Role of Genome Sequences of Major and Minor Millets in Strengthening Food and Nutritional Security for Future Generations
Abstract
:1. Introduction
2. Nutritional Profile and Health Benefits of Millets
3. Germplasm Resources of Millets
4. Evolution of Millets
5. Ploidy Level of Millets
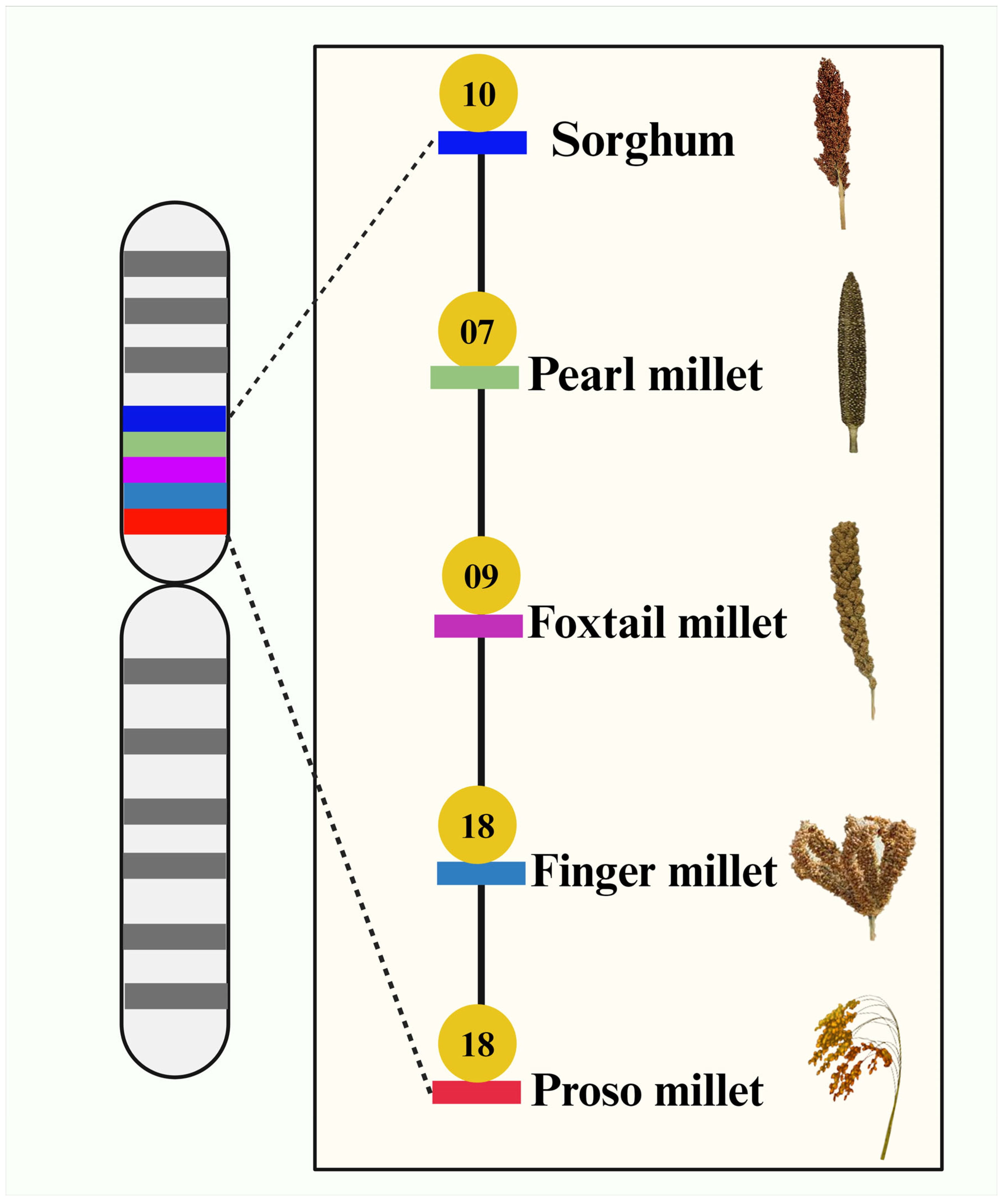
6. Genome Sequences of Millets
6.1. Major Millets
6.1.1. Sorghum
6.1.2. Pearl Millet
6.2. Minor Millets
6.2.1. Foxtail Millet
6.2.2. Finger Millet
6.2.3. Proso Millet
6.2.4. Barnyard Millet
6.2.5. Other Minor Millets
7. Pan-Genomic and Telomere-to-Telomere Genome Resources of Major and Minor Millets
8. QTL Associated with Various Traits in Major and Minor Millets
9. Genome-Wide Association Studies (GWASs) in Millets
10. Identification and Characterization of Candidate Genes from Native Genome Sequences of Millets
11. Will Genome Sequences of Millets Help Improve Food and Nutritional Security by 2050?
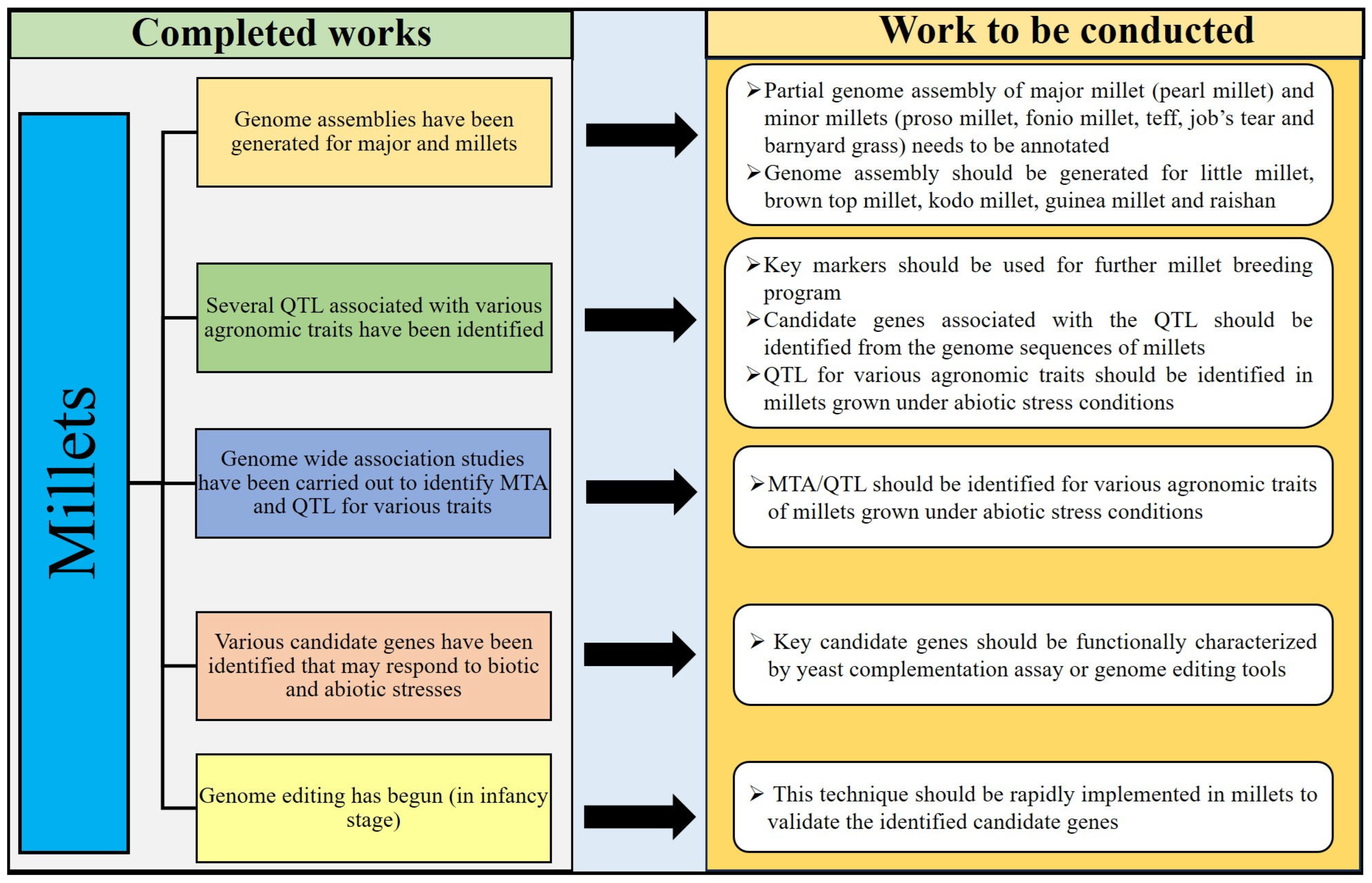
12. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gowda, N.N.; Siliveru, K.; Prasad, P.V.; Bhatt, Y.; Netravati, B.P.; Gurikar, C. Modern processing of Indian millets: A perspective on changes in nutritional properties. Foods 2022, 11, 499. [Google Scholar] [CrossRef]
- Kheya, S.A.; Talukder, S.K.; Datta, P.; Yeasmin, S.; Rashid, M.H.; Hasan, A.K.; Islam, A.K.M.M. Millets: The future crops for the tropics-Status, challenges and future prospects. Heliyon 2023, 9, e22123. [Google Scholar] [CrossRef]
- Ceasar, S.A.; Maharajan, T. The role of millets in attaining United Nation’s sustainable developmental goals. Plants People Planet 2022, 4, 345–349. [Google Scholar] [CrossRef]
- Kumar, A.; Tomer, V.; Kaur, A.; Kumar, V.; Gupta, K. Millets: A solution to agrarian and nutritional challenges. Agric. Food Secur. 2018, 7, 31. [Google Scholar] [CrossRef]
- Vinoth, A.; Ravindhran, R. Biofortification in millets: A sustainable approach for nutritional security. Front. Plant Sci. 2017, 8, 29. [Google Scholar] [CrossRef]
- Budhwar, S.; Sethi, K.; Chakraborty, M. Efficacy of germination and probiotic fermentation on underutilized cereal and millet grains. Food Prod. Process. Nutr. 2020, 2, 12. [Google Scholar] [CrossRef]
- Pramitha, L.J.; Ganesan, J.; Francis, N.; Rajasekharan, R.; Thinakaran, J. Revitalization of small millets for nutritional and food security by advanced genetics and genomics approaches. Front. Genet. 2023, 13, 1007552. [Google Scholar] [CrossRef]
- Hassan, Z.M.; Sebola, N.A.; Mabelebele, M. The nutritional use of millet grain for food and feed: A review. Agric. Food Secur. 2021, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Devi, P.B.; Vijayabharathi, R.; Sathyabama, S.; Malleshi, N.G.; Priyadarisini, V.B. Health benefits of finger millet (Eleusine coracana L.) polyphenols and dietary fiber: A review. J. Food Sci. Technol. 2014, 51, 1021–1040. [Google Scholar] [CrossRef]
- Rai, S.; Kaur, A.; Chopra, C.S. Gluten-free products for celiac susceptible people. Front. Nutr. 2018, 5, 116. [Google Scholar] [CrossRef]
- Liang, S.; Liang, K. Millet grain as a candidate antioxidant food resource: A review. Int. J. Food Prop. 2019, 22, 1652–1661. [Google Scholar] [CrossRef]
- Maharajan, T.; Ceasar, S.A.; Krishna, T.P.A.; Ignacimuthu, S. Finger millet [Eleusine coracana (L.) Gaertn]: An orphan crop with a potential to alleviate the calcium deficiency in the semi-arid tropics of Asia and Africa. Front. Sustain. Food Syst. 2021, 5, 684447. [Google Scholar] [CrossRef]
- Anitha, S.; Kane-Potaka, J.; Botha, R.; Givens, D.I.; Sulaiman, N.L.B.; Upadhyay, S.; Bhandari, R.K. Millets can have a major impact on improving iron status, hemoglobin level, and in reducing iron deficiency anemia—A systematic review and meta-analysis. Front. Nutr. 2021, 8, 725529. [Google Scholar] [CrossRef] [PubMed]
- Singhal, T.; Tara Satyavathi, C.; Singh, S.P.; Mallik, M.; Anuradha, N.; Sankar, S.M.; Singh, N. Achieving nutritional security in India through iron and zinc biofortification in pearl millet (Pennisetum glaucum (L.) R. Br.). Physiol. Mol. Biol. Plants 2022, 28, 849–869. [Google Scholar] [CrossRef] [PubMed]
- Anitha, S.; Govindaraj, M.; Kane-Potaka, J. Balanced amino acid and higher micronutrients in millets complements legumes for improved human dietary nutrition. Cereal Chem. 2020, 97, 74–84. [Google Scholar] [CrossRef]
- Saleh, A.S.; Zhang, Q.; Chen, J.; Shen, Q. Millet grains: Nutritional quality, processing, and potential health benefits. Compr. Rev. Food Sci. Food Saf. 2013, 12, 281–295. [Google Scholar] [CrossRef]
- Longvah, T.; Anantan, I.; Bhaskarachary, K.; Venkaiah, K.; Longvah, T. Indian Food Composition Tables; Longvah, T., Ed.; National Institute of Nutrition, Indian Council of Medical Research: Hyderabad, India, 2017; pp. 2–58. [Google Scholar]
- Shobana, S.; Krishnaswamy, K.; Sudha, V.; Malleshi, N.G.; Anjana, R.M.; Palaniappan, L.; Mohan, V. Finger millet (Ragi, Eleusine coracana L.): A review of its nutritional properties, processing, and plausible health benefits. Adv. Food Nutr. Res. 2013, 69, 1–39. [Google Scholar] [PubMed]
- Anitha, S.; Botha, R.; Kane-Potaka, J.; Givens, D.I.; Rajendran, A.; Tsusaka, T.W.; Bhandari, R.K. Can millet consumption help manage hyperlipidemia and obesity? a systematic review and meta-analysis. Front. Nutr. 2021, 8, 700778. [Google Scholar] [CrossRef] [PubMed]
- Sabuz, A.A.; Rana, M.R.; Ahmed, T.; Molla, M.M.; Islam, N.; Khan, H.H.; Shen, Q. Health-promoting potential of millet: A review. Separations 2023, 10, 80. [Google Scholar] [CrossRef]
- Agrawal, P.; Singh, B.R.; Gajbe, U.; Kalambe, M.A.; Bankar, M. Managing diabetes mellitus with millets: A new solution. Cureus 2023, 15, e44908. [Google Scholar] [CrossRef] [PubMed]
- Uttara, B.; Singh, A.V.; Zamboni, P.; Mahajan, R. Oxidative stress and neurodegenerative diseases: A review of upstream and downstream antioxidant therapeutic options. Curr. Neuropharmacol. 2009, 7, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xian, F.; Guan, X.; Huang, K.; Yu, W.; Liu, D. Neural protective effects of millet and millet polyphenols on high-fat diet-induced oxidative stress in the brain. Plant Foods Hum. Nutr. 2020, 75, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Rani, M.; Mani, S.; Shah, P.; Singh, D.B.; Kudapa, H.; Varshney, R.K. Nutritional significance and antioxidant-mediated antiaging effects of finger millet: Molecular insights and prospects. Front. Sustain. Food Syst. 2021, 5, 684318. [Google Scholar] [CrossRef]
- Chauhan, M.; Sonawane, S.K.; Arya, S.S. Nutritional and nutraceutical properties of millets: A review. Am. J. Clin. Nutr. 2018, 1, 1–10. [Google Scholar]
- Anitha, S.; Givens, D.I.; Botha, R.; Kane-Potaka, J.; Sulaiman, N.L.B.; Tsusaka, T.W.; Bhandari, R.K. Calcium from finger millet—A systematic review and meta-analysis on calcium retention, bone resorption, and in vitro bioavailability. Sustainability 2021, 13, 8677. [Google Scholar] [CrossRef]
- Das, S.; Khound, R.; Santra, M.; Santra, D.K. Beyond bird feed: Proso millet for human health and environment. Agriculture 2019, 9, 64. [Google Scholar] [CrossRef]
- Bunkar, D.S.; Goyal, S.K.; Meena, K.K.; Kamalvanshi, V. Nutritional, functional role of kodo millet and its processing: A review. Int. J. Curr. Microbiol. Appl. Sci. 2021, 10, 1972–1985. [Google Scholar] [CrossRef]
- Singh, N.; Kumari, P.; Bassi, J.G.N. Nutrient rich Kodo millet, importance and value addition: An overview. Pharma Innov. J. 2023, 12, 3713–3719. [Google Scholar]
- Dwivedi, S.L.; Upadhyaya, H.D.; Senthilvel, S.; Hash, C.T.; Fukunaga, K.; Diao, X.; Prasad, M. Millets: Genetic and genomic resources. Genetic and Genomic Resources. In Plant Breeding Reviews; Wiley-Blackwell: Hoboken, NJ, USA, 2012; pp. 247–375. [Google Scholar]
- Vetriventhan, M.; Azevedo, V.C.; Upadhyaya, H.D.; Nirmalakumari, A.; Kane-Potaka, J.; Anitha, S.; Tonapi, V.A. Genetic and genomic resources, and breeding for accelerating improvement of small millets: Current status and future interventions. Nucleus 2020, 63, 217–239. [Google Scholar] [CrossRef]
- Bramel, P.; Giovannini, P.; Eshan Dulloo, M. Global Strategy for the Conservation and Use of Genetic Resources of Selected Millets; Paula, B., Peter, G., Eshan Dulloo, M., Eds.; Global Crop Diversity Trust: Bonn, Germany, 2022; pp. 1–88. [Google Scholar]
