Research on the Identification Method of Maize Seed Origin Using NIR Spectroscopy and GAF-VGGNet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Material
2.2. Instruments and Equipment
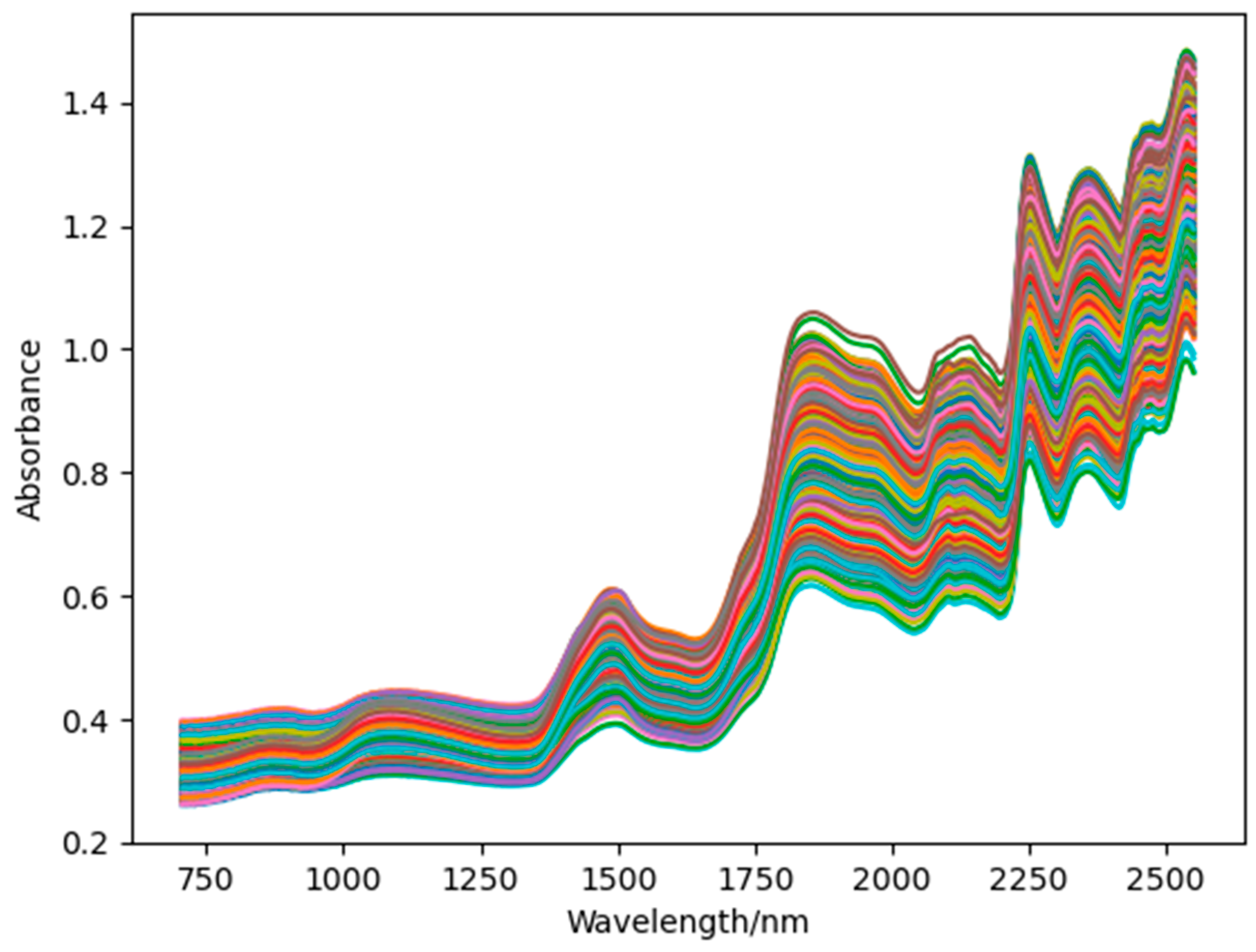
2.3. Spectral Information Acquisition
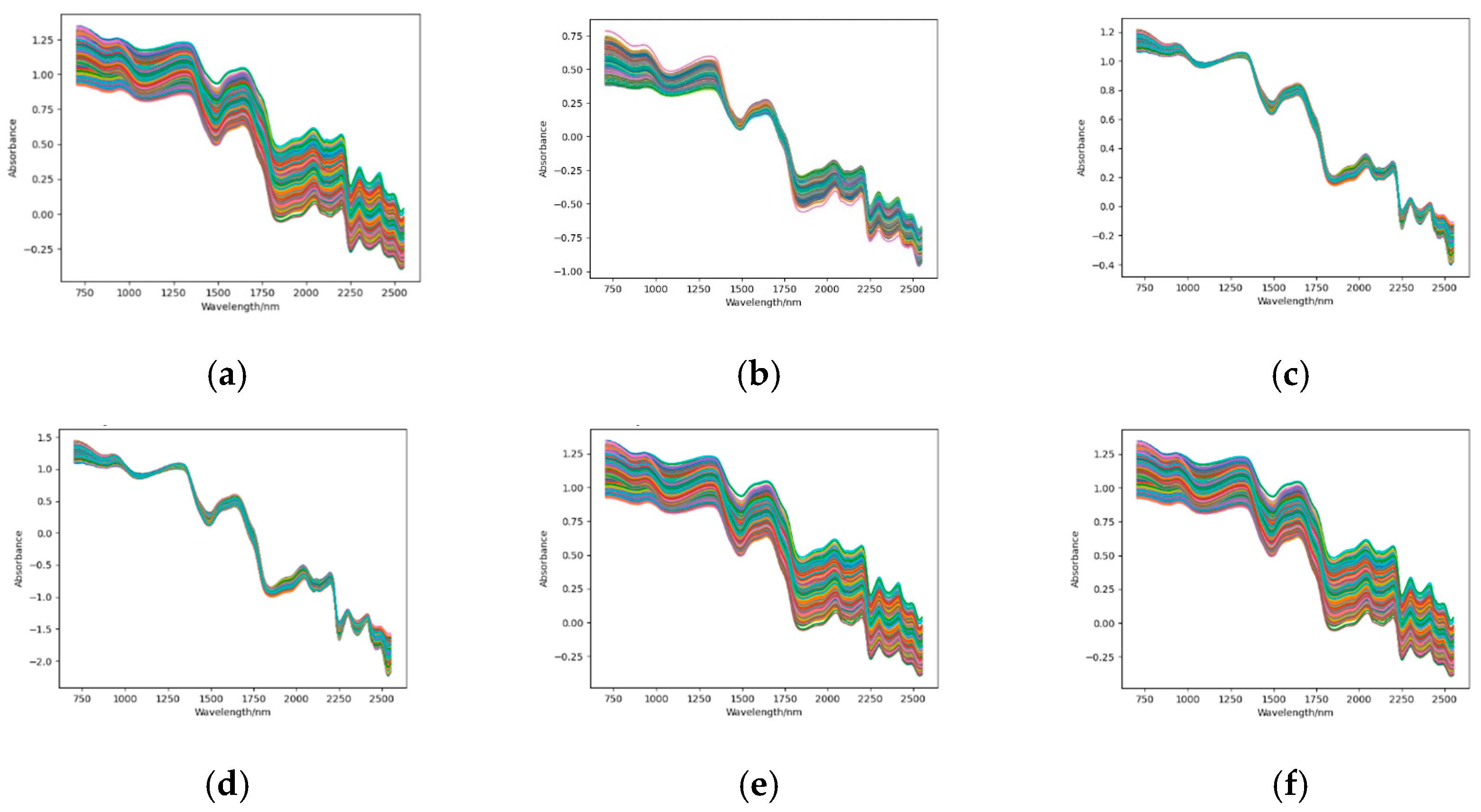
2.4. Spectral Preprocessing
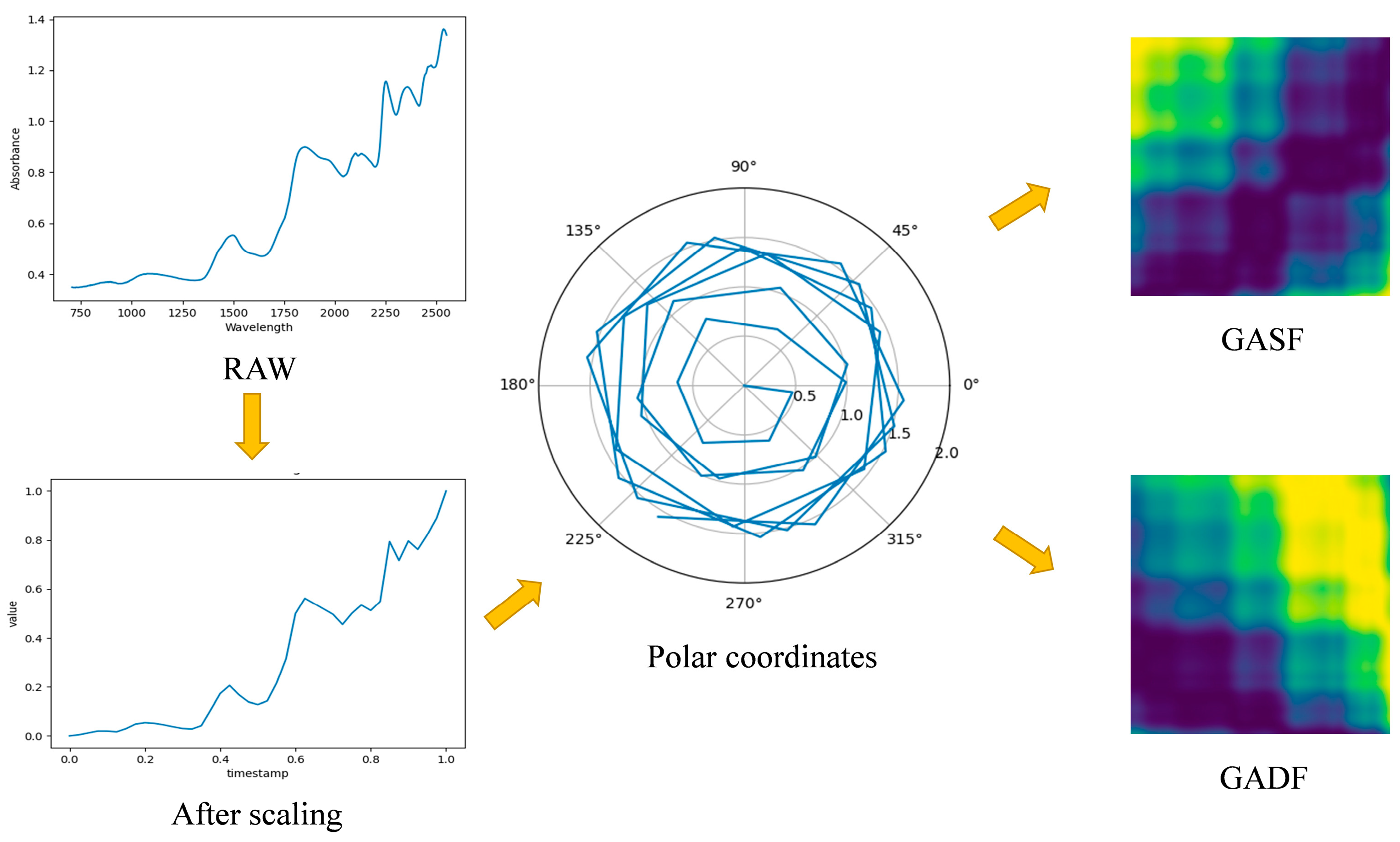
2.5. Near-Infrared Spectral Feature Map Conversion
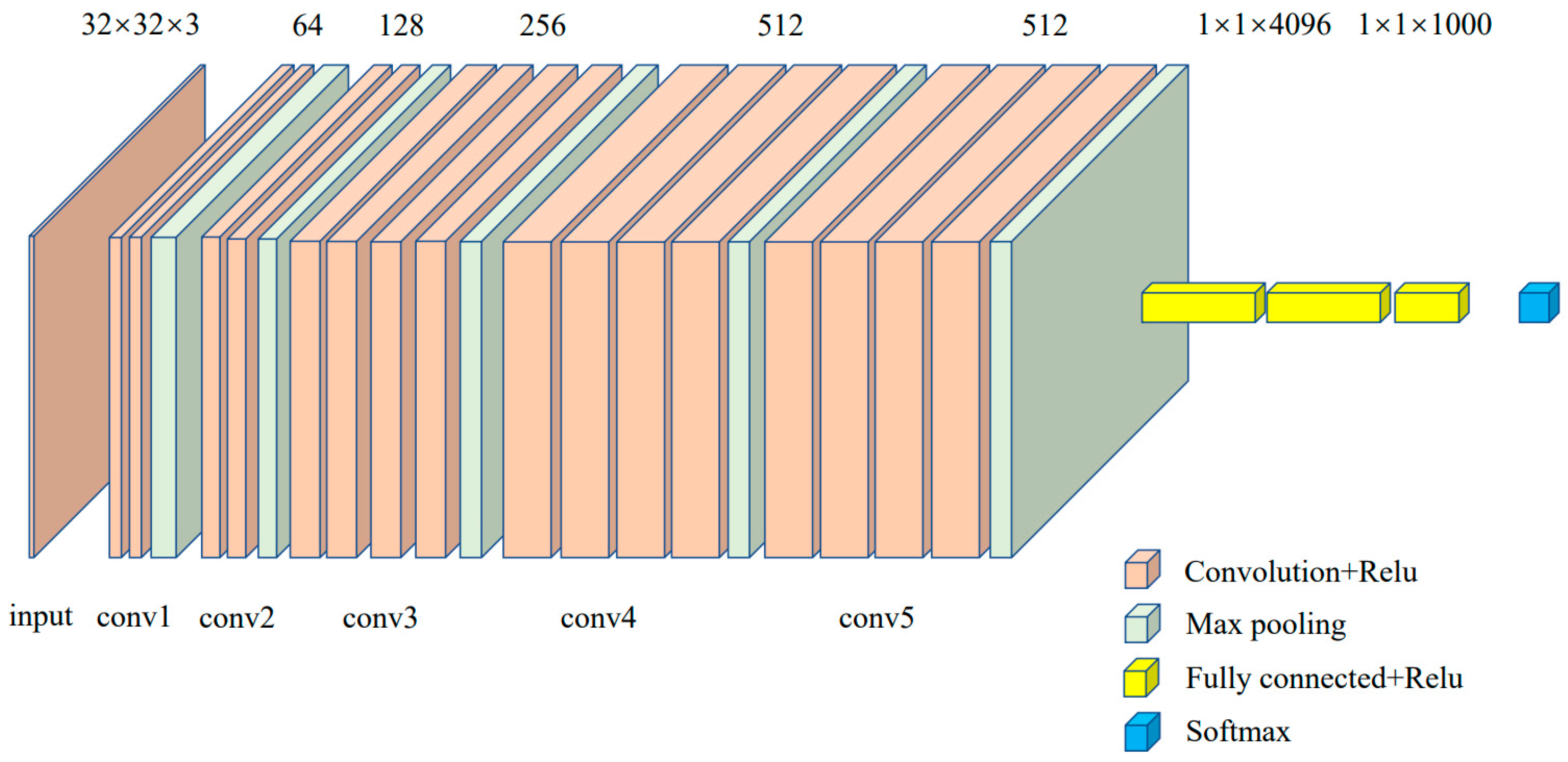
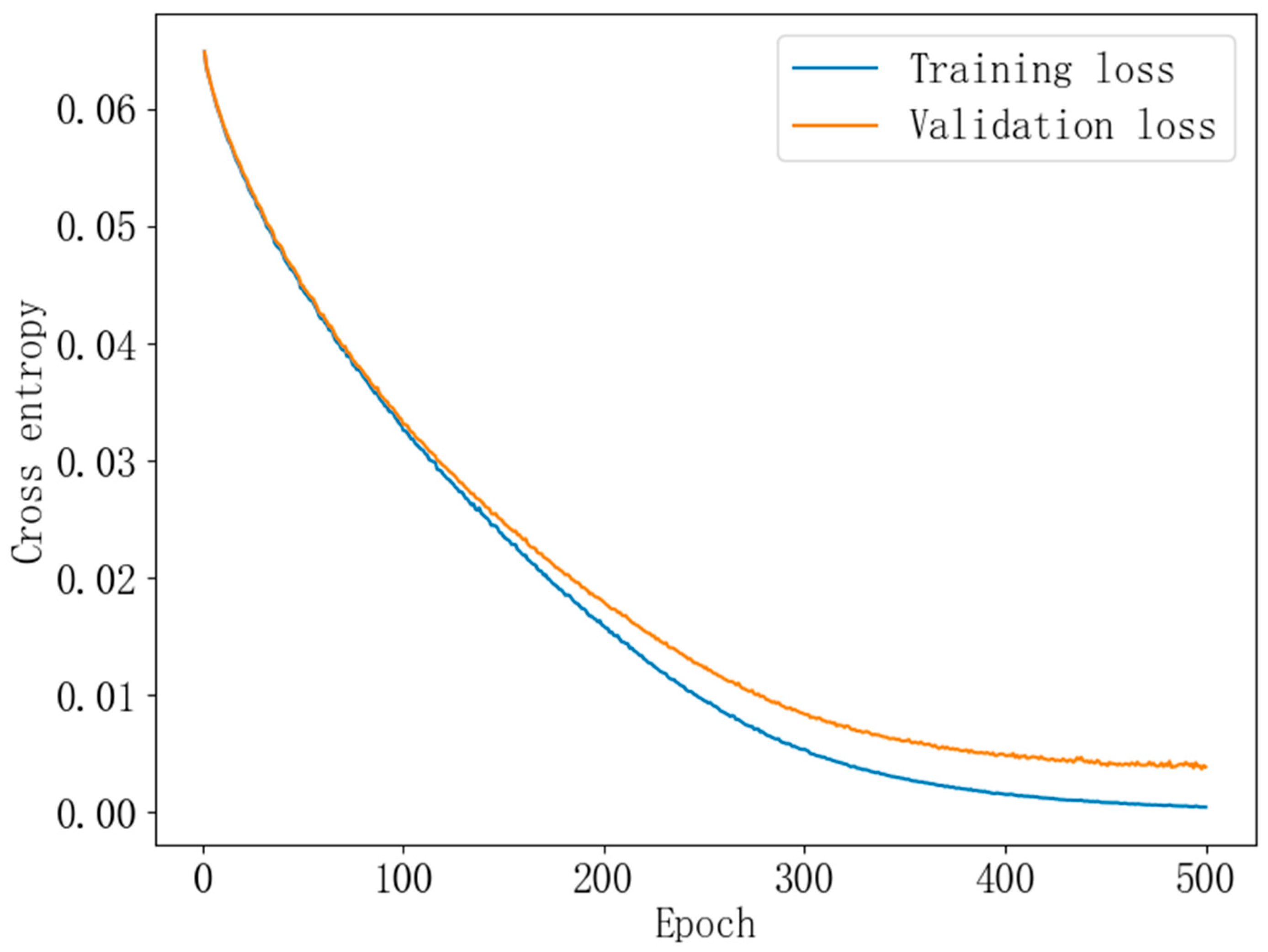
2.6. Model Building
2.7. Model Evaluation Criteria
3. Results and Discussion
3.1. Spectral Acquisition and Preprocessing
3.2. Building Datasets
3.3. Effect of Batch Size on Modeling
3.4. Impact of Learning Rate on the Model
3.5. Impact of Dropout on the Model
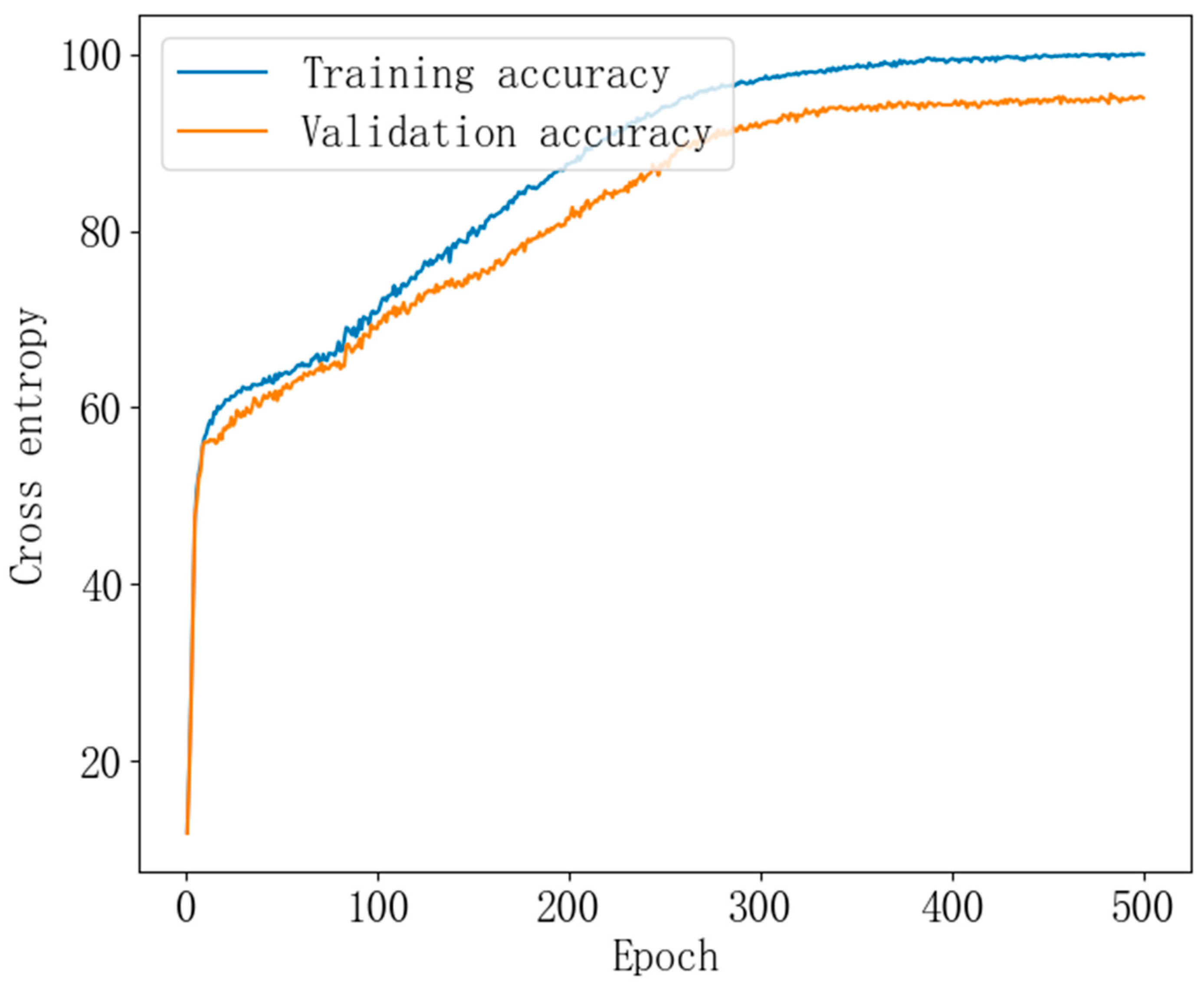
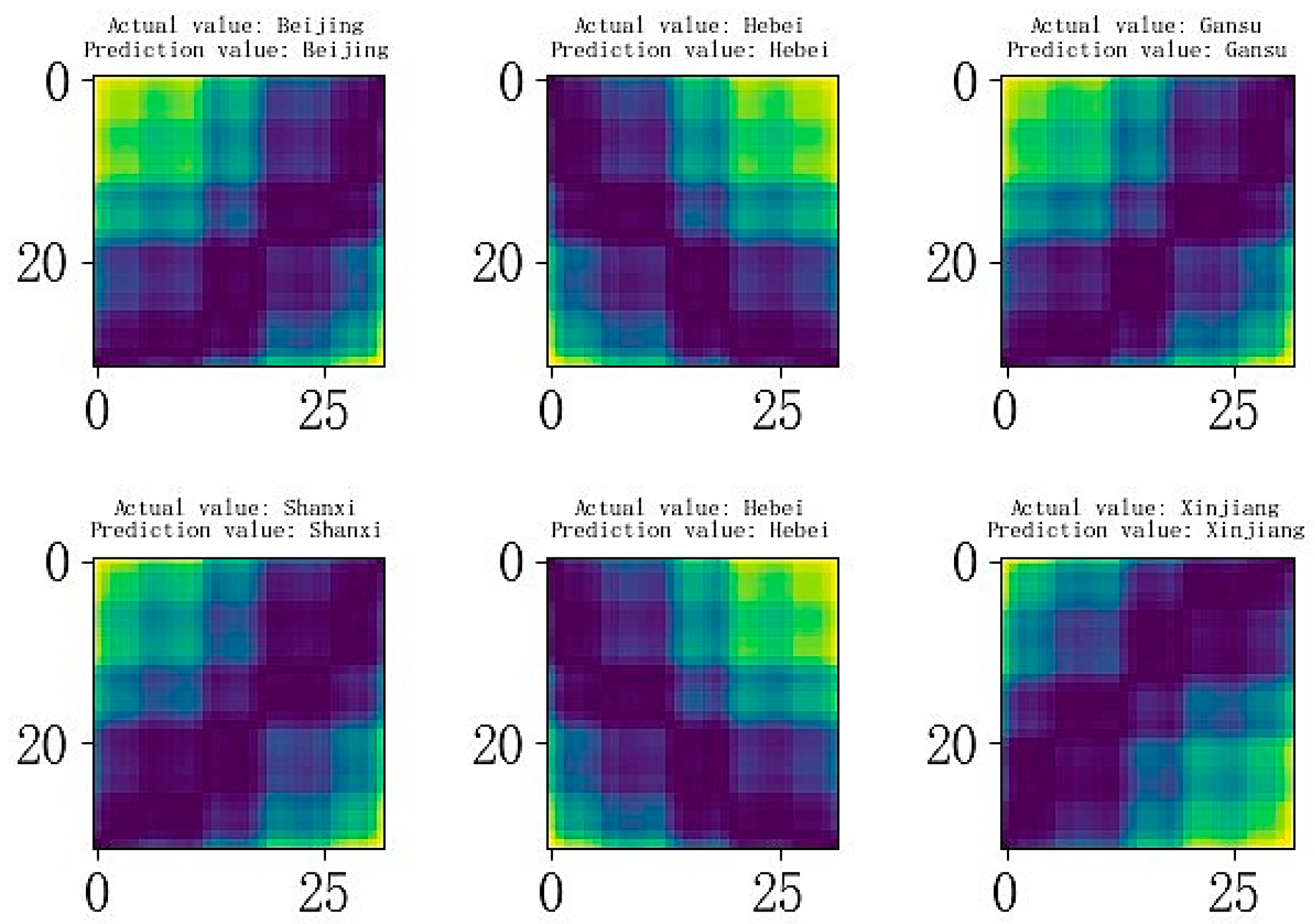
3.6. Maize Seed Origin Identification Model Prediction Results
3.7. Model Comparison
4. Conclusions
- (1)
- GAF leverages the correlation between the one-dimensional NIR spectrum and the time series to enhance the informative content, effectively extracting data from the one-dimensional