Abstract
Leaf spot disease is an extremely common disease in the growth process of maize in Northern China and its degree of harm is quite significant. Therefore, the rapid and accurate identification of maize leaf spot disease is crucial for reducing economic losses in maize. In complex field environments, traditional identification methods are susceptible to subjective interference and cannot quickly and accurately identify leaf spot disease through color or shape features. We present an advanced disease identification method utilizing YOLOv8. This method utilizes actual field images of diseased corn leaves to construct a dataset and accurately labels the diseased leaves in these images, thereby achieving rapid and accurate identification of target diseases in complex field environments. We have improved the model based on YOLOv8 by adding Slim-neck modules and GAM attention modules and introducing them to enhance the model’s ability to identify maize leaf spot disease. The enhanced YOLOv8 model achieved a precision (P) of 95.18%, a recall (R) of 89.11%, an average recognition accuracy (mAP50) of 94.65%, and an mAP50-95 of 71.62%, respectively. Compared to the original YOLOv8 model, the enhanced model showcased enhancements of 3.79%, 4.65%, 3.56%, and 7.3% in precision (P), recall (R), average recognition accuracy (mAP50), and mAP50-95, respectively. The model can effectively identify leaf spot disease and accurately calibrate its location. Under the same experimental conditions, we compared the improved model with the YOLOv3, YOLOv5, YOLOv6, Faster R-CNN, and SSD models. The results show that the improved model not only enhances performance, but also reduces parameter complexity and simplifies the network structure. The results indicated that the improved model enhanced performance, while reducing experimental time. Hence, the enhanced method proposed in this study, based on YOLOv8, exhibits the capability to identify maize leaf spot disease in intricate field environments, offering robust technical support for agricultural production.
1. Introduction
As one of the major staple crops in China, maize holds a significant position in the country’s agricultural landscape. It serves not only as a cornerstone of food production, but also plays a crucial role in maintaining social stability and ensuring national food security. Its importance is evident not only in meeting the daily dietary needs of people, but also in contributing to economic development, agricultural sustainability, and rural livelihoods. The cultivation of maize not only addresses food security issues but also supports the development of rural communities, fosters agricultural innovation, and strengthens the foundation of the national food system. Its strong adaptability to diverse growing conditions has resulted in widespread cultivation in many regions of China [1]. Originating from the south-central region of the Americas, maize was introduced to China from Europe in the first half of the 16th century and has shown a strong ability to survive under harsh growing conditions. With its high yield, versatile applications, cold and drought resistance, and adaptability, maize stands as one of China’s most productive food crops [2]. Not only is it an important source of feed for animal husbandry, farming, aquaculture, etc., but maize is also one of the indispensable raw materials in many fields such as food, healthcare, light industry, chemical industry, etc. However, in recent years, serious outbreaks of corn diseases have seriously threatened the yield and quality of corn [3]. To address this issue, researchers have introduced technologies in the field of machine vision to implement a fast and effective identification of maize diseases based on their characteristics. Through this process, researchers were able to quickly understand the development of the disease, providing reliable data to support the adoption of appropriate protective and management measures. The introduction of this technology has greatly reduced the negative impact of corn diseases on yield and quality. In general, the application of machine vision technology to corn diseases offers a novel scientific and technological tool for agricultural production. This contributes to maintaining food security and advancing sustainable agricultural development in China.
China is among the most severely affected countries globally when it comes to crop pests and diseases, with over 1700 perennial species occurring annually [4]. Hence, the management of crop diseases has become one of the primary solutions to address the food issue. The traditional manual methods of detecting diseases are totally dependent on the observation experience of farmers or asking experts to come to their homes for guidance, which are slow, inefficient, expensive, subjective, inaccurate, and not time-sensitive processes [5]. In recent years, the convergence of machine vision and image processing technology has introduced a novel approach to address these challenges. By adopting the method of segmenting images and extracting image features for corn disease recognition, this technique overcomes the defects of traditional manual disease detection methods, such as their low recognition rates, as well as being time-consuming and labor-intensive [6]. However, despite the progress made using this method, the final results do not achieve the desired accuracy, due to the tedious and time-consuming feature extraction process.
With the continuous evolution of the Internet, researchers, including Hinton [7], introduced the concept of deep learning, leading to swift research and development in the field. Currently, deep learning has been successfully applied across various fields, yielding remarkable results. The advent of deep learning introduces fresh ideas and methodologies for identifying agricultural diseases. Integrating deep learning technology into agricultural disease identification not only reduces workload, but also effectively improves the accuracy of disease recognition. This is pivotal for the precise identification of crop diseases. Researchers have successfully developed a convolutional neural network model, which has achieved remarkable results in crop disease recognition [8]. The convolutional neural network model has made significant strides in image feature extraction, capable of automatically extracting image features with good adaptability and high recognition accuracy for enhanced images [9]. However, in complex field environments, the generalization ability and recognition accuracy of these models are still somewhat degraded and they cannot fully adapt to the changing environments in the field. Subsequently, new algorithms, including the YOLO series [10], Fast R-CNN [11], and others, have emerged, one after another. These algorithms have achieved significant improvements in accuracy and computational speed. Currently, most deep learning models for detecting maize leaf spot disease focus more on accuracy, with long detection times that cannot achieve real-time detection. Moreover, these models are mostly tested in laboratory settings and cannot effectively simulate real-field conditions, thus limiting their practical application in production. Hence, the application of deep learning technology to address issues in the field of crop disease identification holds not only significant theoretical research significance, but also crucial practical value.
To tackle the issues of diminished recognition accuracy and efficiency in identifying corn leaf spot disease, an enhanced method, rooted in YOLOv8, is proposed in this paper. The key improvements of this method include the following:
(1) The incorporation of the Slim-neck module replaces the original backbone network in YOLOv8, rendering the model more lightweight and consequently reduces computational costs.
(2) The GAM attention mechanism is introduced to enhance the computational cost of important areas and to obtain more effective information to assist the model to more accurately locate disease-generating areas.
(3) Introducing the CIoU on top of the , by employing an auxiliary bounding box, in order to accelerate the convergence of the model and, thus, reduce the experimental time consumed.