- Tripathi, T.; Vyas, S. From ancient grains to modern solutions: A history of millets and their significance in agriculture and food security. Int. J. Home Sci. 2023, 9, 72–78. [Google Scholar]
- Yang, X.; Wan, Z.; Perry, L.; Lu, H.; Wang, Q.; Zhao, C.; Ge, Q. Early millet use in northern China. Proc. Natl. Acad. Sci. USA 2012, 109, 3726–3730. [Google Scholar] [CrossRef] [PubMed]
- Kangama, C.O. Pearl millet (Pennisetum glaucum) perspectives in Africa. Int. J. Sci. Res. 2021, 2, 001–007. [Google Scholar]
- Burgarella, C.; Cubry, P.; Kane, N.A.; Varshney, R.K.; Mariac, C.; Liu, X.; Vigouroux, Y. A western Sahara centre of domestication inferred from pearl millet genomes. Nat. Ecol. Evol. 2018, 2, 1377–1380. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; Vanga, S.K.; Wang, J.; Orsat, V.; Raghavan, V. Millets for food security in the context of climate change: A review. Sustainability 2018, 10, 2228. [Google Scholar] [CrossRef]
- Lu, H.; Zhang, J.; Liu, K.B.; Wu, N.; Li, Y.; Zhou, K.; Li, Q. Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc. Natl. Acad. Sci. USA 2009, 106, 7367–7372. [Google Scholar] [CrossRef] [PubMed]
- Animasaun, D.A.; Morakinyo, J.A.; Mustapha, O.T.; Krishnamurthy, R. Genome size and ploidy variations in pearl millet (Pennisetum glaucum) and napier grass (Pennisetum purpureum) genotypes. Acta Agron. 2019, 68, 299–305. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Devos, K.M. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 2012, 30, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, X.; Quan, Z.; Cheng, S.; Xu, X.; Pan, S.; Wang, J. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 2012, 30, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Hittalmani, S.; Mahesh, H.B.; Shirke, M.D.; Biradar, H.; Uday, G.; Aruna, Y.R.; Mohanrao, A. Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genom. 2017, 18, 465. [Google Scholar] [CrossRef]
- Hatakeyama, M.; Aluri, S.; Balachadran, M.T.; Sivarajan, S.R.; Patrignani, A.; Grüter, S.; Shimizu, K.K. Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res. 2018, 25, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Devos, K.M.; Qi, P.; Bahri, B.A.; Gimode, D.M.; Jenike, K.; Manthi, S.J.; Odeny, D.A. Genome analyses reveal population structure and a purple stigma color gene candidate in finger millet. Nat. Commun. 2023, 14, 3694. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Qiu, J.; Ye, C.; Jin, G.; Mao, L.; Zhang, H.; Fan, L. Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 2017, 8, 1031. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Li, L.; Miki, D.; Li, D.; Tang, Q.; Xiao, L.; Zhang, H. The genome of broomcorn millet. Nat. Commun. 2019, 10, 436. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Deshpande, S.; Vetriventhan, M.; Upadhyaya, H.D.; Wallace, J.G. Genome-wide population structure analyses of three minor millets: Kodo millet, little millet, and proso millet. Plant Genome 2019, 12, 190021. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Rokhsar, D.S. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Shi, C.; Thudi, M.; Mariac, C.; Wallace, J.; Qi, P.; Xu, X. Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat. Biotechnol. 2017, 35, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Sebastin, R.; Lee, G.A.; Lee, K.J.; Shin, M.J.; Cho, G.T.; Lee, J.R.; Chung, J.W. The complete chloroplast genome sequences of little millet (Panicum sumatrense Roth ex Roem. and Schult.) (Poaceae). Mitochondrial DNA Part B 2018, 3, 719–720. [Google Scholar] [CrossRef] [PubMed]
- Cannarozzi, G.; Plaza-Wüthrich, S.; Esfeld, K.; Larti, S.; Wilson, Y.S.; Girma, D.; Tadele, Z. Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef). BMC Genom. 2014, 15, 581. [Google Scholar] [CrossRef] [PubMed]
- Abrouk, M.; Ahmed, H.I.; Cubry, P.; Šimoníková, D.; Cauet, S.; Pailles, Y.; Krattinger, S.G. Fonio millet genome unlocks African orphan crop diversity for agriculture in a changing climate. Nat. Commun. 2020, 11, 4488. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H.; Kim, B.; Choi, B.S.; Lee, H.O.; Lee, S.J.; Oh, T.J.; Kim, C.K. Genome assembly and annotation of soft-shelled adlay (Coix lacryma-jobi Variety ma-yuen), a cereal and medicinal crop in the Poaceae family. Front. Plant Sci. 2020, 11, 523041. [Google Scholar] [CrossRef]
- Li, X.D.; Pan, H.; Lu, X.J.; Wei, X.Y.; Shi, M.; Lu, P. Complete chloroplast genome sequencing of Job’s tears (Coix l.): Genome structure, comparative analysis, and phylogenetic relationships. Mitochondrial DNA Part B 2021, 6, 1399–1405. [Google Scholar] [CrossRef] [PubMed]
- Naithani, S.; Deng, C.H.; Sahu, S.K.; Jaiswal, P. Exploring pan-genomes: An overview of resources and tools for unraveling structure, function, and evolution of crop genes and genomes. Biomolecules 2023, 13, 1403. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Luo, H.; Xu, J.; Cruickshank, A.; Zhao, X.; Teng, F.; Mace, E. Extensive variation within the pan-genome of cultivated and wild sorghum. Nat. Plants 2021, 7, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Ruperao, P.; Thirunavukkarasu, N.; Gandham, P.; Selvanayagam, S.; Govindaraj, M.; Nebie, B.; Rathore, A. Sorghum pan-genome explores the functional utility for genomic-assisted breeding to accelerate the genetic gain. Front. Plant Sci. 2021, 12, 666342. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Sun, M.; Zhang, Z.; Jin, Y.; Zhang, A.; Lin, C.; Huang, L. Pangenomic analysis identifies structural variation associated with heat tolerance in pearl millet. Nat. Genet. 2023, 55, 507–518. [Google Scholar] [CrossRef]
- He, Q.; Tang, S.; Zhi, H.; Chen, J.; Zhang, J.; Liang, H.; Diao, X. A graph-based genome and pan-genome variation of the model plant Setaria. Nat. Genet. 2023, 55, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, Y.; Liu, M.; Guo, W.; Wang, Y.; He, Q.; Diao, X. Pangenome analysis reveals genomic variations associated with domestication traits in broomcorn millet. Nat. Genet. 2023, 55, 2243–2254. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, Y.; Xu, J.; Jiang, F.; Li, W.; Zhang, Q.; Zhang, L. A telomere-to-telomere genome assembly of Hongyingzi, a sorghum cultivar used for Chinese Baijiu production. Crop J. 2024, 1–6. [Google Scholar] [CrossRef]
- Tian, B.; Zhang, L.; Liu, Y.; Wu, P.; Wang, W.; Zhang, Y.; Li, H. Identification of QTL for resistance to leaf blast in foxtail millet by genome re-sequencing analysis. Theor. Appl. Genet. 2021, 134, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Ambawat, S.; Senthilvel, S.; Hash, C.T.; Nepolean, T.; Rajaram, V.; Eshwar, K.; Srivastava, R.K. QTL mapping of pearl millet rust resistance using an integrated DArT-and SSR-based linkage map. Euphytica 2016, 209, 461–476. [Google Scholar] [CrossRef]
- Zoclanclounon, Y.A.B.; Kanfany, G.; Kane, A.; Fonceka, D.; Ehemba, G.L.; Ly, F. Current status of pearl millet downy mildew prevalence across agroecological zones of Senegal. Sci. World J. 2019, 2019, 1252653. [Google Scholar] [CrossRef] [PubMed]
- Chelpuri, D.; Sharma, R.; Durga, K.K.; Katiyar, P.; Mahendrakar, M.D.; Singh, R.B.; Srivastava, R.K. Mapping quantitative trait loci (QTLs) associated with resistance to major pathotype-isolates of pearl millet downy mildew pathogen. Eur. J. Plant Pathol. 2019, 154, 983–994. [Google Scholar] [CrossRef]
- Kumar, S.; Hash, C.T.; Nepolean, T.; Mahendrakar, M.D.; Satyavathi, C.T.; Singh, G.; Srivastava, R.K. Mapping grain iron and zinc content quantitative trait loci in an iniadi-derived immortal population of pearl millet. Genes 2018, 9, 248. [Google Scholar] [CrossRef] [PubMed]
- Singhal, T.; Satyavathi, C.T.; Singh, S.P.; Kumar, A.; Sankar, S.M.; Bhardwaj, C.; Singh, N. Multi-environment quantitative trait loci mapping for grain iron and zinc content using bi-parental recombinant inbred line mapping population in pearl millet. Front. Sci. Plant Sci. 2021, 12, 659789. [Google Scholar] [CrossRef] [PubMed]
- Maharajan, T.; Ajeesh Krishna, T.P.; Rakkammal, K.; Ramakrishnan, M.; Ceasar, S.A.; Ramesh, M.; Ignacimuthu, S. Identification of QTL Associated with Agro-Morphological and Phosphorus Content Traits in Finger Millet under Differential Phosphorus Supply via Linkage Mapping. Agriculture 2023, 13, 262. [Google Scholar] [CrossRef]