NIR spectrum. The GAF method solely requires converting the NIR spectrum into an image without involving feature extraction, enabling a more intuitive analysis of NIR spectral data and efficiently addressing the issue of laborious characteristic wavelength extraction. By integrating this converted three-dimensional image with VGG, extensively utilized for large-scale image analysis, we can further discern distinctive features of maize seeds originating from diverse sources. Only adjustments to inputs and specific parameters of the VGG network are necessary to achieve superior results compared to traditional methods, thereby simplifying complexity in NIR spectral modeling.
- (2)
- The combination of preprocessing and PCA cannot achieve high-precision identification analysis of maize seeds from different origins. However, the GAF-VGG network can perform feature extraction under complex conditions with both high and stable prediction accuracy. This network is capable of identifying maize seeds that do not possess the characteristics of their respective origins, providing a new perspective for origin identification and traceability analysis in maize seeds. The results achieved using the GAF-VGG network model outperformed those of Schütz et al. [32], who accurately predicted the origin of maize seeds with 95% accuracy using Fourier Transform NIR spectroscopy and SVM methods, thus emphasizing the advantages of integrating GAF with VGG network for identifying maize seed origins.
- (3)
- The quality and characteristics of a seed can be influenced by its origin. By promptly identifying the origin of maize seeds, growers are able to exercise better control over seed quality, select seeds that are suitable for local climate and soil conditions, enhance crop adaptability and resistance, as well as reduce the occurrence of pests and diseases. Ultimately, this leads to improved crop yield and quality. Certain regions may have specific pests or diseases prevalent in their agricultural systems. Identifying the origin of a seed enables tracing back to its source location, facilitating timely detection and monitoring of pest and disease spread. This aids in implementing appropriate control measures to ensure healthy crop growth. In the