Oil Quality Prediction in Olive Oil by Near-Infrared Spectroscopy: Applications in Olive Breeding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Olive Oil Extraction
2.3. Oxidative Stability Index
2.4. Fatty Acid Composition
2.5. FT-NIR MPA and MicroNIR Measurements
2.6. Data Analysis and Chemometrics
3. Results
3.1. Reference Data
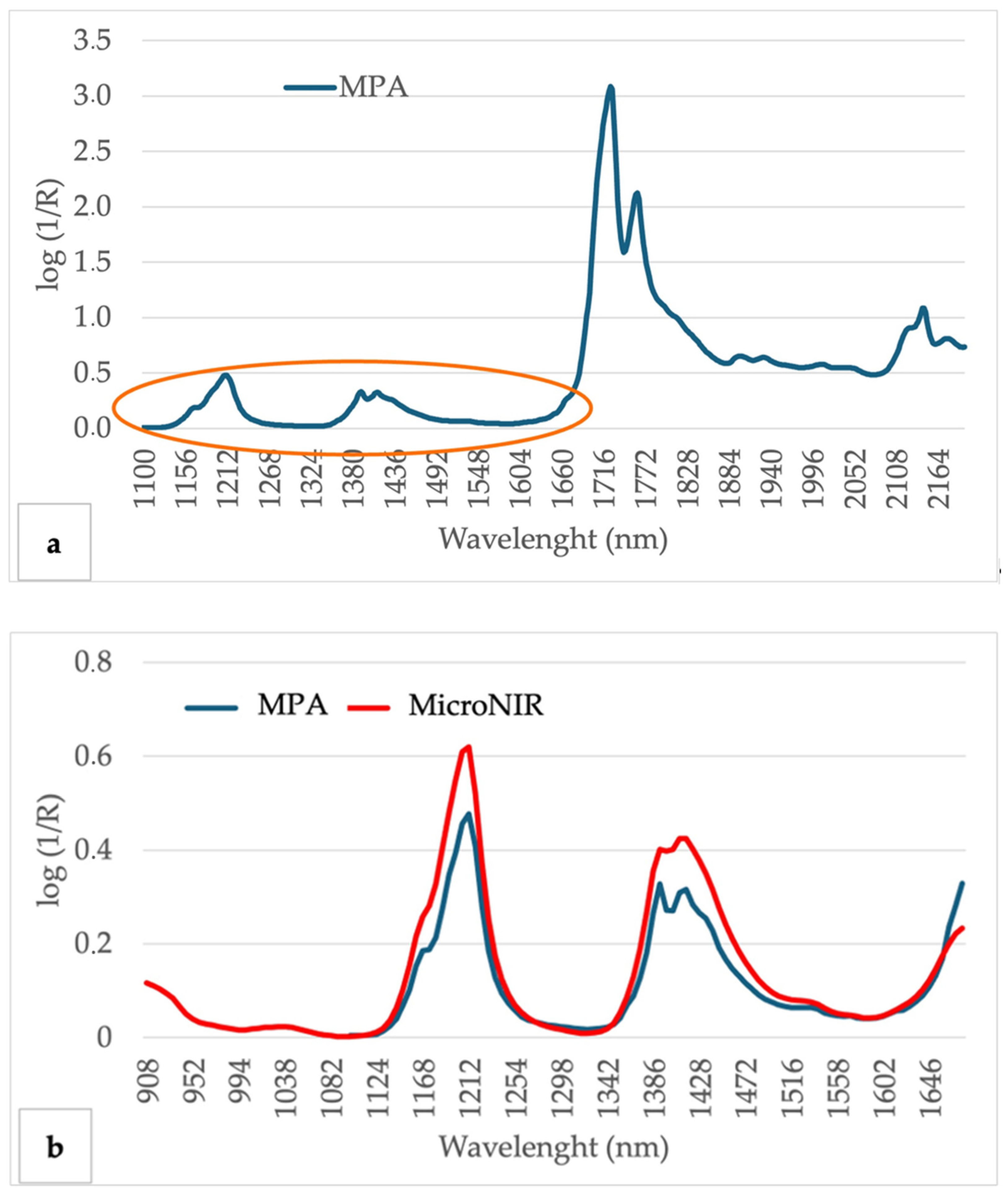
3.2. Olive Oil Spectrum
3.3. Principal Component Analysis (PCA)
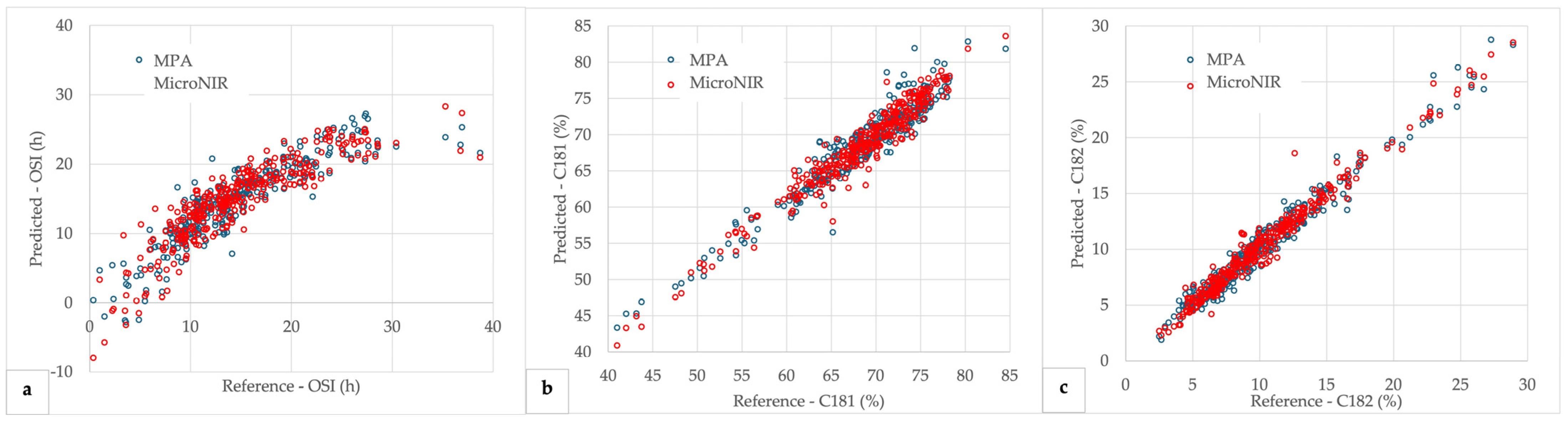
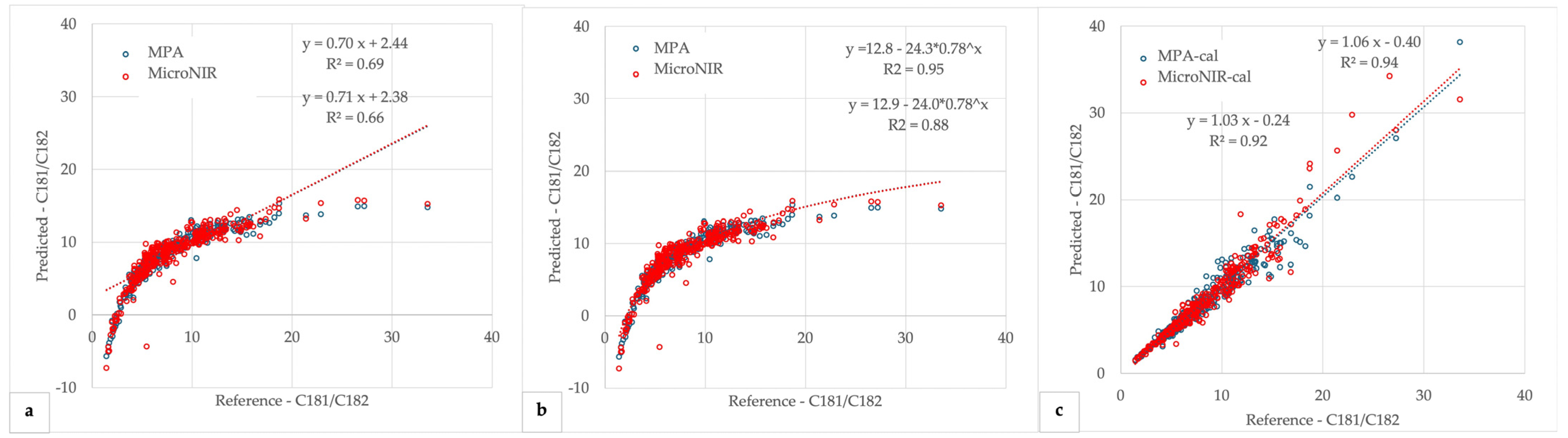
3.4. Partial Least Squares (PLS) Regression Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Servili, M.; Sordini, B.; Esposto, S.; Urbani, S.; Veneziani, G.; Di Maio, I.; Selvaggini, R.; Taticchi, A. Biological Activities of Phenolic Compounds of Extra Virgin Olive Oil. Antioxidants 2013, 3, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Gouvinhas, I.; Machado, N.; Sobreira, C.; Domínguez-Perles, R.; Gomes, S.; Rosa, E.; Barros, A. Critical Review on the Significance of Olive Phytochemicals in Plant Physiology and Human Health. Molecules 2017, 22, 1986. [Google Scholar] [CrossRef]