- Ramakrishnan, M.; Ceasar, S.A.; Vinod, K.K.; Duraipandiyan, V.; Ajeesh Krishna, T.P.; Upadhyaya, H.D.; Ignacimuthu, S. Identification of putative QTLs for seedling stage phosphorus starvation response in finger millet (Eleusine coracana L. Gaertn.) by association mapping and cross species synteny analysis. PLoS ONE 2017, 12, e0183261. [Google Scholar] [CrossRef] [PubMed]
- David, R.H.A.; Ramakrishnan, M.; Maharajan, T.; BarathiKannan, K.; Babu, G.A.; Daniel, M.A.; Ignacimuthu, S. Mining QTL and genes for root traits and biochemical parameters under vegetative drought in South Indian genotypes of finger millet (Eleusine coracana (L.) Gaertn) by association mapping and in silico comparative genomics. Biocatal. Agric. Biotechnol. 2021, 32, 101935. [Google Scholar]
- Kumar, S.; Hash, C.T.; Nepolean, T.; Satyavathi, C.T.; Singh, G.; Mahendrakar, M.D.; Srivastava, R.K. Mapping QTLs controlling flowering time and important agronomic traits in pearl millet. Front. Sci. Plant Sci. 2017, 8, 1731. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Hash, C.T.; Singh, G.; Nepolean, T.; Srivastava, R.K. Mapping QTLs for important agronomic traits in an Iniadi-derived immortal population of pearl millet. Biotechnol. Notes 2021, 2, 26–32. [Google Scholar] [CrossRef]
- Vengadessan, V.; Rai, K.N.; Kannan Bapu, J.R.; Hash, C.T.; Bhattacharjee, R.; Senthilvel, S.; Nepolean, T. Construction of genetic linkage map and QTL analysis of sink-size traits in pearl millet (Pennisetum glaucum). Int. Sch. Res. Not. 2013, 2013, 471632. [Google Scholar] [CrossRef]
- Nepolean, T.; Blummel, M.; Raj, A.B.; Rajaram, V.; Senthilvel, S.; Hash, C.T. QTLs controlling yield and stover quality traits in pearl millet. J. SAT Agric. Res. 2006, 2, 1–4. [Google Scholar]
- Gulia, S.K.; Hash, C.T.; Thakur, R.P.; Breese, W.A.; Sangwan, R.S. Mapping new QTLs for improvement of downy mildew resistance in pearl millet. In Crop Production in Stress Environments: Genetic and Management Options; Agrobios International: Jodhpur, India, 2007; pp. 373–386. [Google Scholar]
- Bidinger, F.R.; Nepolean, T.; Hash, C.T.; Yadav, R.S.; Howarth, C.J. Quantitative trait loci for grain yield in pearl millet under variable postflowering moisture conditions. Crop Sci. 2007, 47, 969–980. [Google Scholar] [CrossRef]
- Liu, T.; He, J.; Dong, K.; Wang, X.; Wang, W.; Yang, P.; Yang, T. QTL mapping of yield component traits on bin map generated from resequencing a RIL population of foxtail millet (Setaria italica). BMC Genom. 2020, 21, 141. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhi, H.; Tang, S.; Xing, L.; Wang, S.; Wang, H.; Diao, X. QTL mapping for foxtail millet plant height in multi-environment using an ultra-high density bin map. Theor. Appl. Genet. 2021, 134, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, J.; Peng, J.; Du, X.; Jiang, M.; Li, Y.; Guo, E. QTL mapping for 11 agronomic traits based on a genome-wide Bin-map in a large F 2 population of foxtail millet (Setaria italica (L.) P. Beauv). Mol. Breed. 2019, 39, 18. [Google Scholar] [CrossRef]
- Guo, S.; Chai, S.; Guo, Y.; Shi, X.; Han, F.; Qu, T.; Yang, P. Mapping of major QTL and candidate gene analysis for hull colour in foxtail millet (Setaria italica (L.) P. Beauv.). BMC Genom. 2023, 24, 458. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Fan, G.; Zhang, X.; Zhao, F.; Wei, W.; Du, G.; Zhao, Z. Identification of QTLs for 14 agronomically important traits in Setaria italica based on SNPs generated from high-throughput sequencing. G3 Genes Genom. Genet. 2017, 7, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; He, J.; Dong, K.; Wang, X.; Zhang, L.; Ren, R.; Zhang, Z. Genome-wide identification of quantitative trait loci for morpho-agronomic and yield-related traits in foxtail millet (Setaria italica) across multi-environments. Mol. Genet. Genom. 2022, 297, 873–888. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Z.; Du, X.; Yang, H.; Han, F.; Han, Y.; Guo, E. A high-density genetic map and QTL analysis of agronomic traits in foxtail millet [Setaria italica (L.) P. Beauv.] using RAD-seq. PLoS ONE 2017, 12, e0179717. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Hou, J.; Fu, N.; Wei, M.; Li, Y.; Yu, K.; Liu, J. Identification of QTL related to anther color and hull color by RAD sequencing in a RIL population of Setaria italica. BMC Genom. 2021, 22, 556. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, M.; Antony Ceasar, S.; Duraipandiyan, V.; Vinod, K.K.; Kalpana, K.; Al-Dhabi, N.A.; Ignacimuthu, S. Tracing QTLs for leaf blast resistance and agronomic performance of finger millet (Eleusine coracana (L.) Gaertn.) genotypes through association mapping and in silico comparative genomics analyses. PLoS ONE 2016, 11, e0159264. [Google Scholar] [CrossRef] [PubMed]
- Pendergast, T.H., IV; Qi, P.; Odeny, D.A.; Dida, M.M.; Devos, K.M. A high-density linkage map of finger millet provides QTL for blast resistance and other agronomic traits. Plant Genom. 2022, 15, e20175. [Google Scholar] [CrossRef] [PubMed]
- Kalyana Babu, B.; Agrawal, P.K.; Pandey, D.; Jaiswal, J.P.; Kumar, A. Association mapping of agro-morphological characters among the global collection of finger millet genotypes using genomic SSR markers. Mol. Biol. Rep. 2014, 41, 5287–5297. [Google Scholar] [CrossRef] [PubMed]
- Rajput, S.G.; Santra, D.K.; Schnable, J. Mapping QTLs for morpho-agronomic traits in proso millet (Panicum miliaceum L.). Mol. Breed. 2016, 36, 37. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Y.; Hou, S.; Men, Y.; Han, Y. The Integration of Genome-Wide Association Study and Homology Analysis to Explore the Genomic Regions and Candidate Genes for Panicle-Related Traits in Foxtail Millet. Int. J. Mol. Sci. 2022, 23, 14735. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, V.; Gupta, S.; Gahlaut, V.; Muthamilarasan, M.; Bandyopadhyay, T.; Ramchiary, N.; Prasad, M. Genome-wide association study of major agronomic traits in foxtail millet (Setaria italica L.) using ddRAD sequencing. Sci. Rep. 2019, 9, 5020. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, V.; Bandyopadhyay, T.; Gahlaut, V.; Gupta, S.; Dhaka, A.; Ramchiary, N.; Prasad, M. Genome-wide association study (GWAS) delineates genomic loci for ten nutritional elements in foxtail millet (Setaria italica L.). J. Cereal Sci. 2019, 85, 48–55. [Google Scholar] [CrossRef]
- Pujar, M.; Gangaprasad, S.; Govindaraj, M.; Gangurde, S.S.; Kanatti, A.; Kudapa, H. Genome-wide association study uncovers genomic regions associated with grain iron, zinc and protein content in pearl millet. Sci. Rep. 2020, 10, 19473. [Google Scholar] [CrossRef] [PubMed]
- Diack, O.; Kanfany, G.; Gueye, M.C.; Sy, O.; Fofana, A.; Tall, H.; Kane, N.A. GWAS unveils features between early-and late-flowering pearl millets. BMC Genom. 2020, 21, 777. [Google Scholar] [CrossRef] [PubMed]
- Yadav, C.B.; Srivastava, R.K.; Beynon, S.; Englyst, K.; Gangashetty, P.I.; Yadav, R.S. Genetic variability and genome-wide marker association studies for starch traits contributing to low glycaemic index in pearl millet. Food Energy Sec. 2022, 11, e341. [Google Scholar] [CrossRef]
- Boukail, S.; Macharia, M.; Miculan, M.; Masoni, A.; Calamai, A.; Palchetti, E.; Dell’Acqua, M. Genome wide association study of agronomic and seed traits in a world collection of proso millet (Panicum miliaceum L.). BMC Plant Biol. 2021, 21, 330. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Beynon, S.; Srivastava, R.K.; Sehgal, D.; Ojulong, H.; Yadav, R. Genome-wide association mapping and comparative genomics identifies genomic regions governing grain nutritional traits in finger millet (Eleusine coracana L. Gaertn.). Plants People Planet 2020, 2, 649–662. [Google Scholar] [CrossRef]