marketplace, information regarding the origin of maize seeds is crucial for both consumers and traders alike. Swift identification of seed origins ensures market credibility by enhancing product quality standards and safety while also boosting market competitiveness. To summarize, rapid identification of maize seed origins significantly contributes to quality control measures, epidemic monitoring efforts, market traceability initiatives, as well as improving production efficiency levels while ensuring stable agricultural development.
- (4)
- Future work should focus on further improving identification techniques and methods, such as enhancing spectrogram conversion and exploring the combination of different spectral preprocessing techniques and conversion methods to reduce the number of features in the generated spectral images that do not meet the requirements. Additionally, efforts should be made to enhance the accuracy and speed of identification, reduce costs, and improve anti-interference capabilities. This may involve innovations in sensor technology, image processing algorithms, machine learning models, etc. Furthermore, it is important to develop portable identification devices that can be easily used in the field to provide growers with instant information about seed origin. This will offer growers more flexibility and convenience in seed selection and management. Moreover, it is necessary to apply seed origin identification technology to seed quality testing and origin traceability for other crops like wheat and soybean in order to cater to the needs of growers from various agricultural sectors. Establishing a data-sharing platform for seed origin identification is crucial for promoting the exchange and sharing of seed information. Simultaneously, promoting formulation and unification of relevant standards is important for improving the standardization level and universality of seed origin identification technology. In summary, future advancements in rapid maize seed origin identification will focus on technological improvements, the development of portable equipment, application expansion, and data sharing to provide more reliable and efficient services for seed quality management and origin tracing in agricultural production.