2. Data and Methods
2.1. Data Collection and Dataset Construction
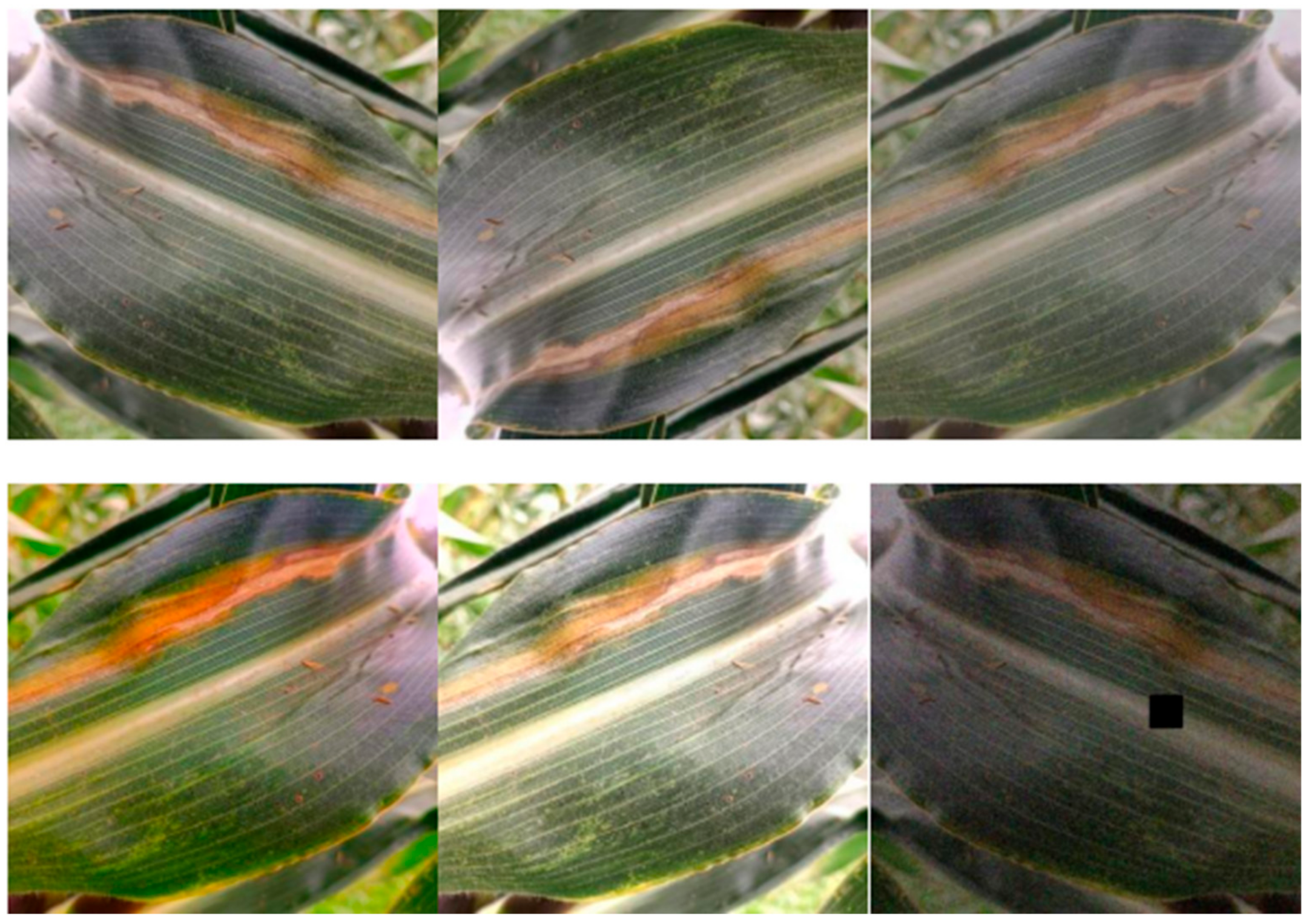
The images were collected from a corn planting field in Beishifo Village, Laiyuan County, Baoding City, Hebei Province, China. In order to ensure that the collected data were representative, we collected the diseased plants at different time periods and under different shooting backgrounds, as shown in Figure 1. As can be observed through Figure 1A,B, under low light conditions, the disease showed obvious features with clear shapes that were easy to identify, while, under strong light conditions, the overall features of the disease became blurred and the color was similar to that of the light, which was not easy to identify. There are many disturbing conditions in the actual field environment, such as leaf shading and soil, as shown in Figure 1C. In total, we acquired 3120 images with a resolution of 640 × 640 pixels. Given that some of the collected images were not usable, we selected 1000 images of corn leaf spot disease from publicly available datasets to add to the dataset to ensure data completeness and diversity.

Figure 1.
Data image. (A) Less light; (B) plenty of light; and (C) shaded by leaves.
To effectively enhance data quality, improve the generalization ability of model training, and prevent overfitting phenomena, we conducted data augmentation on some of the acquired images. By means of rotating, mirroring, brightness adjustment, adding Gaussian noise, and random masking, we expanded the data to 5521 images to further enhance the representativeness of the data. The enhanced data are shown in Figure 2. This series of operations aims to ensure that the model can accurately perform leaf spot disease recognition in various contexts, creating more favorable conditions for model training and performance enhancement.
Figure 2.
Enhanced data.
To achieve a more efficient identification of corn leaf spot disease, the data were first manually labeled. This series of operations aims to ensure that the model can accurately perform leaf spot disease recognition in various contexts, creating more favorable conditions for model training and performance enhancement. Afterward, the labeled data were divided into training, validation, and test sets, maintaining an 8:1:1 ratio. Ultimately, we obtained 4416, 552, and 553 images to fulfill the requirements for comprehensive training and model evaluation. This labeling and division process aims to provide sufficient and accurate training data for the model to improve its performance in real applications.
2.2. Methodology Research
2.2.1. YOLOv8 Network Models
The YOLO family of algorithms is a class of single-stage target detection algorithms, i.e., only one feature extraction is required to accomplish target detection. These algorithms redefine target detection as a way of solving regression problems. Since its inception in 2015 by Joseph Redmon et al., the YOLO family of algorithms has seen multiple iterations and improvements, including YOLOv1 [12], YOLOv2 [13], YOLOv3 [14], YOLOv4 [15], and the most recent version, YOLOv8 [16]. The architecture of YOLOv8 is predominantly composed of inputs, the backbone network, Backbone, the Neck module, and outputs [17]. In the Backbone and Neck sections, YOLOv8 utilizes the C2f structure, enabling the amalgamation of diverse gradient features and fine-tuning to notably enhance the overall model performance. Within the Head segment, YOLOv8 incorporates the widely used decoupled head structure, effectively segregating the classification and detection functions. Simultaneously, the model’s performance is further enhanced by incorporating the Distribution Focal Loss function into the loss function. These optimizations make YOLOv8 perform well in the target detection task.
2.2.2. Slim-Neck Module
The original YOLOv8 model consumes a significant amount of time in the process of detecting maize leaf spot disease, but, due to the real-time nature of detecting the disease, so to address the serious time-consumption problem, the introduction of the Slim-neck lightweight network structure in the YOLOv8 model is considered to reduce the complexity of the network results, so as to reduce the serious time-consumption problem of the model in the process of detecting the target.
The Slim-neck module is a lightweight network structure widely used in real-time detection [18]. Currently, many lightweight networks, such as Xception [19] and MobileNets [20], are using depth-wise separable convolution (DSC) to effectively reduce the time-consuming problem of target detection. While these methods have shown notable success in decreasing computation time, they have also led to a decrease in the overall target detection accuracy of the model.