- Adeyanju, A.; Little, C.; Yu, J.; Tesso, T. Genome-wide association study on resistance to stalk rot diseases in grain sorghum. G3 Genes Genomes Genet. 2015, 5, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Boyles, R.E.; Cooper, E.A.; Myers, M.T.; Brenton, Z.; Rauh, B.L.; Morris, G.P.; Kresovich, S. Genome-wide association studies of grain yield components in diverse sorghum germplasm. Plant Genome 2016, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Mantilla Perez, M.B.; Hu, J.; Salas Fernandez, M.G. Genome-wide association study for nine plant architecture traits in Sorghum. Plant Genome 2016, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chopra, R.; Burow, G.; Burke, J.J.; Gladman, N.; Xin, Z. Genome-wide association analysis of seedling traits in diverse Sorghum germplasm under thermal stress. BMC Plant Biol. 2017, 17, 12. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chopra, R.; Hayes, C.; Morris, G.; Marla, S.; Burke, J.; Burow, G. Genome-wide association study of developing leaves’ heat tolerance during vegetative growth stages in a sorghum association panel. Plant Genome 2017, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Parra-Londono, S.; Fiedler, K.; Kavka, M.; Samans, B.; Wieckhorst, S.; Zacharias, A.; Uptmoor, R. Genetic dissection of early-season cold tolerance in sorghum: Genome-wide association studies for seedling emergence and survival under field and controlled environment conditions. Theor. Appl. Genet. 2018, 131, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tang, W.; Zhang, Y.W.; Chen, K.N.; Wang, C.; Liu, Y.; Wang, L. Genome-wide association studies for five forage quality-related traits in sorghum (Sorghum bicolor L.). Front. Plant Sci. 2018, 9, 1146. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, H.E.; Fermin-Pérez, R.A.; Prom, L.K.; Cooper, E.A.; Bean, S.; Rooney, W.L. Genome-wide association mapping of grain mold resistance in the US sorghum association panel. Plant Genome 2019, 12, 180070. [Google Scholar] [CrossRef] [PubMed]
- Girma, G.; Nida, H.; Seyoum, A.; Mekonen, M.; Nega, A.; Lule, D.; Mengiste, T. A large-scale genome-wide association analyses of Ethiopian sorghum landrace collection reveal loci associated with important traits. Front. Plant Sci. 2019, 10, 691. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.R.; Wang, C.Y.; Wang, P.; Zhu, Z.X.; Ning, X.U.; Shi, G.S.; Huang, R.D. Genome-wide association study for starch content and constitution in sorghum (Sorghum bicolor (L.) Moench). J. Integr. Agric. 2019, 18, 2446–2456. [Google Scholar] [CrossRef]
- Tao, Y.; Zhao, X.; Wang, X.; Hathorn, A.; Hunt, C.; Cruickshank, A.W.; Jordan, D.R. Large-scale GWAS in sorghum reveals common genetic control of grain size among cereals. Plant Biotechnol. J. 2020, 18, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Cruet-Burgos, C.; Cox, S.; Ioerger, B.P.; Perumal, R.; Hu, Z.; Herald, T.J.; Rhodes, D.H. Advancing provitamin A biofortification in sorghum: Genome-wide association studies of grain carotenoids in global germplasm. Plant Genome 2020, 13, e20013. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Upadhyaya, H.D.; Zheng, J.; Liu, Y.; Singh, S.K.; Gowda, C.L.L.; Li, J. Genome-Wide association mapping identifies novel panicle morphology loci and candidate genes in sorghum. Front. Plant Sci. 2021, 12, 743838. [Google Scholar] [CrossRef] [PubMed]
- Maina, F.; Harou, A.; Hamidou, F.; Morris, G.P. Genome-wide association studies identify putative pleiotropic locus mediating drought tolerance in sorghum. Plant Dir. 2022, 6, e413. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Y.; Gao, L.; Yang, X.; Zhang, X.; Xie, S.; Shen, Y. Identification of candidate forage yield genes in sorghum (Sorghum bicolor L.) using integrated genome-wide association studies and RNA-seq. Front. Plant Sci. 2022, 12, 788433. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Yang, B.; Kim, W.J.; Kim, J.; Kwon, S.J.; Kim, J.H.; Ryu, J. Genome-Wide Association Study (GWAS) of the Agronomic Traits and Phenolic Content in Sorghum (Sorghum bicolor L.) Genotypes. Agronomy 2023, 13, 1449. [Google Scholar] [CrossRef]
- Xin, Y.; Gao, L.; Hu, W.; Gao, Q.; Yang, B.; Zhou, J.; Xu, C. Genome-wide association study based on plant height and drought-tolerance indices reveals two candidate drought-tolerance genes in sweet sorghum. Sustainability 2022, 14, 14339. [Google Scholar] [CrossRef]
- Ahn, E.; Botkin, J.; Ellur, V.; Lee, Y.; Poudel, K.; Prom, L.K.; Magill, C. Genome-wide association study of seed morphology traits in senegalese sorghum cultivars. Plants 2023, 12, 2344. [Google Scholar] [CrossRef] [PubMed]
- Yadav, C.B.; Srivastava, R.K.; Gangashetty, P.I.; Yadav, R.; Mur, L.A.; Yadav, R.S. Metabolite Diversity and Metabolic Genome-Wide Marker Association Studies (Mgwas) for Health Benefiting Nutritional Traits in Pearl Millet Grains. Cells 2021, 10, 3076. [Google Scholar] [CrossRef] [PubMed]
- Yadav, C.B.; Tokas, J.; Yadav, D.; Winters, A.; Singh, R.B.; Yadav, R.; Yadav, R.S. Identifying anti-oxidant biosynthesis genes in pearl millet [Pennisetum glaucum (L.) R. Br.] using genome—Wide association analysis. Front. Sci. Plant. Sci. 2021, 12, 599649. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Huang, X.; Zhi, H.; Zhao, Y.; Zhao, Q.; Li, W.; Chai, Y.; Yang, L.; Liu, K.; Lu, H.; et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 2013, 45, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, V.; Bandyopadhyay, T.; Singh, R.K.; Gahlaut, V.; Muthamilarasan, M.; Prasad, M. Multi-environment GWAS identifies genomic regions underlying grain nutrient traits in foxtail millet (Setaria italica). Plant Cell Rep. 2024, 43, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Sun, S.; Jin, C.; Su, J.; Wei, J.; Luo, X.; Wen, J.; Wei, T.; Sahu, S.K.; et al. GWAS, MWAS and mGWAS provide insights into precision agriculture based on genotype-dependent microbial effects in foxtail millet. Nat. Commun. 2022, 13, 5913. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Chen, Y.; Liu, Z.; Sun, M.; Han, F.; Wang, T.; Chen, L. Genome-wide association mapping for agronomic and quality traits in foxtail millet. 2019; preprint. [Google Scholar] [CrossRef]
- Sharma, D.; Tiwari, A.; Sood, S.; Jamra, G.; Singh, N.K.; Meher, P.K.; Kumar, A. Genome wide association mapping of agro-morphological traits among a diverse collection of finger millet (Eleusine coracana L.) genotypes using SNP markers. PLoS ONE 2018, 13, e0199444. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Sharma, D.; Sood, S.; Jaiswal, J.P.; Pachauri, S.P.; Ramteke, P.W.; Kumar, A. Genome-wide association mapping for seed protein content in finger millet (Eleusine coracana) global collection through genotyping by sequencing. J. Cereal Sci. 2020, 91, 102888. [Google Scholar] [CrossRef]
- Fan, Y.; Lai, D.; Yang, H.; Xue, G.; He, A.; Chen, L.; Cheng, J. Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet (Setaria italica L.). BMC Genom. 2021, 22, 778. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hao, D.; Wang, X.; Zhang, H.; Yang, P.; Zhang, L.; Zhang, B. Genome-wide identification and expression analysis of the SNARE genes in Foxtail millet (Setaria italica) reveals its roles in drought stress. Plant Growth Reg. 2021, 95, 355–369. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, B.; Zheng, G.; Zhang, A.; Li, R. Genome-wide characterization of the SiDof gene family in foxtail millet (Setaria italica). Biosystems 2017, 151, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shu, H.; Zhang, A.Y.; Liu, B.L.; Xing, G.F.; Xue, J.A.; Li, R.Z. Foxtail millet WRKY genes and drought stress. J. Agric. Sci. 2017, 155, 777–790. [Google Scholar] [CrossRef]
- Ceasar, S.A.; Hodge, A.; Baker, A.; Baldwin, S.A. Phosphate concentration and arbuscular mycorrhizal colonisation influence the growth, yield and expression of twelve PHT1 family phosphate transporters in foxtail millet (Setaria italica). PLoS ONE 2014, 9, e108459. [Google Scholar] [CrossRef] [PubMed]
- Ceasar, S.A.; Baker, A.; Ignacimuthu, S. Functional characterization of the PHT1 family transporters of foxtail millet with development of a novel Agrobacterium-mediated transformation procedure. Sci. Rep. 2017, 7, 14064. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xiao, W.; Yu, W.; Yao, L.; Li, L.; Wei, J.; Li, R. Foxtail millet SiHAK1 excites extreme high-affinity K+ uptake to maintain K+ homeostasis under low K+ or salt stress. Plant Cell Rep. 2018, 37, 1533–1546. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, J.; Liang, Y.; Wang, X.; Li, K.; Chen, L.; Hou, S. Natural Resistance-Associated Macrophage Protein (Nramp) Family in Foxtail Millet (Setaria italica): Characterization, Expression Analysis and Relationship with Metal Content under Cd Stress. Agronomy 2023, 13, 2000. [Google Scholar] [CrossRef]