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shakiba, N.; Gerdes, A.; Holz, N.; Wenck, S.; Bachmann, R.; Schneider, T.; Seifert, S.; Fischer, M.; Hackl, T. Determination of the geographical origin of hazelnuts (Corylus avellana L.) by Near-Infrared spectroscopy (NIR) and a Low-Level Fusion with nuclear magnetic resonance (NMR). Microchem. J. 2022, 174, 107066. [Google Scholar] [CrossRef]
- Varrà, M.O.; Ghidini, S.; Ianieri, A.; Zanardi, E. Near infrared spectral fingerprinting: A tool against origin-related fraud in the sector of processed anchovies. Food Control 2021, 123, 107778. [Google Scholar] [CrossRef]
- Leiva, S.F.; Sandoval, J.L.; Abascal-Ponciano, G.A.; Flees, J.J.; Calderon, A.J.; Pacheco, W.J.; Starkey, C.W. Improper sample preparation negatively affects near infrared reflectance spectroscopy (NIRS) nutrient analysis of ground corn. Anim. Feed. Sci. Technol. 2022, 293, 115472. [Google Scholar] [CrossRef]
- Song, C.; Peng, B.; Wang, H.; Zhou, Y.; Sun, L.; Suo, X.; Fan, X. Maize seed appearance quality assessment based on improved Inception-ResNet. Front. Plant Sci. 2023, 14, 1249989. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Liu, L.; Liu, C.; Li, W.; Sun, J.; Li, H.; He, Y.; Yang, L.; Zhang, D.; Cui, T.; et al. Discriminant analysis of maize haploid seeds using near-infrared hyperspectral imaging integrated with multivariate methods. Biosyst. Eng. 2022, 222, 142–155. [Google Scholar] [CrossRef]
- Febrianto, N.A.; Zhu, F. Composition of methylxanthines, polyphenols, key odorant volatiles and minerals in 22 cocoa beans obtained from different geographic origins. LWT 2022, 153, 112395. [Google Scholar] [CrossRef]
- Wei, X.; Zhou, Y.; Jiang, Y.; Tsang, D.C.; Zhang, C.; Liu, J.; Zhou, Y.; Yin, M.; Wang, J.; Shen, N.; et al. Health risks of metal (loid) s in maize (Zea mays L.) in an artisanal zinc smelting zone and source fingerprinting by lead isotope. Sci. Total Environ. 2020, 742, 140321. [Google Scholar] [CrossRef]
- Moghaddam, H.N.; Tamiji, Z.; Lakeh, M.A.; Khoshayand, M.R.; Mahmoodi, M.H. Multivariate analysis of food fraud: A review of NIR based instruments in tandem with chemometrics. J. Food Compos. Anal. 2022, 107, 104343. [Google Scholar] [CrossRef]
- Mansuri, S.M.; Chakraborty, S.K.; Mahanti, N.K.; Pandiselvam, R. Effect of germ orientation during Vis-NIR hyperspectral imaging for the detection of fungal contamination in maize kernel using PLS-DA, ANN and 1D-CNN modelling. Food Control 2022, 139, 109077. [Google Scholar] [CrossRef]
- Arena, E.; Campisi, S.; Fallico, B.; Maccarone, E. Distribution of fatty acids and phytosterols as a criterion to discriminate geographic origin of pistachio seeds. Food Chem. 2007, 104, 403–408. [Google Scholar] [CrossRef]
- de Oliveira Salles, R.C.; Muniz, M.P.; Nunomura, R.D.C.S.; Nunomura, S.M. Geographical origin of guarana seeds from untargeted UHPLC-MS and chemometrics analysis. Food Chem. 2022, 371, 131068. [Google Scholar] [CrossRef]
- Zheng, Y.; Cao, Y.; Yang, J.; Xie, L. Enhancing model robustness through different optimization methods and 1-D CNN to eliminate the variations in size and detection position for apple SSC determination. Postharvest Biol. Technol. 2023, 205, 112513. [Google Scholar] [CrossRef]
- Vitale, R.; Bevilacqua, M.; Bucci, R.; Magrì, A.D.; Magrì, A.L.; Marini, F. A rapid and non-invasive method for authenticating the origin of pistachio samples by NIR spectroscopy and chemometrics. Chemom. Intell. Lab. Syst. 2013, 121, 90–99. [Google Scholar] [CrossRef]
- Jin, X.; Zhou, J.; Rao, Y.; Zhang, X.; Zhang, W.; Ba, W.; Zhou, X.; Zhang, T. An innovative approach for integrating two-dimensional conversion of Vis-NIR spectra with the Swin Transformer model to leverage deep learning for predicting soil properties. Geoderma 2023, 436, 116555. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z.; Zhang, F.; Wei, Z.; Huang, Y.; Chen, C.; Zheng, Y.; Wei, Q.; Sun, H.; Chen, F. Research on detection of potato varieties based on spectral imaging analytical algorithm. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2024, 311, 123966. [Google Scholar] [CrossRef]
