]
- Baltes, N.J.; Hummel, A.W.; Konecna, E.; Cegan, R.; Bruns, A.N.; Bisaro, D.M.; Voytas, D.F. Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants 2015, 1, 15145. [Google Scholar] [CrossRef]
- Roy, A.; Zhai, Y.; Ortiz, J.; Neff, M.; Mandal, B.; Mukherjee, S.K.; Pappu, H.R. Multiplexed editing of a begomovirus genome restricts escape mutant formation and disease development. PLoS ONE 2019, 14, e0223765. [Google Scholar] [CrossRef]
- Liu, H.; Soyars, C.L.; Li, J.; Fei, Q.; He, G.; Peterson, B.A.; Meyers, B.C.; Nimchuk, Z.L.; Wang, X. CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant Direct 2018, 2, e00047. [Google Scholar] [CrossRef]
- Kis, A.; Hamar, É.; Tholt, G.; Bán, R.; Havelda, Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 2019, 17, 1004. [Google Scholar] [CrossRef]
- Tripathi, J.N.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2019, 2, 46. [Google Scholar] [CrossRef]
- Mehta, D.; Stürchler, A.; Anjanappa, R.B.; Zaidi, S.S.E.A.; Hirsch-Hoffmann, M.; Gruissem, W.; Vanderschuren, H. Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 2019, 20, 80. [Google Scholar] [CrossRef]
- Honig, A.; Marton, I.; Rosenthal, M.; Smith, J.J.; Nicholson, M.G.; Jantz, D.; Zuker, A.; Vainstein, A. Transient expression of virally delivered meganuclease in planta generates inherited genomic deletions. Mol. Plant 2015, 8, 1292–1294. [Google Scholar] [CrossRef]
- Zhan, X.; Zhang, F.; Zhong, Z.; Chen, R.; Wang, Y.; Chang, L.; Bock, R.; Nie, B.; Zhang, J. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 2019, 17, 1814–1822. [Google Scholar] [CrossRef]
- Noureen, A.; Zuhaib Khan, M.; Amin, I.; Zainab, T.; Ahmad, N.; Haider, S.; Mansoor, S. Broad-spectrum resistance against multiple PVY-strains by CRSIPR/Cas13 system in Solanum tuberosum crop. GM Crops Food 2022, 13, 97–111. [Google Scholar] [CrossRef]
- Ashraf, S.; Ahmad, A.; Khan, S.H.; Jamil, A.; Sadia, B.; Brown, J.K. LbCas12a mediated suppression of Cotton leaf curl Multan virus. Front. Plant Sci. 2023, 14, 1233295. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Faiq, A.; Khan, M.Z.; Mansoor, S.; Amin, I. Fully Transient CRISPR/Cas12f system in plants capable of broad-spectrum resistance against Begomovirus. bioRxiv 2022. [Google Scholar] [CrossRef]
- De Faria, J.C.; Aragão, F.J.L.; Souza, T.L.P.O.; Quintela, E.D.; Kitajima, E.W.; Ribeiro, S.D.G. Golden Mosaic of Common Beans in Brazil: Management with a Transgenic Approach. APSnet Featur. Artic. 2016. [Google Scholar] [CrossRef]
- Liu, Q.; Li, Y.; Xu, K.; Li, D.; Hu, H.; Zhou, F.; Song, P.; Yu, Y.; Wei, Q.; Liu, Q.; et al. Clay nanosheet-mediated delivery of recombinant plasmids expressing artificial miRNAs via leaf spray to prevent infection by plant DNA viruses. Hortic. Res. 2020, 7, 179. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W. Strategies for the development of GM crops in accordance with the environmental risk assessment (I). J. Plant Biotechnol. 2011, 38, 125–129. [Google Scholar] [CrossRef]
- Chiba, S.; Kondo, H.; Tani, A.; Saisho, D.; Sakamoto, W.; Kanematsu, S.; Suzuki, N. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 2011, 7, e1002146. [Google Scholar] [CrossRef] [PubMed]
- Niraula, P.M.; Fondong, V.N. Development and adoption of genetically engineered plants for virus resistance: Advances, opportunities and challenges. Plants 2021, 10, 2339. [Google Scholar] [CrossRef]
- Rego-Machado, C.M.; Nakasu, E.Y.; Silva, J.M.; Lucinda, N.; Nagata, T.; Inoue-Nagata, A.K. siRNA biogenesis and advances in topically applied dsRNA for controlling virus infections in tomato plants. Sci. Rep. 2020, 10, 22277. [Google Scholar] [CrossRef]
- Namgial, T.; Kaldis, A.; Chakraborty, S.; Voloudakis, A. Topical application of double-stranded RNA molecules containing sequences of Tomato leaf curl virus and Cucumber mosaic virus confers protection against the cognate viruses. Physiol. Mol. Plant Pathol. 2019, 108, 101432. [Google Scholar] [CrossRef]
- Delgado-Martín, J.; Ruiz, L.; Janssen, D.; Velasco, L. Exogenous application of dsRNA for the control of viruses in cucurbits. Front. Plant Sci. 2022, 13, 895953. [Google Scholar] [CrossRef]
- Dalakouras, A.; Ganopoulos, I. Induction of promoter DNA methylation upon high-pressure spraying of double-stranded RNA in plants. Agronomy 2021, 11, 789. [Google Scholar] [CrossRef]
- Lau, S.E.; Mazumdar, P.; Hee, T.W.; Song, A.L.A.; Othman, R.Y.; Harikrishna, J.A. Crude extracts of bacterially-expressed dsRNA protect orchid plants against Cymbidium mosaic virus during transplantation from in vitro culture. J. Hortic. Sci. Biotechnol. 2014, 89, 569–576. [Google Scholar] [CrossRef]
- Holeva, M.C.; Sklavounos, A.; Rajeswaran, R.; Pooggin, M.M.; Voloudakis, A.E. Topical application of double-stranded RNA targeting 2b and CP genes of Cucumber mosaic virus protects plants against local and systemic viral infection. Plants 2021, 10, 963. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yu, T.; Zhang, D.; Song, H.; Huang, K.; Wang, Y.; Shen, L.; Li, Y.; Wang, F.; Zhang, S.; et al. Evaluation of the anti-viral efficacy of three different dsRNA nanoparticles against potato virus Y using various delivery methods. Ecotoxicol. Environ. Saf. 2023, 255, 114775. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).