- Maharajan, T.; Krishna, T.P.A.; Shilpha, J.; Ceasar, S.A. Effects of Individual or Combined Deficiency of Phosphorous and Zinc on Phenotypic, Nutrient Uptake, and Molecular Responses of Finger Millet (Eleusine coracana): A Nutri-Rich Cereal Crop. Plants 2023, 12, 3378. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, A.; Mallikarjuna, M.G.; Bansal, S.; Nayaka, S.C.; Rajashekara, H.; Chellapilla, T.S.; Prakash, G. Genome-wide identification and characterization of NBLRR genes in Finger millet (Eleusine coracana L.) and their expression in response to Magnaporthe grisea infection. BMC Plant Biol. 2024, 24, 75. [Google Scholar] [CrossRef]
- Rakkammal, K.; Maharajan, T.; Shriram, R.N.; Ram, P.J.; Ceasar, S.A.; Ramesh, M. Physiological, biochemical and molecular responses of finger millet (Eleusine coracana) genotypes exposed to short-term drought stress induced by PEG-6000. S. Afr. J. Bot. 2023, 155, 45–59. [Google Scholar] [CrossRef]
- Lin, M.; Dong, Z.; Zhou, H.; Wu, G.; Xu, L.; Ying, S.; Chen, M. Genome-Wide Identification and Transcriptional Analysis of the MYB Gene Family in Pearl Millet (Pennisetum glaucum). Int. J. Mol. Sci. 2023, 24, 2484. [Google Scholar] [CrossRef] [PubMed]
- Chanwala, J.; Khadanga, B.; Jha, D.K.; Sandeep, I.S.; Dey, N. MYB transcription factor family in pearl millet: Genome-wide identification, evolutionary progression and expression analysis under abiotic stress and phytohormone treatments. Plants 2023, 12, 355. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Jiang, Y.; Li, H.; Guo, J.; Dong, M.; Zhang, J.; Liu, G. Genome-wide analysis of the NAC transcription factor family in broomcorn millet (Panicum miliaceum L.) and expression analysis under drought stress. BMC Genom. 2020, 21, 96. [Google Scholar] [CrossRef] [PubMed]
- Ceasar, A. Genome-editing in millets: Current knowledge and future perspectives. Mol. Biol. Rep. 2022, 49, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Beyene, G.; Chauhan, R.D.; Villmer, J.; Husic, N.; Wang, N.; Gebre, E.; MacKenzie, D.J. CRISPR/Cas9-mediated tetra-allelic mutation of the ‘Green Revolution’SEMIDWARF-1 (SD-1) gene confers lodging resistance in tef (Eragrostis tef). Plant Biotechnol. J. 2022, 20, 1716–1729. [Google Scholar] [CrossRef] [PubMed]
- Elkonin, L.A.; Gerashchenkov, G.A.; Borisenko, N.V.; Kenzhegulov, O.A.; Sarsenova, S.K.; Rozhnova, N.A.; Panin, V.M. Development of sorghum mutants with improved in vitro protein digestibility by CRISPR/Cas9 editing of kafirin genes. Crop J. 2023, 11, 1411–1418. [Google Scholar] [CrossRef]
- Li, X.; Liu, W.; Wang, G.; Sun, S.S.M.; Yuan, L.; Wang, J. Improving digestibility of sorghum proteins by CRISPR/Cas9-based genome editing. Food Energy Secur. 2024, 13, e506. [Google Scholar] [CrossRef]
- Liang, Z.; Wu, Y.; Ma, L.; Guo, Y.; Ran, Y. Efficient genome editing in Setaria italica using CRISPR/Cas9 and base editors. Front. Plant Sci. 2022, 12, 815946. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Yang, Y.; Futrell, S.; Kelly, E.A.; Lorts, C.M.; Nebie, B.; Schachtman, D.P. CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase (CCD) genes in sorghum alters strigolactone biosynthesis and plant biotic Interactions. Phytobio. J. 2023, 7, 339–351. [Google Scholar] [CrossRef]
- Chanwala, J.; Satpati, S.; Dixit, A.; Parida, A.; Giri, M.K.; Dey, N. Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genom. 2020, 21, 231. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Shinde, H.; Dudhate, A.; Tsugama, D.; Gupta, S.K.; Liu, S.; Takano, T. Genome-wide investigation of SQUAMOSA promoter binding protein-like transcription factor family in pearl millet (Pennisetum glaucum (L) R. Br.). Plant Gene 2021, 27, 100313. [Google Scholar] [CrossRef]
- Dudhate, A.; Shinde, H.; Yu, P.; Tsugama, D.; Gupta, S.K.; Liu, S.; Takano, T. Comprehensive analysis of NAC transcription factor family uncovers drought and salinity stress response in pearl millet (Pennisetum glaucum). BMC Genom. 2021, 22, 70. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Cui, Y.; Zhao, Z.; Li, S.; Liang, D.; Wang, C.; Liu, Z. Genome-wide identification and characterization of the BES/BZR gene family in wheat and foxtail millet. BMC Genom. 2021, 22, 682. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Yan, J.; He, A.; Xue, G.; Yang, H.; Feng, L.; Cheng, J. Genome-wide identification, phylogenetic and expression pattern analysis of MADS-box family genes in foxtail millet (Setaria italica). Sci. Rep. 2022, 12, 4979. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Fan, K.; Guo, X.; Liu, J.; Zhang, K.; Lu, P. Genome-wide identification, molecular evolution and expression analysis of the non-specific lipid transfer protein (nsLTP) family in Setaria italica. BMC Plant Biol. 2022, 22, 547. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Song, H.; Zhang, B.; Lu, Q.; Liu, Z.; Zhang, S.; Peng, R. Genome-wide identification, characterization, and expression analysis of superoxide dismutase (SOD) genes in foxtail millet (Setaria italica L.). 3 Biotech 2018, 8, 486. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Yao, X.; Yan, J.; Gao, A.; Yang, H.; Xiang, D.; Cheng, J. Genome wide identification, phylogenetic and expression pattern analysis of GATA family genes in foxtail millet (Setaria italica). BMC Genom. 2022, 23, 549. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Dai, S.; Qin, N.; Zhu, C.; Qin, J.; Li, J. Genome-wide identification and expression analysis of the SAUR gene family in foxtail millet (Setaria italica L.). BMC Plant Biol. 2023, 23, 31. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Fan, Y.; Xue, G.; He, A.; Yang, H.; He, C.; Cheng, J. Genome-wide identification and characterization of the SPL gene family and its expression in the various developmental stages and stress conditions in foxtail millet (Setaria italica). BMC Genom. 2022, 23, 389. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fu, H.; Zhao, J.; Wang, J.; Dong, S.; Yuan, X.; Chen, M. Genome-Wide Identification and Expression Profiling of Glutathione S-Transferase Gene Family in Foxtail Millet (Setaria italica L.). Plants 2023, 12, 1138. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, J.; Chen, S.; Zhang, H.; Li, L. Genome-Wide Identification and Characterization of Clavata3/Embryo Surrounding Region (CLE) Gene Family in Foxtail Millet (Setaria italica L.). Genes 2023, 14, 2046. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Liu, Y.W.; Zhou, J.M.; Zhao, S.P.; Zhang, X.H.; Min, D.H. Genome-wide analysis of the lectin receptor-like kinase family in foxtail millet (Setaria italica L.). Plant Cell Tissue Organ Cult. 2016, 127, 335–346. [Google Scholar] [CrossRef]
- Li, Y.; Yu, S.; Zhang, Q.; Wang, Z.; Liu, M.; Zhang, A.; Li, C. Genome-wide identification and characterization of the CCT gene family in foxtail millet (Setaria italica) response to diurnal rhythm and abiotic stress. Genes 2022, 13, 1829. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Wei, S.; Guo, Y.; Wu, Y. Genome wide identification of MPK and MKK gene families and their responses to phytohormone treatment and abiotic stress in foxtail millet. Plant Growth Regul. 2023, 99, 85–99. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, C.; Kang, X.; Pei, Z.Q.; Bai, X.; Wang, J.; Zhang, T.G. Identification and expression analysis of MAPK cascade gene family in foxtail millet (Setaria italica). Plant Signal. Behav. 2023, 18, 2246228. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.F.; Zhao, W.Y.; Fu, J.D.; Liu, Y.W.; Chen, M.; Zhou, Y.B.; Xi, Y.J. Genome-wide analysis of CDPK family in foxtail millet and determination of SiCDPK24 functions in drought stress. Front. Sci. Plant. Sci. 2018, 9, 651. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, L.; Li, W.; Zhang, X.; Zhang, Y.; Dong, S.; Yuan, X. Genome-Wide Identification and Expression Profiling of Cytochrome P450 Monooxygenase Superfamily in Foxtail Millet. Int. J. Mol. Sci. 2023, 24, 11053. [Google Scholar] [CrossRef] [PubMed]
- Lata, C.; Mishra, A.K.; Muthamilarasan, M.; Bonthala, V.S.; Khan, Y.; Prasad, M. Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS ONE 2014, 9, e113092. [Google Scholar] [CrossRef]