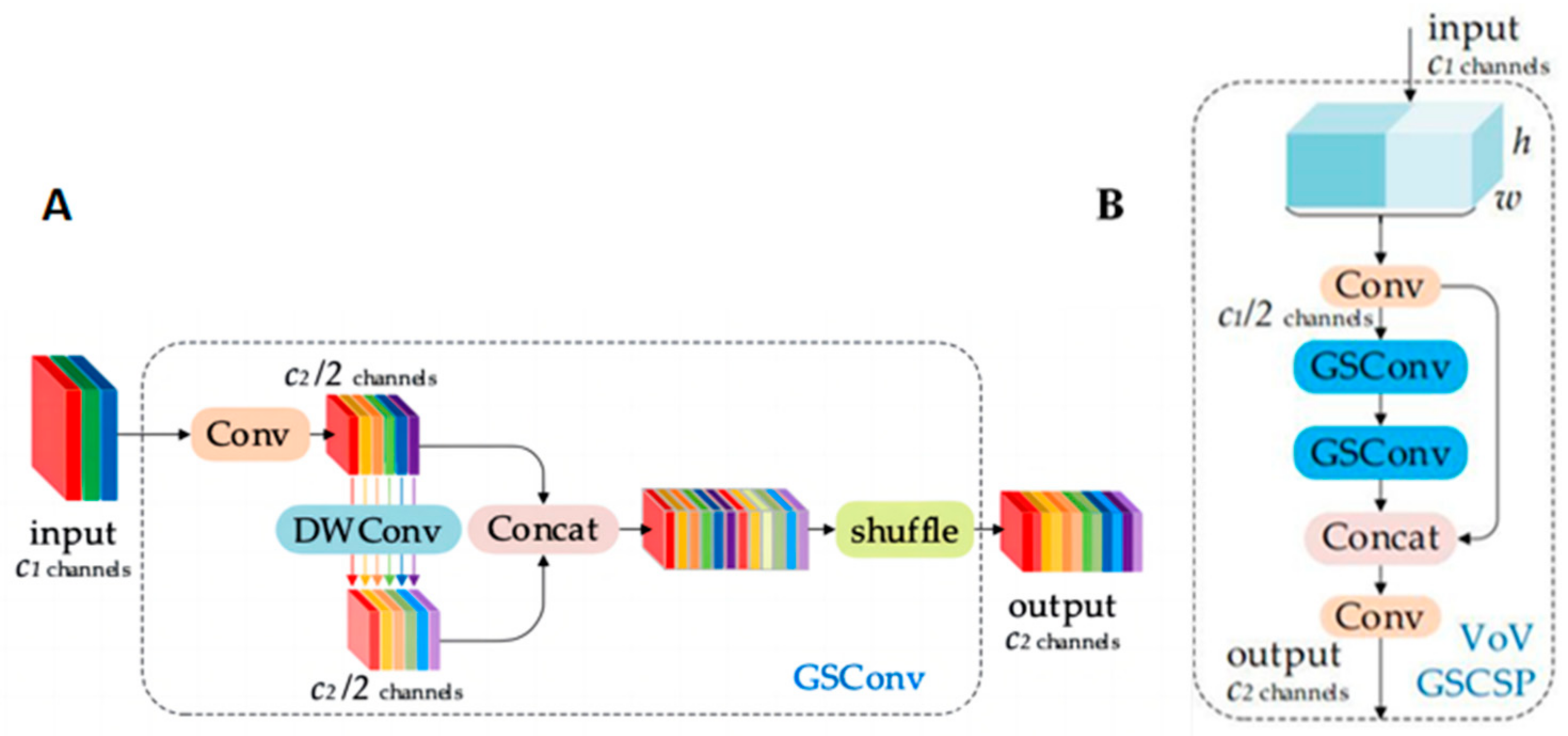
GSConv (Grouped Separable Convolution) is introduced in the Slim-neck module to replace the traditional Conv convolution. GSConv is a hybrid convolution that combines SC, DSC, and Shuffle, as depicted in Figure 3A. GSConv transfers the information generated using SC to the information of each part in DSC by applying Shuffle. This method comprehensively integrates the information generated using SC and DSC, facilitating the even exchange of feature information across different channels. This results in the extraction of richer information about maize leaf spot disease. Additionally, GSConv significantly reduces computational costs when compared to the original model’s Conv convolution, efficiently simplifying the model’s network structure complexity, while maintaining optimal performance.
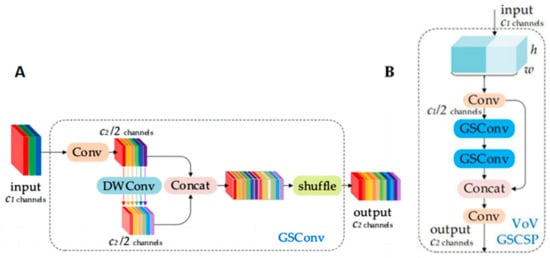
Figure 3.
Slim-neck module. (A) Structure of GSConv; (B) structure of VoV-GSCSP.
The Slim-neck module introduces a one-time aggregation method based on GSConv, forming the VoV-GSCSP module. In this paper, the VoV-GSCSP module is employed to replace the C2f module, further simplifying the network structure of the model, based on the introduction of GSConv. Thus, the purpose of lightweighting the network structure is achieved. The structure of the VoV-GSCSP module is shown in Figure 3B.
The main network of the model has been improved by introducing the Slim-neck module. While ensuring that the model’s accuracy is not compromised, this modification significantly reduces the complexity of the model’s network structure. It addresses the significant time-consumption issue during the identification process and offers effective methodological support for subsequent research.
2.2.3. GAM Attention Mechanisms
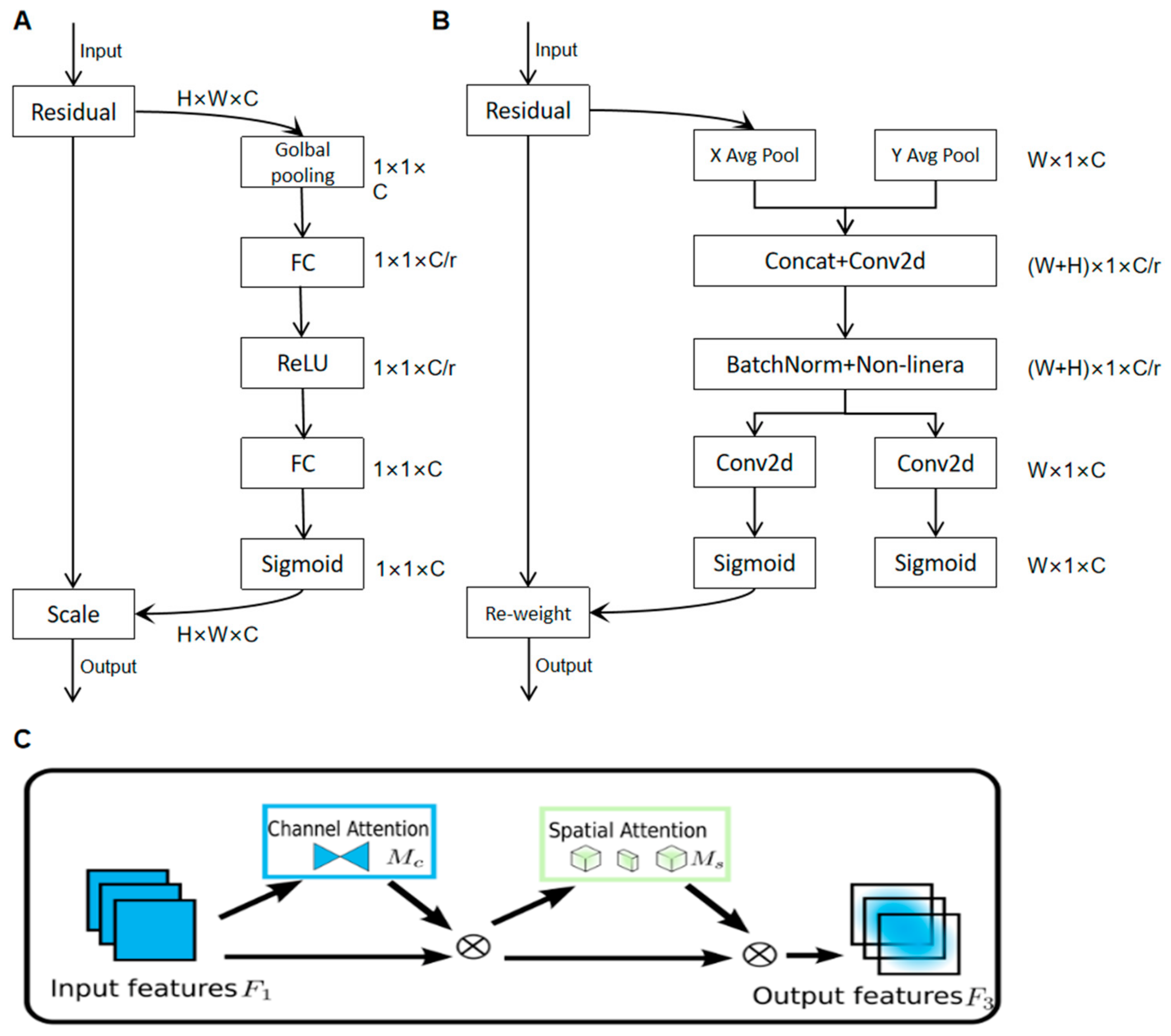
Presently, the most widely employed attentional mechanisms include the SE (Squeeze and Excitation) [21] attentional mechanisms and the CBAM (Convolutional Block Attention Module) [22] attentional mechanisms.
The SE attention mechanism aims to allocate distinct weights to each channel using a weight matrix to acquire more information about key elements. Its internal structure is shown in Figure 4A.The CBAM attention mechanism is a method that combines spatial and channel attention mechanisms, as shown in Figure 4B.