- Tan, A.; Wang, B.; Zhao, Y.; Wang, Y.; Zhao, J.; Wang, A.X. Near-infrared spectroscopy analysis of compound fertilizer based on GAF and quaternion convolution neural network. Chemom. Intell. Lab. Syst. 2023, 240, 104900. [Google Scholar] [CrossRef]
- Carvalho, J.K.; Moura-Bueno, J.M.; Ramon, R.; Almeida, T.F.; Naibo, G.; Martins, A.P.; Santos, L.S.; Gianello, C.; Tiecher, T. Combining different pre-processing and multivariate methods for prediction of soil organic matter by near infrared spectroscopy (NIRS) in Southern Brazil. Geoderma Regional. 2022, 29, e00530. [Google Scholar] [CrossRef]
- An, M.; Cao, C.; Wang, S.; Zhang, X.; Ding, W. Non-destructive identification of moldy walnut based on NIR. J. Food Compos. Anal. 2023, 121, 105407. [Google Scholar] [CrossRef]
- Bian, X.; Wang, K.; Tan, E.; Diwu, P.; Zhang, F.; Guo, Y. A selective ensemble preprocessing strategy for near-infrared spectral quantitative analysis of complex samples. Chemom. Intell. Lab. Syst. 2020, 197, 103916. [Google Scholar] [CrossRef]
- Arianti, N.D.; Saputra, E.; Sitorus, A. An automatic generation of pre-processing strategy combined with machine learning multivariate analysis for NIR spectral data. J. Agric. Food Res. 2023, 13, 100625. [Google Scholar] [CrossRef]
- Wang, M.; Xu, Y.; Yang, Y.; Mu, B.; Nikitina, M.A.; Xiao, X. Vis/NIR optical biosensors applications for fruit monitoring. Biosens. Bioelectron. X 2022, 11, 100197. [Google Scholar] [CrossRef]
- de Almeida, A.G.; Tormena, C.D.; de Aguiar, N.S.; Wendling, I.; Rakocevic, M.; Pauli, E.D.; Scarminio, I.S.; Bruns, R.E.; Marcheafave, G.G. Direct NIR spectral determination of genetic improvement, light availability, and their interaction effects on chemically selected yerba-mate leaves. Microchem. J. 2023, 191, 108828. [Google Scholar] [CrossRef]
- Chen, R.; Li, S.; Cao, H.; Xu, T.; Bai, Y.; Li, Z.; Leng, X.; Huang, Y. Rapid quality evaluation and geographical origin recognition of ginger powder by portable NIRS in tandem with chemometrics. Food Chem. 2024, 438, 137931. [Google Scholar] [CrossRef]
- Schoot, M.; Kapper, C.; van Kollenburg, G.H.; Postma, G.J.; van Kessel, G.; Buydens, L.M.; Jansen, J.J. Investigating the need for preprocessing of near-infrared spectroscopic data as a function of sample size. Chemom. Intell. Lab. Syst. 2020, 204, 104105. [Google Scholar] [CrossRef]
- Lee, H.; Yang, K.; Kim, N.; Ahn, C.R. Detecting excessive load-carrying tasks using a deep learning network with a Gramian Angular Field. Autom. Constr. 2020, 120, 103390. [Google Scholar] [CrossRef]
- Qi, P.; Chiaro, D.; Piccialli, F. FL-FD: Federated learning-based fall detection with multimodal data fusion. Inf. Fusion 2023, 99, 101890. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, X.; Liu, P.; Li, H.; Liu, W. Rice disease identification method based on improved CNN-BiGRU. Artif. Intell. Agric. 2023, 9, 100–109. [Google Scholar] [CrossRef]
- Min, W.; Wang, Z.; Yang, J.; Liu, C.; Jiang, S. Vision-based fruit recognition via multi-scale attention CNN. Comput. Electron. Agric. 2023, 210, 107911. [Google Scholar] [CrossRef]
- Li, J.; Zhu, Z.; Liu, H.; Su, Y.; Deng, L. Strawberry R-CNN: Recognition and counting model of strawberry based on improved faster R-CNN. Ecol. Inform. 2023, 77, 102210. [Google Scholar] [CrossRef]
- Aishwarya, M.P.; Reddy, P. Ensemble of CNN models for classification of groundnut plant leaf disease detection. Smart Agric. Technol. 2023, 6, 100362. [Google Scholar]
- da Silva, G.S.; Canuto, K.M.; Ribeiro, P.R.V.; de Brito, E.S.; Nascimento, M.M.; Zocolo, G.J.; Coutinho, J.P.; de Jesus, R.M. Chemical profiling of guarana seeds (Paullinia cupana) from different geographical origins using UPLC-QTOF-MS combined with chemometrics. Food Res. Int. 2017, 102, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Schütz, D.; Riedl, J.; Achten, E.; Fischer, M. Fourier-transform near-infrared spectroscopy as a fast screening tool for the verification of the geographical origin of grain maize (Zea mays L.). Food Control 2022, 136, 108892. [Google Scholar] [CrossRef]


| Origin Labels | Code of Origin |
|---|---|