- Li, W.; Chen, M.; Wang, E.; Hu, L.; Hawkesford, M.J.; Zhong, L.; Ma, Y. Genome-wide analysis of autophagy-associated genes in foxtail millet (Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice. BMC Genom. 2016, 17, 797. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.J.; He, G.H.; Zheng, W.J.; Lu, P.P.; Chen, M.; Gong, Y.M.; Xu, Z.S. Foxtail millet NF-Y families: Genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front. Sci. Plant. Sci. 2015, 6, 1142. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cheng, J.; Han, H.; Sun, R.; Li, Y.; Zhang, Y.; Li, X. Genome-wide identification of the HKT transcription factor family and their response to salt stress in foxtail millet (Setaria italica). Plant Growth Regul. 2023, 99, 113–123. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Khandelwal, R.; Yadav, C.B.; Bonthala, V.S.; Khan, Y.; Prasad, M. Identification and molecular characterization of MYB transcription factor superfamily in C4 model plant foxtail millet (Setaria italica L.). PLoS ONE 2014, 9, e109920. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wei, Y.; Wang, Y.; Tan, B.; Chen, S.; Li, H. Genome-Wide Identification of LBD Genes in Foxtail Millet (Setaria italica) and Functional Characterization of SiLBD21. Int. J. Mol. Sci. 2023, 24, 7110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yu, Y.; Song, T.; Zhang, M.; Li, N.; Yu, M.; Zhang, X. Genome-wide identification of foxtail millet’s TRX family and a functional analysis of SiNRX1 in response to drought and salt stresses in transgenic Arabidopsis. Front. Sci. Plant. Sci. 2022, 13, 946037. [Google Scholar] [CrossRef] [PubMed]
- Xing, G.; Jin, M.; Qu, R.; Zhang, J.; Han, Y.; Han, Y.; Zhao, X. Genome-wide investigation of histone acetyltransferase gene family and its responses to biotic and abiotic stress in foxtail millet (Setaria italica [L.] P. Beauv). BMC Plant Biol. 2022, 22, 292. [Google Scholar] [CrossRef] [PubMed]
- Chai, W.; Si, W.; Ji, W.; Qin, Q.; Zhao, M.; Jiang, H. Genome-wide investigation and expression profiling of HD-zip transcription factors in foxtail millet (Setaria italica L.). BioMed Res. Int. 2018, 2018, 8457614. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Varshney, V.; Tak, N.; Jha, S. Genome-wide identification and expression analysis of glycogen synthase kinase encoding genes in foxtail millet (Setaria italica L.) under salinity, dehydration, and oxidative stress. Plant Stress 2023, 8, 100165. [Google Scholar] [CrossRef]
- Cheng, J.; Tan, H.; Shan, M.; Duan, M.; Ye, L.; Yang, Y.; Wang, X. Genome-wide identification and characterization of the NPF genes provide new insight into low nitrogen tolerance in Setaria. Front. Sci. Plant. Sci. 2022, 13, 1043832. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, Y.; Pan, J.; Li, Z.; Wang, Q.; Mastouri, F.; Li, Y.; Liu, W. Genome-wide identification of PTI1 family in Setaria italica and salinity-responsive functional analysis of SiPTI1–5. BMC Plant Biol. 2021, 21, 319. [Google Scholar] [CrossRef]
- Gong, L.; Li, B.; Zhu, T.; Xue, B. Genome-wide identification and expression profiling analysis of DIR gene family in Setaria italica. Front. Sci. Plant. Sci. 2023, 14, 1243806. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Guo, Y.; Wang, H.; Wang, X.; Lv, M.; Yang, P.; Zhang, L. Identification and characterization of Shaker K+ Channel Gene Family in Foxtail Millet (Setaria italica) and their role in stress response. Front. Sci. Plant. Sci. 2022, 13, 907635. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chai, Y.; Liu, J.; Zheng, J.; Zhao, Z.; Amo, A.; Hu, Y.G. Amino acid transporter (AAT) gene family in foxtail millet (Setaria italica L.): Widespread family expansion, functional differentiation, roles in quality formation and response to abiotic stresses. BMC Genom. 2021, 22, 519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, Y.; Zhang, J.; Li, X.; Ma, F.; Duan, M.; Li, H. The responses of the lipoxygenase gene family to salt and drought stress in foxtail millet (Setaria italica). Life 2021, 11, 1169. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Li, M.; Li, Y.; Zhao, Z.; Li, C.; Yue, J. Genome-wide characterization of remorin genes in terms of their evolution and expression in response to hormone signals and abiotic stresses in foxtail millet (Setaria italica). Diversity 2022, 14, 711. [Google Scholar] [CrossRef]
- Nadeem, F.; Ahmad, Z.; Wang, R.; Han, J.; Shen, Q.; Chang, F.; Li, X. Foxtail millet [Setaria italica (L.) Beauv.] grown under low nitrogen shows a smaller root system, enhanced biomass accumulation, and nitrate transporter expression. Front. Sci. Plant. Sci. 2018, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tang, S.; Zhi, H.; Xing, L.; Zhang, H.; Tang, C.; Diao, X. The boron transporter SiBOR1 functions in cell wall integrity, cellular homeostasis, and panicle development in foxtail millet. Crop J. 2022, 10, 342–353. [Google Scholar] [CrossRef]
- Alagarasan, G.; Dubey, M.; Aswathy, K.S.; Chandel, G. Genome wide identification of orthologous ZIP genes associated with zinc and iron translocation in Setaria italica. Front. Sci. Plant. Sci. 2017, 8, 775. [Google Scholar] [CrossRef] [PubMed]
- Blume, R.; Yemets, A.; Korkhovyi, V.; Radchuk, V.; Rakhmetov, D.; Blume, Y. Genome-wide identification and analysis of the cytokinin oxidase/dehydrogenase (ckx) gene family in finger millet (Eleusine coracana). Front. Genet. 2022, 13, 963789. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Andreev, K.; Dupre, M.E. Major trends in population growth around the world. China CDC Wkly. 2021, 3, 604. [Google Scholar] [CrossRef] [PubMed]
- Hemathilake, D.M.K.S.; Gunathilake, D.M.C.C. Agricultural productivity and food supply to meet increased demands. In Future Foods Global Trends, Opportunities, and Sustainability Challenges; Academic Press: Cambridge, MA, USA, 2022; pp. 539–553. [Google Scholar]
- Antony Ceasar, S.; Maharajan, T.; Ajeesh Krishna, T.P.; Ramakrishnan, M.; Victor Roch, G.; Satish, L.; Ignacimuthu, S. Finger millet [Eleusine coracana (L.) Gaertn.] improvement: Current status and future interventions of whole genome sequence. Front. Sci. Plant Sci. 2018, 9, 1054. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.M.; Arora, S.; Mirza, N.; Pande, A.; Lata, C.; Puranik, S.; Kumar, A. Finger millet: A “certain” crop for an “uncertain” future and a solution to food insecurity and hidden hunger under stressful environments. Front. Plant Sci. 2017, 8, 643. [Google Scholar] [CrossRef] [PubMed]
| Name of the Millets | Scientific Name | Chromosome Number | Genome Size (Mb) | Ploidy Level | Reference |
|---|---|---|---|---|---|
| Sorghum | Sorghum bicolor | 2x = 2x = 20 | 730 | Diploid | [48] |
| Pearl millet | Cenchrus americanus | 2n = 2x =14 | ~1700 | Diploid | [49] |
| Finger millet | Eleusine coracana | 2n = 4x = 36 | 1593 | Allotetraploid | [42,43,44] |
| Foxtail millet | Setaria italica | 2n = 2x = 18 | ~515 | Diploid | [40,41] |
| Proso millet | Panicum miliaceum | 2n = 4x = 36 | ~900 | Tetraploid | [46] |
| Kodo millet | Paspalum scrobiculatum | 2n = 4x = 40 | ~1900 | Tetraploid | [47] |
| Little millet | Panicum sumatrense | 2n = 4x = 36 | Unknown | Tetraploid | [47,50] |
| Japanese Barnyard millet | Echinochloa crus-galli | 2n = 6x = 54 | ~1270 | Hexaploid | [45] |
| Teff | Eragrostis tef | 2n = 4x = 40 | 672 | Tetraploid | [51] |
| Fonio | Digitaria exilis | 2n = 4x = 36 | 716 | Tetraploid | [52] |
| Job’s tears | Coix lacryma-jobi | 2n = 20 | 1280 | Diploid | [53,54] |
| Name of the Millet | Genotype Name | Sequenced Genome Size | Number of Scaffolds | Scaffold N50 (Mb) | Number of Genes Identified | Bio-Sample ID | Gen-Bank Assembly Accession |
|---|---|---|---|---|---|---|---|
| Sorghum | BTx623 | ~730 Mb * | 867 | 68.7 | 34,118 | SAMN02953738 | GCA_000003195 |
| Hongyingzi | 724.4 Mb * | 10 | 70.9 | - | SAMN38071627 | GCA_033546955 | |
| Huandiaonuo | 726.9 Mb * | 10 | 70.7 | - | SAMN38071628 | GCA_033546955 | |
| Rio | 729.4 Mb * | 3830 | 0.39 | 35,490 | SAMN05444726 | GCA_015952705 | |
| TX2783 | 721.4 Mb # | 13 | 66.1 | - | SAMEA6819231 | GCA_903166285 | |
| TX436 | 722.1 Mb # | 13 | 68.1 | - | SAMEA6819238 | GCA_903166325 | |
| Leoti | 687.3 Mb # | 10 | 68 | - | SAMEA111279259 | GCA_947241725 | |
| Rio | 817.2 Mb # | 622 | 76.7 | - | SAMEA111279266 | GCA_947241735 | |
| Tx430 | 666.2 Mb # | 308 | 32.2 | - | SAMN09228096 | GCA_003482435 | |
| Chinese Amber | 789.4 Mb # | 588 | 78.7 | - | SAMEA111279257 | GCA_947241645 | |
| 655,972 | 792.7 Mb # | 835 | 73.8 | - | SAMEA111279265 | GCA_947241635 | |
| 506,069 | 795.5 Mb # | 957 | 75.3 | - | SAMEA111279263 | GCA_947241665 | |
| 297,155 | 767.9 Mb # | 826 | 74.9 | - | SAMEA111279261 | GCA_947241675 | |
| 229,841 | 757.5 Mb # | 923 | 72.6 | - | SAMEA111279260 | GCA_947241625 | |