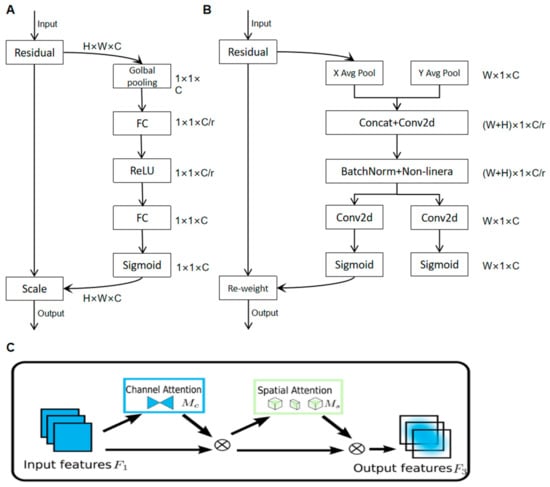
Figure 4.
Diagram of the internal structure of various attention mechanisms. (A) SE internal structure diagram; (B) CBAM internal structure diagram; (C) internal structure of GAM.
Both the SE Attention Mechanism and the CBAM Attention Mechanism perform attention operations in their respective dimensions, not fully considering the interrelationship between the two dimensions of channel and space, neglecting the importance of cross-dimensional information. Therefore, this paper adopts the GAM (Global Attention Module) [23] attention mechanism, i.e., the global attention mechanism, as shown in Figure 4C.
Introducing the GAM module into the model, this innovative design achieved a significant performance improvement, while adding only a small number of parameters, and the GAM attention mechanism can effectively mitigate the issue of information loss through information between the two channels. The GAM attention mechanism proves effective in capturing the small target diseases that are often overlooked in the intricate field environment. It also optimally allocates computational resources, thereby enhancing the network model’s detection capabilities for maize leaf spot disease. The integration of the GAM and Slim-neck modules makes the improved model better able to cope with the task of detecting corn leaf spot disease and provides a more reasonable method to support agricultural development.
2.2.4. Loss Function Improvement
In target detection algorithms, the performance of detection depends largely on the design of the loss function. Within the target detection loss function, the edge loss function holds significance and a well-designed one can markedly elevate the performance of the detection model. In YOLOv8, the CIoU [24] loss function is used. CIoU is one of the most widely used loss functions, which covers the coverage area, centroid distance, and aspect ratio.
The relevant formulas are as follows:
represents the intersection over union between the predicted box and the ground truth box; is the Euclidean distance between the centers of the predicted box and the ground truth box; and are the coordinates of the centers of the predicted box and the ground truth box; is the diagonal distance of the minimum closure region between the predicted box and the ground truth box; and are the width and height of the predicted box, and and are the width and height of the ground truth box.
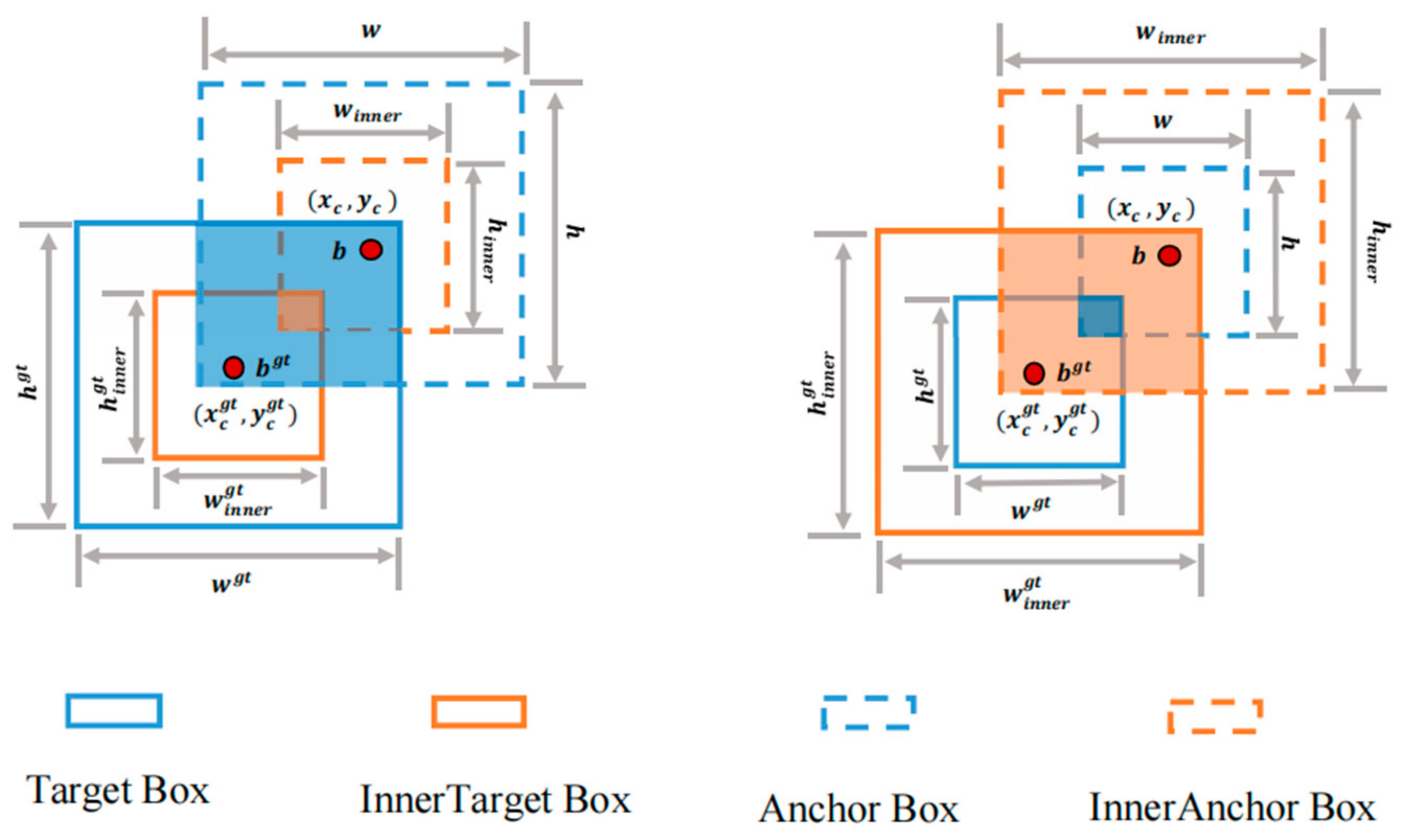
Currently, most of the IoU-based edge loss functions accelerate convergence by introducing new loss terms; however, this approach ignores the limitations of the new loss terms themselves. In practice, this strategy cannot be adjusted according to different detectors and detection tasks and, thus, may exhibit a slower convergence and a poorer generalization ability in different detection tasks. Therefore, this paper adopts Inner-IoU Loss [25], which calculates the IoU Loss through auxiliary edges; its structure is shown in Figure 5. This approach is more adaptable and can be customized for specific detectors and tasks, enhancing the model’s convergence speed and generalization capability.
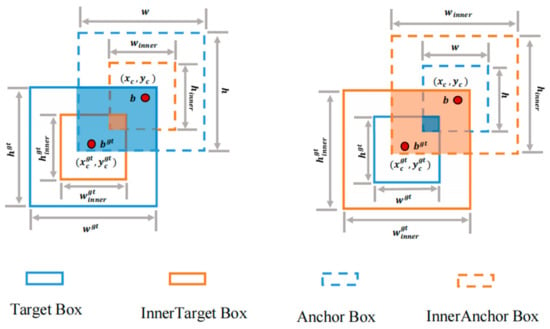
Figure 5.
Inner-IoU Loss architecture diagram.
The Inner-IoU formula is as follows:
Among them, and represent the centers of the GT frame and Inner GT frame, respectively. and represent the center points of the anchor frame and Inner anchor frame, respectively, and represents the scale factor.