| Gansu Suke Sweet 1506 | A1 |
| Shandong Suke Sweet 1506 | A2 |
| Shanxi Hua Nuo 2 | B1 |
| Hebei Hua Nuo 2 | B2 |
| Shandong Star Sweet 230 | C1 |
| Beijing Star Sweet 230 | C2 |
| Shandong Moxidome | D1 |
| Jiangsu Ink Pupil | D2 |
| Xinjiang Tiangui Glutinous 932 | E1 |
| Guangxi Tiangui Glutinous 932 | E2 |
| Beijing Honey Blossom Sweet Glutinous 3 | F |
| Beijing Star Sweet 221 | G |
| Shandong Golden Sweet 13 | H |
| Hebei Zhongnong Sweet 488 | I |
| Shanxi Golden Queen | J |
| Shanxi Black Sticky 301 | K |
| Gansu Huanai color sweet glutinous 102 | L |
| Method | Correction Set | Prediction Set | ||||||
|---|---|---|---|---|---|---|---|---|
| R2c | RMSEC | SEC | Rc | R2p | RMSEP | SEP | Rp | |
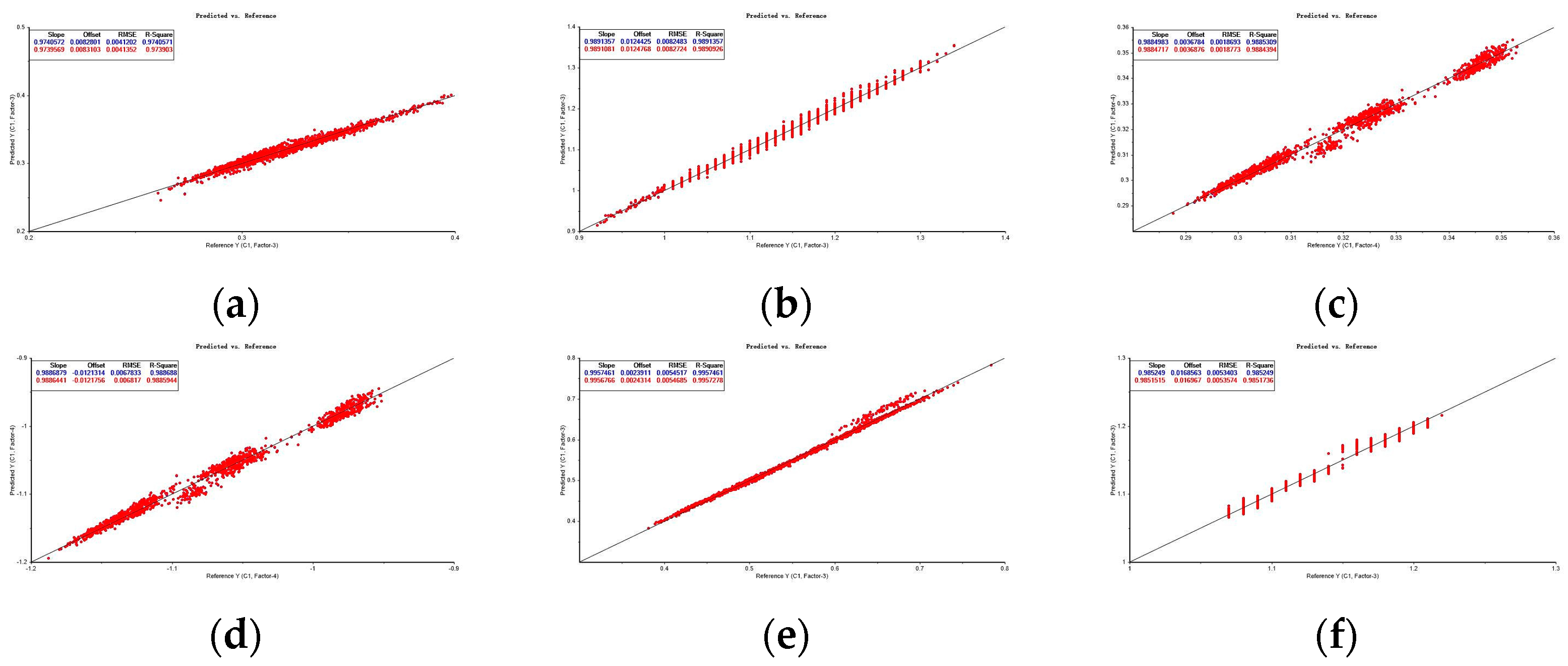
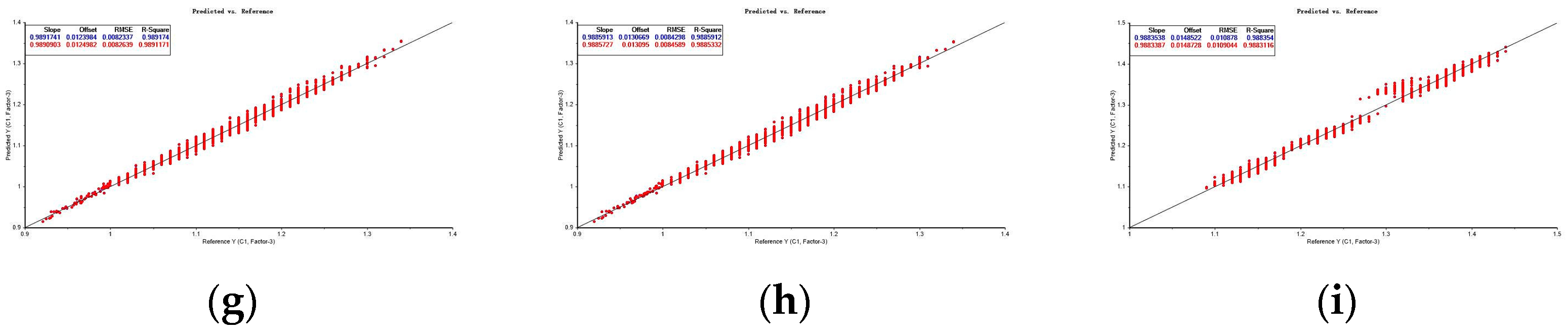
| RAW | 0.974 | 0.004 | 0.008 | 0.987 | 0.974 | 0.004 | 0.008 | 0.987 |
| FD | 0.989 | 0.008 | 0.012 | 0.995 | 0.989 | 0.008 | 0.012 | 0.995 |
| MSC | 0.989 | 0.002 | 0.004 | 0.994 | 0.988 | 0.002 | 0.004 | 0.994 |
| SNV | 0.989 | 0.007 | 0.012 | 0.994 | 0.989 | 0.007 | 0.012 | 0.994 |
| FDSNV | 0.996 | 0.005 | 0.002 | 0.998 | 0.996 | 0.005 | 0.002 | 0.998 |
| FDMSC | 0.985 | 0.005 | 0.017 | 0.993 | 0.985 | 0.005 | 0.017 | 0.993 |
| FDMA | 0.989 | 0.008 | 0.012 | 0.995 | 0.989 | 0.008 | 0.012 | 0.995 |
| FDSG | 0.989 | 0.008 | 0.013 | 0.994 | 0.989 | 0.008 | 0.013 | 0.994 |
| FDCT | 0.988 | 0.011 | 0.015 | 0.994 | 0.988 | 0.011 | 0.015 | 0.994 |
| Labels | Original Code | Train | Test |
|---|---|---|---|
| Gansu Suke Sweet 1506 | A1 | 120 | 40 |
| Gansu huanai color sweet glutinous 102 | L | 120 | 40 |
| Shandong Suke Sweet 1506 | A2 | 60 | 20 |
| Shandong Star Sweet 230 | C1 | 60 | 20 |
| Shandong Moxidome | D1 | 60 | 20 |
| Shandong Golden Sweet 13 | H | 60 | 20 |
| Beijing Star Sweet 230 | C2 | 80 | 26 |
| Beijing Honey Blossom Sweet Glutinous 3 | F | 80 | 27 |
| Beijing Star Sweet 221 | G | 80 | 27 |
| Shanxi Golden Queen | J | 80 | 26 |
| Shanxi Black Sticky 301 | K | 80 | 27 |
| Shanxi Huagnuo 2 | B1 | 80 | 27 |
| Hebei Zhongnong Sweet 488 | I | 120 | 40 |
| Hebei Huagnuo 2 | B2 | 120 | 40 |
| Jiangsu Ink Pupil | D2 | 240 | 80 |
| Xinjiang Tiangui Glutinous 932 | E1 | 240 | 80 |
| Guangxi Tiangui Glutinous 932 | E2 | 240 | 80 |
| Batch_Size | Train/% | Test/% | Time/mins |
|---|---|---|---|
| 16 | 90.59 | 69.85 | 54.26 |
| 32 | 96.93 | 89.06 | 57.15 |
| 48 | 98.7 | 93.75 | 56.35 |
| 64 | 97.55 | 71.41 | 66.83 |
| 128 | 96.81 | 81.63 | 72.98 |
| Learning Rate | Train/% | Test/% | Time/mins |
|---|---|---|---|
| 10−3 | 51.04 | 42.03 | 55.7 |
| 10−4 | 83.33 | 79.53 | 58.52 |
| 10−5 | 97.11 | 92.66 | 53.57 |
| 10−6 | 95.74 | 93.91 | 53.99 |
| 10−7 | 100 | 97.03 | 54.18 |
| 5−7 | 100 | 95.94 | 57.7 |
| 10−8 | 100 | 92.97 | 53.7 |
| 10−9 | 64.69 | 62.97 | 59.8 |
| Dropout | Train/% | Test/% |
|---|---|---|
| 0.3 | 100 | 92.97 |
| 0.4 | 100 | 94.38 |
| 0.5 | 100 | 95.61 |
| 0.6 | 100 | 95.31 |
| 0.7 | 100 | 94.69 |
| Method | Accuracy | Recall/ Sensitivity | Specificity | Precision |
|---|---|---|---|---|
| RAW | 40.08 | 88.65 | 34.53 | 37.35 |
| PCA | 40.65 | 90.47 | 36.28 | 39.28 |
| GAF-VGG | 96.81 | 97.23 | 95.35 | 95.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Fu, C.; Gao, Y.; Kang, Y.; Zhang, W. Research on the Identification Method of Maize Seed Origin Using NIR Spectroscopy and GAF-VGGNet. Agriculture 2024, 14, 466. https://doi.org/10.3390/agriculture14030466
Xu X, Fu C, Gao Y, Kang Y, Zhang W. Research on the Identification Method of Maize Seed Origin Using NIR Spectroscopy and GAF-VGGNet. Agriculture. 2024; 14(3):466. https://doi.org/10.3390/agriculture14030466
Chicago/Turabian StyleXu, Xiuying, Changhao Fu, Yingying Gao, Ye Kang, and Wei Zhang. 2024. "Research on the Identification Method of Maize Seed Origin Using NIR Spectroscopy and GAF-VGGNet" Agriculture 14, no. 3: 466. https://doi.org/10.3390/agriculture14030466
APA StyleXu, X., Fu, C., Gao, Y., Kang, Y., & Zhang, W. (2024). Research on the Identification Method of Maize Seed Origin Using NIR Spectroscopy and GAF-VGGNet. Agriculture, 14(3), 466. https://doi.org/10.3390/agriculture14030466