| 329,311 | 789.8 Mb # | 1761 | 74.8 | - | SAMEA111279262 | GCA_947241655 | |
| Grassl | 717.4 Mb # | 648 | 70.2 | - | SAMEA111279258 | GCA_947241715 | |
| BTx623 | 374.3 Mb # | 2657 | 0.2 | - | SAMN12341013 | GCA_008000285 | |
| 510757 | 768.2 Mb # | 1800 | 74.3 | - | SAMEA111279264 | GCA_947241685 | |
| Pearl millet | Tift 23D2B1-P1-P5 | 1.8 Gb * | 52,033 | 240.6 | 40,658 | SAMN04124419 | GCA_002174835 |
| Tift 23D2B1-P1-P5 | 1.8 Gb * | 7 | 259.2 | - | SAMEA112192700 | GCA_947561735 | |
| PI537069 | 1.9 Gb * | 98 | 266.8 | - | SAMN20372178 | GCA_020739565 | |
| PI526529 | 2 Gb * | 839 | 287 | - | SAMN20372180 | GCA_020739535 | |
| Tifleaf 3 | 2 Gb * | 69 | 279.2 | - | SAMN20372183 | GCA_020739585 | |
| PI343841 | 2 Gb * | 912 | 263.7 | - | SAMN28616536 | GCA_027745475 | |
| PI521612 | 1.9 Gb * | 331 | 278.5 | - | SAMN20372179 | GCA_020739525 | |
| PI587025 | 1.9 Gb * | 4064 | 257.5 | - | SAMN20372182 | GCA_021560375.1 | |
| PI186338 | 2 Gb * | 139 | 284.6 | - | SAMN28616529 | GCA_027789755 | |
| PI583800 | 1.9 Gb * | 138 | 261.4 | - | SAMN20372181 | GCA_020739575 | |
| Foxtail millet | Yugu1 | 405.7 Mb * | 327 | 47.3 | >34,584 | SAMN02981383 | GCA_000263155 |
| Zhang gu | ~423 Mb * | 2689 | 1.0 | >38,801 | SAMN04534922 | GCA_001652605 | |
| Finger millet | KNE 796-S | 1.12 Gb * | 1058 | 12.1 | 73,012 | SAMN35346668 | GCA_032690845 |
| PR202 | 1.5 Gb # | 1196 | 23.9 | 62,348 | SAMD00076255 | GCA_021604985 | |
| ML365 | 1.19 Gb # | 525,759 | 23.7 | 85,243 | SAMN04849255 | GCA_002180455 | |
| Proso millet | Pm_0390 | 923 Mb * | 1305 | 46.7 | 61,631 | SAMN08389585 | GCA_003046395 |
| jinshu7 | 862 Mb * | - | SAMN30451036 | GCA_026771285 | |||
| BC332 | 856.2 Mb * | 580 | 48.5 | - | SAMN13925972 | GCA_032594955 | |
| BC475 | 870.7 Mb * | 843 | 48.1 | - | SAMN13926115 | GCA_032595135 | |
| Longmi4 | 846 Mb * | 441 | 48.2 | - | SAMN08335224 | GCA_002895445 | |
| BC477 | 877.2 Mb * | 1031 | 48.2 | - | SAMN13926117 | GCA_032595115 | |
| BC494 | 863.2 Mb * | 722 | 48.1 | - | SAMN13926134 | GCA_032595125 | |
| BC404 | 870.4 Mb * | 813 | 48.1 | - | SAMN13926044 | GCA_032595235 | |
| BC498 | 869.9 Mb * | 753 | 48.2 | - | SAMN13926138 | GCA_032595105 | |
| BC328 | 867.1 Mb * | 799 | 48.2 | - | SAMN13925968 | GCA_032595055 | |
| BC27 | 907.3 Mb * | 1520 | 45 | - | SAMN13925667 | GCA_032594635 | |
| BC426 | 882.1 Mb * | 1166 | 48.3 | - | SAMN13926066 | GCA_032595225 | |
| BC398 | 896.4 Mb * | 1451 | 48.1 | - | SAMN13926038 | GCA_032595255 | |
| BC382 | 861 Mb * | 799 | 48.4 | - | SAMN13926022 | GCA_032594965 | |
| BC418 | 867.8 Mb * | 874 | 48.2 | - | SAMN13926058 | GCA_032595215 | |
| BC407 | 874.9 Mb * | 1052 | 48.2 | - | SAMN13926047 | GCA_032595245 | |
| BC434 | 872.2 Mb * | 994 | 48.5 | - | SAMN13926074 | GCA_032595155 | |
| BC362 | 870 Mb * | 1068 | 48 | - | SAMN13926002 | GCA_032594995 | |
| BC311 | 890.6 Mb * | 1408 | 48.1 | - | SAMN13925951 | GCA_032594875 | |
| BC264 | 892.1 Mb * | 1166 | 48.4 | - | SAMN13925904 | GCA_032594675 | |
| BC350 | 867.7 Mb * | 945 | 48.1 | - | SAMN13925990 | GCA_032594985 | |
| BC360 | 860.2 Mb * | 594 | 48.2 | - | SAMN13926000 | GCA_032594975 | |
| BC292 | 891 Mb * | 1075 | 48.3 | - | SAMN13925932 | GCA_032594685 | |
| BC315 | 891.8 Mb * | 1343 | 48.4 | - | SAMN13925955 | GCA_032594835 | |
| BC310 | 872.1 Mb * | 970 | 48.1 | - | SAMN13925950 | GCA_032594855 | |
| BC136 | 867.8 Mb * | 889 | 48.1 | - | SAMN13925776 | GCA_032594655 | |
| BC235 | 871.9 Mb * | 934 | 48.2 | - | SAMN13925875 | GCA_032594705 | |
| BC217 | 872.5 Mb * | 1137 | 49.3 | - | SAMN13925857 | GCA_032594775 | |
| BC170 | 875.8 Mb * | 929 | 48.1 | - | SAMN13925810 | GCA_032594555 | |
| BC48 | 878.1 Mb * | 1260 | 48.2 | - | SAMN13925688 | GCA_032594585 | |
| BC188 | 861.7 Mb * | 752 | 47.9 | - | SAMN13925828 | GCA_032594795 | |
| BC204 | 871 Mb * | 935 | 49.2 | - | SAMN13925844 | GCA_032594715 | |
| BC244 | 890.4 Mb * | 1443 | 47.8 | - | SAMN13925884 | GCA_032594695 | |
| BC100 | 864.3 Mb * | 883 | 47.8 | - | SAMN13925740 | GCA_032594575 | |
| BC40 | 860.4 Mb * | 846 | 48.6 | - | SAMN13925680 | GCA_032594595 | |
| Barnyard Grass | STB08 | 1.27 Gb # | 19,699 | 1.8 | 108,771 | SAMN03246123 | GCA_025118225 |
| - | 1.5 Gb # | 4534 | 1.8 | - | SAMEA104207156 | GCA_900205405 |
| Name of the Millets | Number of RILs | Number and Types of Markers Used | Number of QTL Identified | Targeting Traits | References |
|---|---|---|---|---|---|
| Pearl millet | 106 | 95 SSR, 2 STS, and 208 DArT | 44 | FT, PH, PL, and GW | [71] |
| 168 | 256 DArT and 70 SSR | 1 | Rust disease | [63] | |
| 317 | 235 DArT and 33 SSR | 23 | FL, PH, PL, and TGW | [72] | |
| 188 | 96 SSCP-SNP, 96 SSR, 96 EST-SSR, and 43 STS | 18 | PL, PD, and GS | [73] | |
| 187 | 88 SSR | 5 | Downy mildew disease | [65] | |
| 317 | 258 DArT and 63 SSR | 19 | Grain iron and zinc content | [66] | |
| 210 | 372 SSR | 22 | Grain iron and zinc content | [67] | |
| 149 | 95 RFLP, SSR, TRAP, and EST-SSR | 24 | Dry stover yield and GY | [74] | |
| 172 | 26 SSR and 20 RFLP | 24 | Downy mildew disease | [75] | |
| 50 RFLP and 29 SSR | 3 | GY | [76] | ||
| Foxtail millet | 305 | 35,065 SNP | 3 | Leaf blast | [62] |
| 164 | 1047 SNP | 47 | SW, PW, GW, and TGW | [77] | |
| 333 | 3744 SNP | 26 | PH | [78] | |
| 543 | 48,790 SNP | 57 | PH, PL, PD, PNL, FID, SID, PW, GW, and TGW | [79] | |
| 215 | 20,748 SNP | 39 | Hull color traits | [80] | |
| 439 | 33,579 SNP | 59 | HD, PL, TN, PW, PD, FLL, FLD, PH, SD, SNN, code number, CGN, TGW, and NL | [81] | |
| 164 | 2297 bin and 74 SSR | 221 | SL, SD, SNN, PDL, TN, FLL, FLW, PL, PD, SDY, GN, bristle length, SW, PW, GW, and TGW | [82] | |
| 124 | 9968 SNP | 11 | PH, PDL, PD, FID, SID, and TID | [83] | |
| 400 | 43,001 SNP | 5 | Anther and hull color | [84] | |
| Finger millet | 100 | 101 SSR | 92 | SDW, RDW, ShL, RHL, RHD, and SPC and RPC | [68] |
| - | 87 SSR | 15 | Leaf blast, PH, TN, NPT, NF, RL, and GY | [85] | |
| - | 87 SSR | 8 | RDW, SDW, and RL | [69] | |
| - | 87 SSR | 11 | RL, RDW, and biochemical traits | [70] | |
| 151 | 5422 SNP | 8 | DF, PH, PN, LBS, and PBI | [86] | |
| 190 | 46 SSR | 2 | DF, FLW, and PH | [87] | |
| Proso millet | 93 | 833 SNP | 18 | HD, PH, PDL, lodging, PL, grain shattering, TGW, and GPP | [88] |
| Name of the Millet | Number of Genotype | Number of SNPs | Number of QTL/MTA Identified | Targeting Traits | Reference |
|---|---|---|---|---|---|
| Sorghum | 300 | 79,132 | 14 | Stalk rot diseases in grain | [97] |
| 390 | 268,830 | 108 | Yield per panicle, grain number per panicle, and 1000-grain weight | [98] | |
| 315 | 136,285 | 101 | Plant height, panicle length, panicle exsertion, stem circumference, tiller number, internode number, flowering time, leaf angle, and seed number | [99] | |
| 300 | 265,487 | 42 | Shoot and root weight, shoot and root length, chlorophyll contents, and anthocyanin content in shoot under cold and heat stress | [100] | |
| 374 | 265,487 | 14 | Leaf firing and leaf blotching under heat stress | [101] | |
| 194 | 44,515 | 21 | Final emergence percentage, seedling survival, and seedling vigor under cold stress | [102] | |
| 245 | 85,585 | 42 | Crude protein, neutral detergent fiber, acid detergent fiber, hemicellulose, and cellulose contents | [103] | |
| 212 | 268,289 | 2 | Low seed deterioration and emergence rate | [104] | |
| 1425 | 72,190 | 102 | Plant height, presence or absence of awns, glume cover, pericarp color, panicle compactness and shape, panicle exsertion, smut resistance, and male sterility | [105] | |
| 634 | 260,000 | 70 | Amylose and amylopectin contents in grain | [106] | |
| 2000 | 142,567 | 81 | Seed size | [107] | |
| 403 | 341,514 | 52 | Grain carotenoids (β-Carotene and Zeaxanthin) | [108] | |
| 242 | 6,094,317 | 19 | Panicle length and width, panicle compactness, and peduncle recurving | [109] | |
| 219 | 73,730 | >80 | Plant height, flowering time, forage biomass, grain weight, and water use efficiency under drought stress | [110] | |
| 245 | 85,585 | 338 | Plant height, tiller number, stem diameter, and fresh weight per plant | [111] | |
| 96 | 192,040 | 40 | Heading date, plant height, dry yield, and phenolic compounds | [112] | |
| 354 | 6186 | 79 | Plant height and drought-tolerance indices | [113] | |
| 162 | 193,727 | 100 | Seed area size, length, width, length-to-width ratio, perimeter, circularity, distance between intersection of length and width, center of gravity, and seed darkness and brightness | [114] | |