In this study, the integration of Inner-IoU Loss with CIoU results in a significant performance enhancement, while concurrently simplifying the network model’s complexity. The improved model well solves the overfitting problem during training and accelerates the model convergence by detecting the task to adjust the appropriate auxiliary edge scale, thus improving the overall performance of the model.
The formula is as follows:
2.2.5. Improved Network Structure
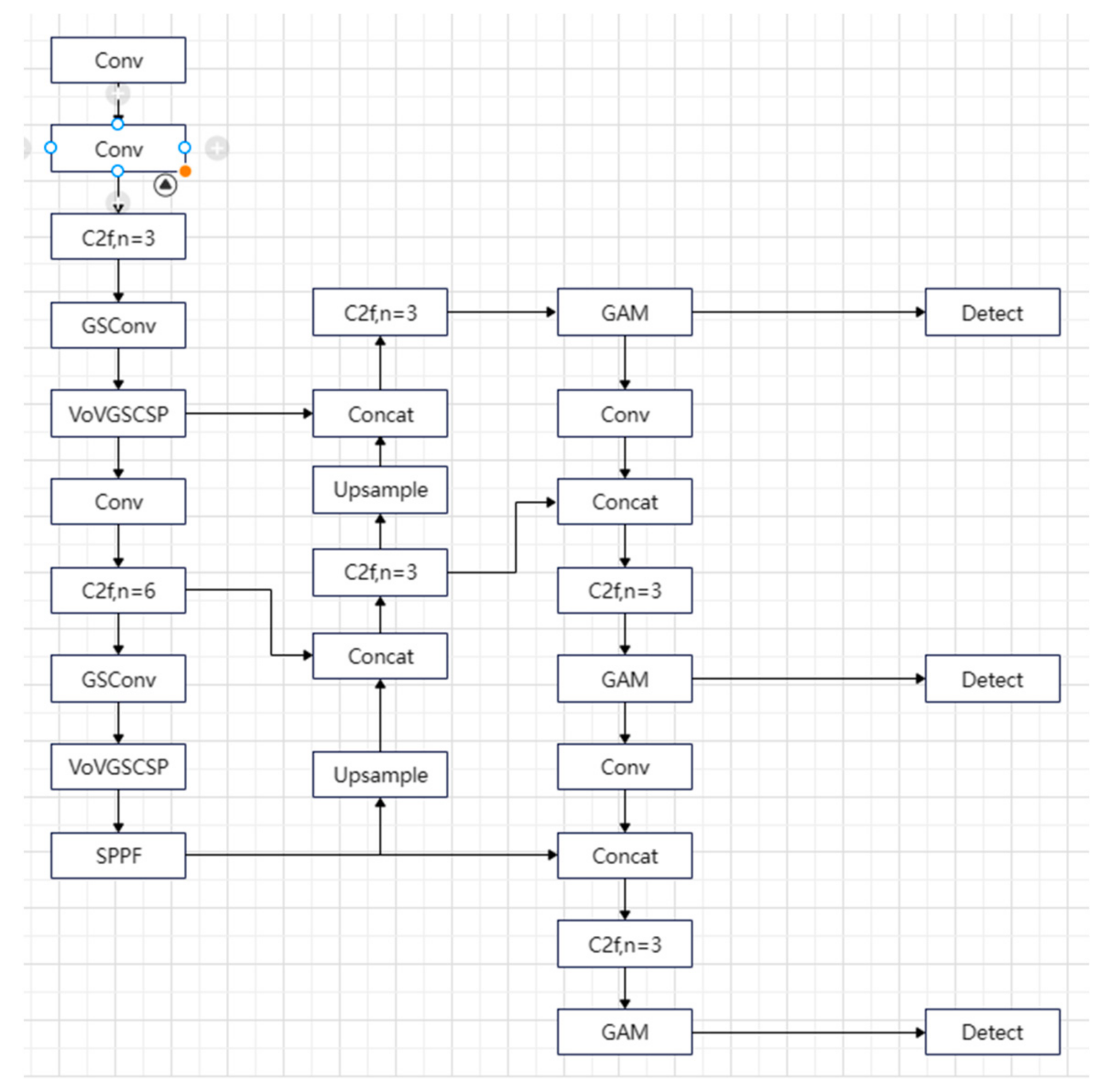
In this paper, YOLOv8n is used as the base model to be improved to adapt to the recognition and detection of maize leaf spot disease in the complex environment of a real field. Based on the above description, the improvement of YOLOv8n is as follows: in the backbone network and Neck part, GSConv and VoV-GSCSP are used to replace the Conv convolution and C2f module in the base model, reducing the complexity of the model network structure. The GAM attention mechanism is introduced at the end of the Neck module to enhance feature extraction capabilities by integrating information from each dimension. This aids the model in more effectively pinpointing the location of the disease, thereby preventing unnecessary computational resource wastage. Finally, Inner-IoU Loss is introduced on the basis of CIoU, so that the model can adjust the auxiliary edge scale in a timely manner that is most suitable for the detection task, accelerating the model convergence and improving the generalization ability of the model, as shown in Figure 6.
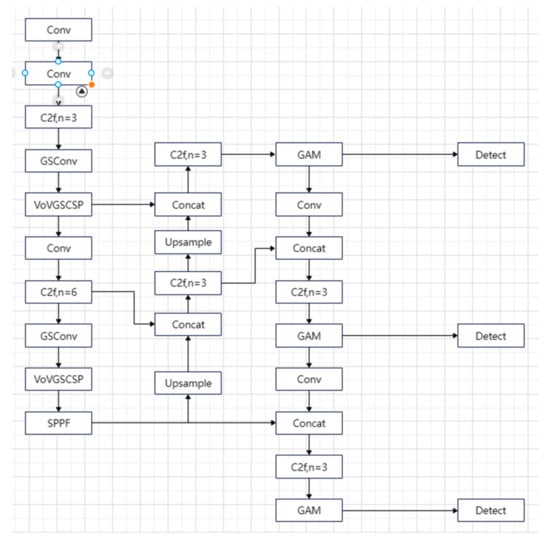
Figure 6.
Improved YOLOv8 network structure diagram.
3. Experiments and Analysis of Results
3.1. Experimental Environment
The experimental environment is set up in the Autodl workstation and the main hardware configuration is shown in Table 1:

Table 1.
Experimental environment.
The initial learning rate is 0.01, the momentum silver is 0.937, the image input size is 640 × 640 pixels, the batch size is set to 16, and the training rounds are set to 800.
3.2. Evaluation Indicators
In this paper, Recall, Precision, mean average precision (mAP50), mAP50-95, total parameter count, and inference time are primarily used as the evaluation criteria for assessing and comparing the improved model with other control models. In this case, Recall indicates the probability of being correctly recognized in positive samples and Cha-accuracy indicates the probability of being correctly recognized in all samples.
The relevant formulas are as follows:
where TP is True positives, i.e., positive samples are correctly recognized as positive samples. FN is False negatives, i.e., positive samples are incorrectly recognized as negative samples. FP is False Positives, i.e., negative samples are incorrectly recognized as positive samples. represents the area under the precision–recall curve. mAP stands for mean Average Precision.
3.3. Analysis of Results
3.3.1. Performance Analysis of Algorithm
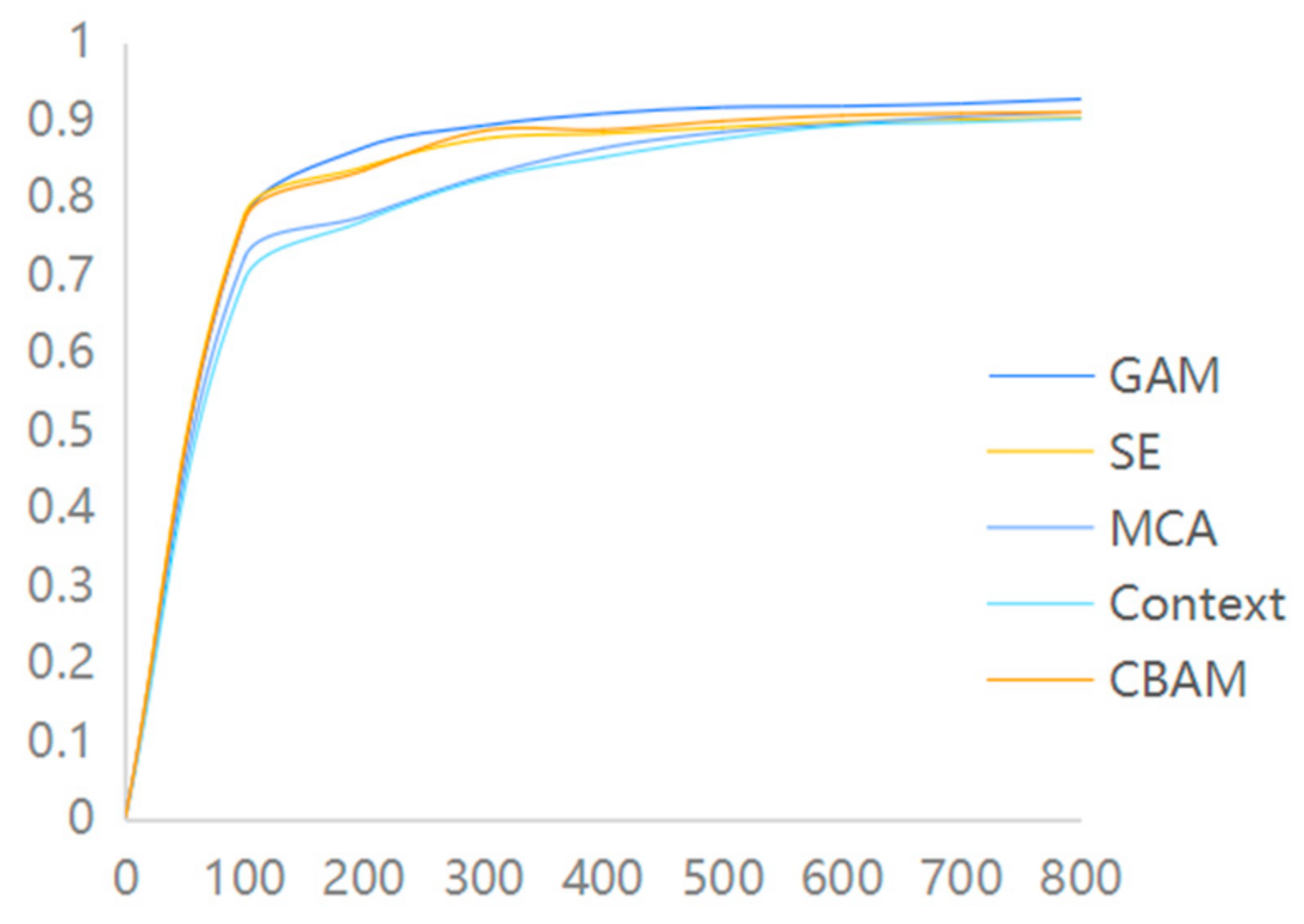
To further assess the performance of each algorithm, several comparative experiments were conducted during the experiment to identify the most suitable algorithm for this study. To investigate the performance of different attention mechanisms, various commonly used attention mechanism modules were selected for comparison in the experiments. These included the SE attention mechanism module, MCA attention module, Context attention module, and other attention modules, along with the GAM attention module used in this paper. The experiments were conducted on the same dataset, following the aforementioned experimental parameter settings. The experimental results reveal that the GAM attention module employed in this study exhibits a superior recognition performance compared to other attention modules on the same dataset, as depicted in Figure 7.
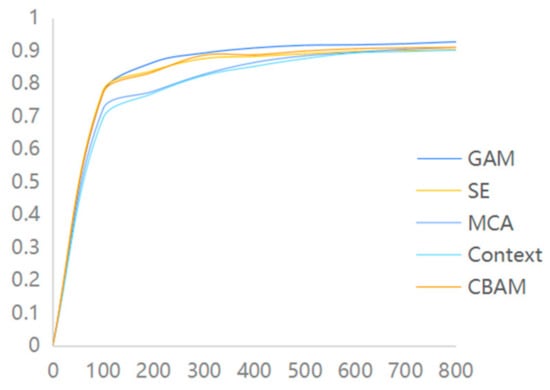
Figure 7.
Attention mechanism accuracy comparison chart.
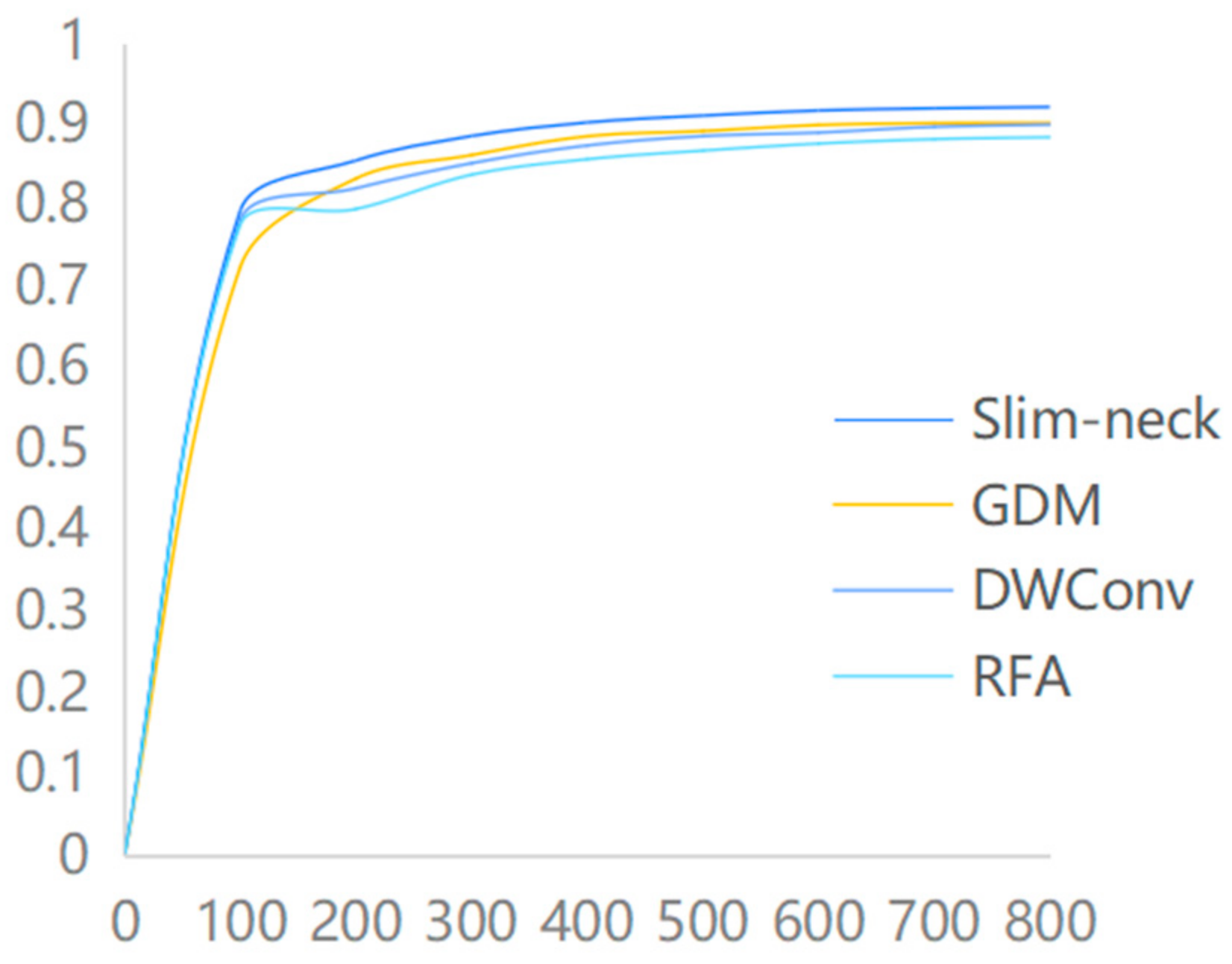
Based on the introduction of the GAM attention module, this paper introduces the Slim-neck module, which replaces the traditional Conv convolution and C2f module at the backbone network and Neck side of YOLOv8. At the same time, a comparison is made with the introduction of the GDM module, the DWConv module, and the RFA module. The experimental results demonstrate that the Slim-neck module utilized in this study outperforms other modules significantly, in terms of recognition effectiveness, as illustrated in Figure 8.