| Pearl millet | 392 | 21,663 | 18 | Biomass, flowering time, plant height, and tillering | [93] |
| 281 | 58,719 | 78 | Iron, zinc, and protein content | [92] | |
| 197 | 76,000 | 897 | Starch-, lipid-, antioxidant-, vitamin-, sucrose-, and flavone-related traits | [115] | |
| 222 | 67,000 | 218 | Antioxidant biosynthesis | [116] | |
| 166 | 78,000 | 1132 | Readily digested starch, slowly digested starch, resistant starch, total starch, and available starch | [94] | |
| Foxtail millet | 916 | 2,584,083 | 512 | Morphology characteristics, yield components, growth time, disease resistance, and coloration | [117] |
| 104 | 30,000 | 67 | Eleven nutritional-related traits | [118] | |
| 407 | 706,646 | 87 | Panicle length, main panicle diameter, panicle weight per panicle, grain weight per panicle, and thousand-grain weight | [89] | |
| 827 | 161,562 | 257 | Top second leaf width, main stem width, panicle diameter of main stem, panicle length of main stem, per plant grain weight, and main stem panicle weight | [119] | |
| 107 | 72,181 | 53 | Plant height, stem diameter, leaf length, leaf width, chlorophyll SPD value, spike length, spike weight, spike diameter, grain length, grain width, and grain length/width ratio | [120] | |
| 93 | 10,000 | 74 | Grain nutritional elements such as potassium, nickel, calcium, boron, magnesium, phosphorus, sulfur, zinc manganese, and iron | [91] | |
| 142 | 10,000 | 81 | Flag leaf length, flag leaf width, peduncle length, panicle length, tiller maturity, grain yield, and thousand-grain weight and plant height | [90] | |
| Finger millet | 190 | 169,365 | 418 | Grain nutrition traits (calcium, iron, sodium, potassium, magnesium, zinc) and protein content | [96] |
| 113 | 23,000 | 109 | Basal tiller number, culm thickness, days to 50% flowering, days to 50% maturity, ear length, ear width, flag leaf blade length, flag leaf blade width, fingers per head, grain yield, length of longest finger, plant height, peduncle length, and width of longest finger | [121] | |
| 113 | 2977 | 40 | Seed protein content, grain yield, and days to maturity | [122] | |
| Proso millet | 88 | 494,200,000 | 13 | Plant height, leaf number, seed length, seed width, seed perimeter, seed length-to-width ratio, and seed color | [95] |
| Name of the Millet | Name of the Genes | Name and Total Number of Genes Identified | Treatments | Reference |
|---|---|---|---|---|
| Pearl millet | MYB | 208 (PgMYB1–PgMYB208) | Cold, high temperature, osmotic stress, drought, and salinity | [134] |
| MYB | 279 (PgMYB1-PgMYB279) | Dehydration and salinity stress, abscisic acid, salicylic acid, and methyl jasmonate | [135] | |
| WRKY | 97 (PgWRKY1-PgWRKY97) | Dehydration and salinity stress | [143] | |
| SBP | 18 (PgSBP1-PgSBP18) | Drought, salinity, and abscisic acid | [144] | |
| NAC | 151 (PgNAC1-PgNAC151) | Drought and salinity stress | [145] | |
| Foxtail millet | BZR | 7 (SiBZR1 to SiBZR7) | Abscisic acid and salinity stress | [146] |
| MADS-box | 89 (SiMADS1–SiMADS89) | Acid, alkali, salt, drought, flooding, dark, heat and cold stresses | [147] | |
| bHLH | 187 (SibHLH1-SibHLH187) | Acid, alkali, drought, salinity, heat, cold, flooding, and darkness conditions | [123] | |
| nsLTP | 45 (SinsLTP1-SinsLTP45) | Drought, salt, and cold stress | [148] | |
| SOD | 8 (SiSOD1-SiSOD8) | Drought and salinity | [149] | |
| GATA | 28 (SiGATA1-SiGATA28) | Acid, alkali, salinity, drought, dark, flooding, heat, and cold | [150] | |
| SAUR | 72 (SiSAUR1-SiSAUR72) | Drought, salinity, abscisic acid, salicylic acid and gibberellic acid | [151] | |
| SNARE | 52 (SiSNARE1-SiSNARE52) | Drought stress | [124] | |
| SPL | 18 (SiSPL1 to SiSPL18) | Acid, alkali, salinity, drought, flooding, dark, heat, and cold | [152] | |
| GST | 73 (SiGST1-SiGST73) | Osmotic, salinity, cold stress, and abscisic acid | [153] | |
| CLE | 41 (SiCLE1-SiCLE41) | Gibberellic acid treatment | [154] | |
| Dof | 35 (SiDof1-SiDof35) | Drought stress | [125] | |
| LecRLK | 113 (SiLecRLK1- SiLecRLK113) | Drought and high temperature | [155] | |
| CCT | 19 (SiCCT1-SiCCT19) | Abscisic acid, drought, and salinity | [156] | |
| MAPK | 16 (SiMAPK1-SiMAPK16) | Abscisic acid, gibberellic acid, jasmonic acid, drought, salinity, cold, and heat | [157] | |
| MKK | 11 (SiMKK1-SiMKK11) | Abscisic acid, Gibberellic acid, jasmonic acid, drought, salinity, cold, and heat | [157] | |
| MAPK | 93 | Cold, salinity, and drought and abscisic acid and jasmonic acid | [158] | |
| CDPK | 29 (SiCDPK1-SiCDPK29) | Drought and abscisic acid | [159] | |
| CYP450 | 331 | Drought, salinity, abscisic acid, low-temperature, and herbicide treatments | [160] | |
| AP2/ERF | 171 (SiAP2/ERF1-SiAP2/ERF171) | Dehydration and salinity stress | [161] | |
| ATG | 37 | Drought, salt and cold, and nitrogen and carbon starvation | [162] | |
| NF-Y | 39 (10 NF-YA, 13 NF-YB, and 13 NF-YC) | Drought, salinity, osmotic, and oxidative stress | [163] | |
| HAK | 7 (SiHKT1-SiHKT7) | Salt stress | [164] | |
| MYB | 209 (SiMYB1-SiMYB209) | Salinity, dehydration, abscisic methyl jasmonate, and salicylic acid | [165] | |
| LBD | 33 (SiLBD1-SiLBD33) | Drought, salinity, and abscisic acid | [166] | |
| TRX | 35 (SiTRX1-SiTRX35) | Drought and salinity stress | [167] | |
| HAT | 24 (SiHAT1-SiHAT24) | Nitrate and phosphate deficiency, salinity, and drought | [168] | |
| HD-Zip | 47 (SiHD-ZIP1-SiHD-ZIP47) | Dehydration, salinity, and abscisic acid | [169] | |
| GSK | 8 (SiGSK21, 23, 24, 11, 12, 13, 31 and 41) | Dehydration, salt, and oxidative stress | [170] | |
| NPF | 92 | Low-nitrate stress | [171] | |
| PTI1 | 12 (SiPTI1–1 to SiPTI1–12) | Salinity stress | [172] | |
| DIR | 38 (SiDIR1-SiDIR38) | Salinity, drought, and higher concentrations of calcium and cadmium stress | [173] | |
| SPCP | 10 | Cold, heat, salinity, drought, and various phytohormones | [174] | |
| AAT | 94 | Drought and salinity | [175] | |
| LOX | 13 (SiLOX1-SiLOX13) | Salt and drought | [176] | |
| REM | 21 (SiREM1-SiREM21) | Abscisic acid, gibberellic acid, methyl jasmonate, drought, and salinity | [177] | |
| WRKY | 103 (SiWRKY1-SiWRKY103) | Drought stress | [126] | |
| PHT1 | 12 (SiPHT1;1-SiPHT1;12) | Phosphate stress | [127] | |
| NRAMP | 12 (SiNRAMP1-SiNRAMP12) | Cadmium stress | [130] | |
| HAK | 29 (SiHAK1-SiHAK29) | Potassium deficiency and salt stress | [129] | |
| NRT1 and NRT2 | 3 NRT1 (NRT1;1, 1;11 and 1;12) and 1 NRT2 (SiNRT2;1) | Low N stress | [178] | |
| BOR | 1 (SiBOR1) | Boron stress | [179] | |
| ZIP | 7 (SiZIP1-SiZIP7) | Drought stress | [180] | |
| Finger millet | PHT1 | 12 (EcPHT1;1-EcPHT1;12) | Phosphate and zinc stress | [131] |
| ZIP | 6 (EcZIP1-EcZIP6) | Phosphate and zinc stress | [131] | |
| ckx | 20 (EcCKK1-EcCKK10) | Various biotic and abiotic stress | [181] | |
| NBLRR | 116 (EcNBLRR1-EcNBLRR116) | Blast disease Magnaporthe grisea infection | [132] | |
| Proso millet | NAC | 180 (PmNAC1-PmNAC180) | Drought stress | [136] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maharajan, T.; Krishna, T.P.A.; Krishnakumar, N.M.; Vetriventhan, M.; Kudapa, H.; Ceasar, S.A. Role of Genome Sequences of Major and Minor Millets in Strengthening Food and Nutritional Security for Future Generations. Agriculture 2024, 14, 670. https://doi.org/10.3390/agriculture14050670
Maharajan T, Krishna TPA, Krishnakumar NM, Vetriventhan M, Kudapa H, Ceasar SA. Role of Genome Sequences of Major and Minor Millets in Strengthening Food and Nutritional Security for Future Generations. Agriculture. 2024; 14(5):670. https://doi.org/10.3390/agriculture14050670
Chicago/Turabian StyleMaharajan, Theivanayagam, Thumadath Palayullaparambil Ajeesh Krishna, Neenthamadathil Mohandas Krishnakumar, Mani Vetriventhan, Himabindu Kudapa, and Stanislaus Antony Ceasar. 2024. "Role of Genome Sequences of Major and Minor Millets in Strengthening Food and Nutritional Security for Future Generations" Agriculture 14, no. 5: 670. https://doi.org/10.3390/agriculture14050670
APA StyleMaharajan, T., Krishna, T. P. A., Krishnakumar, N. M., Vetriventhan, M., Kudapa, H., & Ceasar, S. A. (2024). Role of Genome Sequences of Major and Minor Millets in Strengthening Food and Nutritional Security for Future Generations. Agriculture, 14(5), 670. https://doi.org/10.3390/agriculture14050670