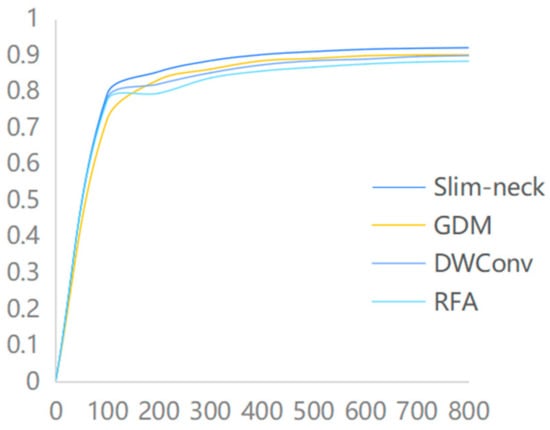
Figure 8.
Comparison of the accuracy of different modules.
3.3.2. Ablation Experiments
To assess the impact of the enhanced modules on the recognition accuracy of YOLOv8, an ablation test is employed for comparison, as shown in Table 2.

Table 2.
Ablation experiment results.
Through a meticulous examination of the data presented in Table 2, it becomes evident that the introduction of the GAM attention module has yielded noteworthy advancements in comparison to the baseline model. Despite a noticeable increase in the overall parameter count, there has been a commendable improvement in key performance metrics. Specifically, the precision (P), recall (R), and mean average precision at 50 (mAP50) have demonstrated enhancements of 2.07%, 2.47%, and 1.56%, respectively.
This notable improvement underscores the effectiveness of the GAM attention module in directing the model’s focus towards pertinent areas of interest, resulting in a marked enhancement in precision. Moreover, following its integration, there have been discernible increments of 0.27%, 1.57%, and 1.38% in P, R, and mAP50, respectively, indicating an augmented recognition accuracy and generalization capability of the model.
Upon the incorporation of the Slim-neck module, further improvements are observed, with P, R, and mAP50 experiencing increases of 0.84%, 0.08%, and 0.56%, respectively. Notably, this enhancement is achieved alongside a substantial reduction in the model’s total parameter count, suggesting that the Slim-neck module effectively streamlines the model’s architecture, while preserving its performance integrity.
In summary, the introduction of the GAM attention module, Slim-neck module, and their associated enhancements have demonstrated their efficacy in enhancing the performance of the YOLOv8 model. This corroborates the viability of the algorithmic refinements proposed in this study, as depicted in Figure 9.
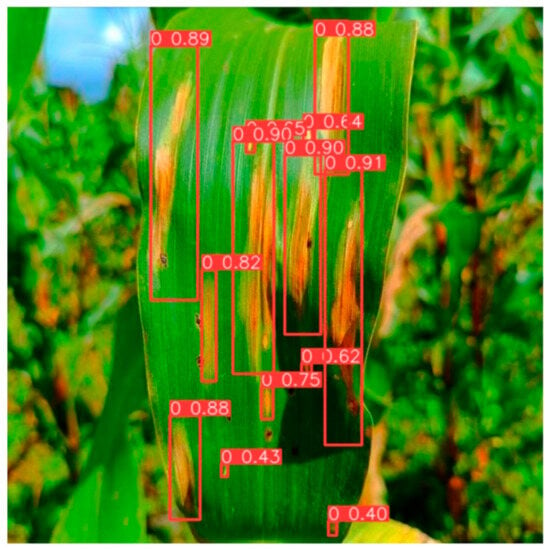
Figure 9.
Validation results graph.
3.3.3. Comparison Test
To assess the performance of the enhanced YOLOv8 model, a comparative analysis is conducted with contemporary mainstream neural network models. The experiments use the YOLOv3, YOLOv5, YOLOv6, YOLOv8, and Faster R-CNN models, using the above experimental parameters with the same dataset, training, and testing; the final experimental results are shown in Table 3.

Table 3.
Comparison results of accuracy of different models.
According to the data in Table 3, compared with the current mainstream neural network model, the improved YOLOv8 model in this paper achieves a 95.18% check accuracy and 89.11% recall rate, under the same parameter settings and dataset conditions, and its mean average precision (mAP50) is 94.65%. The better performance indicates that the improved YOLOv8 model is able to capture the key information more accurately, fuse the dimensional features more adequately, allocate the computational resources more reasonably, and prevent the wastage of computational resources as much as possible, which improves the recognition accuracy of the model. This enables the model to quickly and accurately localize disease locations and identify disease types in complex field environments.
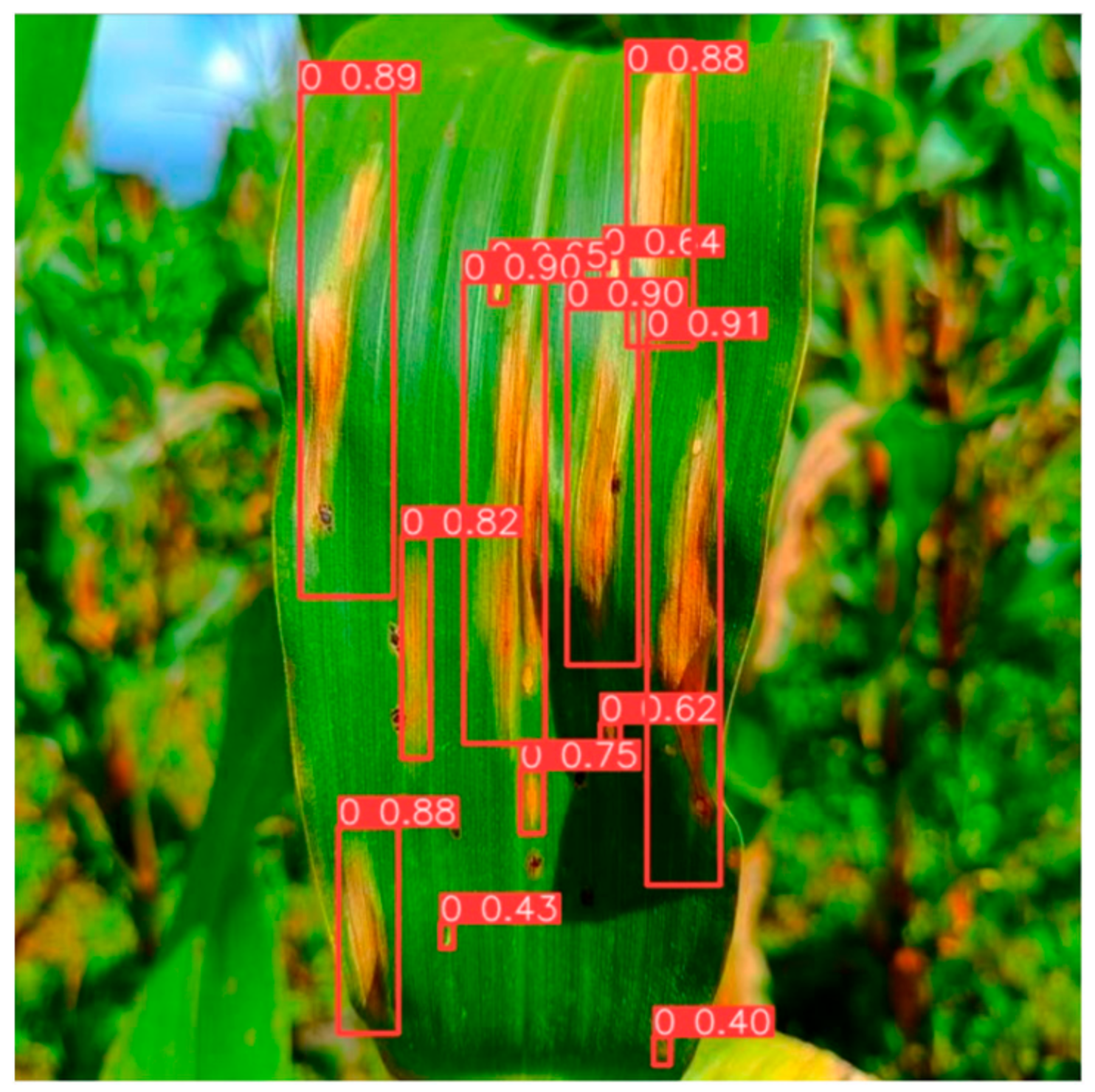
To enhance the credibility of the detection capabilities of the upgraded YOLOv8 model, we employed the optimized weight file of the improved YOLOv8 model to analyze and authenticate images depicting maize leaf spot disease, sourced from the PlantVillage public dataset. The outcomes of this validation process are illustrated in Figure 10. As depicted in Figure 10, it becomes apparent that the enhanced YOLOv8 model adeptly discerns the precise locations of maize leaf spot disease occurrences, free from any instances of False Positives. Consequently, when considering these findings alongside our experimental data, the maize leaf spot disease recognition methodology proposed in this study, anchored in YOLOv8, demonstrates its practical utility for meeting production demands.

Figure 10.
The inference validation graph of the PlantVillage public dataset.
4. Conclusions
In this study, we present an enhancement strategy for the YOLOv8 model. Building upon the original YOLOv8 architecture, we introduce the GAM attention module. This module guides the network to prioritize the relevant region by amplifying global information interaction and amalgamating multidimensional information features. Consequently, this minimizes the inefficient utilization of computational resources in complex field environments and mitigates the impact on recognition outcomes. Additionally, we introduced the Slim-neck module to replace the original Conv and C2f modules, thereby maintaining the model’s recognition accuracy, while simplifying the model structure. On top of CIoU, we introduced Inner-IoU Loss to accelerate model convergence and enhance the model’s generalization capability. By training and testing the model on the corn leaf spot disease dataset, we obtained satisfactory results, as shown in Table 2. The check accuracy and recall were 95.18% and 89.11%, while the mAP50 and mAP50-95 were 94.65% and 71.62%, respectively. Compared with the original YOLOv8 model, they are improved by 3.79%, 4.65%, 3.56%, and 7.3%, respectively. Compared with existing methods, this model has made significant improvements in both accuracy and computational efficiency. By introducing new enhancement strategies, we have successfully reduced detection time, achieving almost real-time detection. Additionally, our model has demonstrated an outstanding performance in complex field environments, reliably completing the task of maize leaf spot disease detection. Therefore, these enhancement strategies not only simplify the model structure, but also effectively improve the model’s recognition accuracy. Overall, our research meets the urgent demand for the rapid localization and identification of maize leaf spot disease in practical applications.
Author Contributions
S.Y.: Situational analysis, data management, original draft writing, review, and editing. J.Y.: Formal analysis, Data curation, Methodology, Writing—review & editing. G.T.: Conceptualization Ideas, Investigation, Supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant number U20A20180), the Agricultural Science and Technology Achievement Transformation Fund Project of Hebei Province (grant number V1705309944504), the Key Research Program of Hebei Province (grant number 21327405D), and the China University Industry Research Innovation Fund (grant number 2021LDA10005).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors upon request.
Acknowledgments
We appreciate the technical support provided by the team we are a part of.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Fan, X.; Zhou, J.; Xu, Y.; Peng, X. Improved convolutional neural network-based maize disease recognition in complex background. JAM 2021, 52, 210–217. [Google Scholar]
- Xu, J.; Shao, M.; Wang, Y.; Han, W. Convolutional neural network corn disease image recognition based on migration learning. JAM 2020, 51, 230–236+253. [Google Scholar]
- Liao, J.; Tao, W.; Zang, Y.; Zeng, H.; Wang, P.; Luo, X. Progress and Prospect of Key Technologies for Remote Sensing Monitoring of Crop Pests and Diseases. JAM 2023, 54, 1–19. [Google Scholar]
- Wu, K. The development direction of crop pest control science and technology in China. J. Agron. 2018, 8, 35–38. [Google Scholar]
- Zhang, S.; Zhang, C. Corn disease identification method based on local discriminant mapping algorithm. J. Agric. Eng-Italy 2014, 30, 167–172. [Google Scholar]
- Mu, J.; Ma, B.; Wang, Y.; Ren, Z.; Liu, S.; Wang, J. A review of crop pest and disease detection algorithms based on deep learning. JAM 2023, 54, 301–313. [Google Scholar]
- Hinton, G.E.; Osindero, S.; The, Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Duan, X.; Ye, Z. Research on corn disease detection with improved YOLOv5 algorithm. Comput. Sci. Explor. 2023, 17, 933–941. [Google Scholar]
- Sun, H.; Chen, J.; Lei, L.; Ji, K.; Kuang, G. A review of techniques for adversarial robustness of deep convolutional neural network image recognition models. Radar J. 2021, 10, 571–594. [Google Scholar]
- Tagarelli, A.; Zumpano, E.; Anastasiu, D.C.; Andrea, C.; Gottfried, V. Managing, mining and learning in the legal data domain. Inf. Syst. 2022, 106, 101981. [Google Scholar] [CrossRef]
- Wang, F.; Wang, Q.; Chen, J.; Liu, F. Improved FasterR-CNN target detection algorithm based on attention mechanism and Soft-NMS. Adv. Lasers Optoelectron. 2021, 58, 405–416. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), San Francisco, CA, USA, 18–20 June 2016; pp. 779–788. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. In Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6517–6525. [Google Scholar]
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. Yolov4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Terven, J.; Cordova-Esparza, D. A comprehensive review of YOLO: From YOLOv1 to YOLOv8 and beyond. arXiv 2023, arXiv:2304.00501. [Google Scholar]
- Liu, R.; He, Z. A fast target detection method for satellite remote sensing images based on YOLOv8. Space Control. Technol. Appl. 2023, 49, 89–97. [Google Scholar]
- Liu, M.; Chu, Z.; Cui, M.; Yang, Q.; Wang, J.; Yang, H. Red-ripe strawberry identification and stalk detection based on improved YOLO v8-Pose. JAM 2023, 54, 244–251. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Sepa- rable Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Piscataway, NJ, USA, 21–26 July 2016; IEEE Press: Piscataway, NJ, USA, 2017; pp. 1257–1258. [Google Scholar]
- Yuan, P.; Ouyang, L.; Zhai, Z.; Ding, Y. Research on lightweight identification of rice diseases based on MobileNetV3Small-ECA. JAM 2024, 55, 253–262. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Li, L.; Lu, S.; Ren, H.; Xu, G.; Zhou, Y. A method for recognizing and localizing mulberry branches and trunks in complex environments based on improved YOLO v5. JAM 2024, 55, 249–257. [Google Scholar]
- Liu, Y.; Shao, Z.; Hoffmann, N. Global attention mechanism: Retain information to enhance channel-spatial interactions. arXiv 2021, arXiv:2112.05561. [Google Scholar]
- Zheng, Z.; Wang, P.; Liu, W.; Li, J.; Ye, R.; Ren, D. Distance-IoU loss: Faster and better learning for bounding box regression. AAAI Conf. Artif. Intell. 2020, 34, 12993–13000. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, C.; Zhang, S. Inner-IoU: More Effective Intersection over Union Loss with Auxiliary Bounding Box. arXiv 2023, arXiv:2311.02877. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).