- Kris-Etherton, P.M.; Pearson, T.A.; Wan, Y.; Hargrove, R.L.; Moriarty, K.; Fishell, V.; Etherton, T.D. High–Monounsaturated Fatty Acid Diets Lower Both Plasma Cholesterol and Triacylglycerol Concentrations. Am. J. Clin. Nutr. 1999, 70, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Visioli, F.; Galli, C. Olive Oil: More than Just Oleic Acid. Am. J. Clin. Nutr. 2000, 72, 853. [Google Scholar] [CrossRef] [PubMed]
- Ceci, L.N.; Carelli, A.A. Relation Between Oxidative Stability and Composition in Argentinian Olive Oils. J. Am. Oil Chem. Soc. 2010, 87, 1189–1197. [Google Scholar] [CrossRef]
- Milinovic, J.; Garcia, R.; Rato, A.E.; Cabrita, M.J. Rapid Assessment of Monovarietal Portuguese Extra Virgin Olive Oil’s (EVOO’s) Fatty Acids by Fourier-Transform Near-Infrared Spectroscopy (FT-NIRS). Eur. J. Lipid Sci. Technol. 2019, 121, 1800392. [Google Scholar] [CrossRef]
- Emmanouilidou, M.G.; Koukourikou-Petridou, M.; Gerasopoulos, D.; Kyriacou, M.C. Oxidative Stability, Fatty-Acid and Phenolic Composition of Cypriot Monovarietal Virgin Olive Oils with Progressive Fruit Maturity. J. Food Compos. Anal. 2021, 104, 104191. [Google Scholar] [CrossRef]
- Williams, P.; Norris, K. (Eds.) Near-Infrared Technology in the Agricultural and Food Industries; American Association of Cereal Chemists, Inc.: St. Paul, MN, USA, 1987; 330p. [Google Scholar]
- León, L.; Downey, G. Preliminary studies by visible and near–infrared reflectance spectroscopy of juvenile and adult olive (Olea europaea L.) leaves. J. Sci. Food Agric. 2006, 86, 999–1004. [Google Scholar] [CrossRef]
- Aouidi, F.; Dupuy, N.; Artaud, J.; Roussos, S.; Msallem, M.; Perraud-Gaime, I.; Hamdi, M. Discrimination of Five Tunisian Cultivars by Mid InfraRed Spectroscopy Combined with Chemometric Analyses of Olive Olea Europaea Leaves. Food Chemistry 2012, 131, 360–366. [Google Scholar] [CrossRef]
- León, L.; Garrido-Varo, A.; Downey, G. Parent and Harvest Year Effects on Near-Infrared Reflectance Spectroscopic Analysis of Olive (Olea Europaea L.) Fruit Traits. J. Agric. Food Chem. 2004, 52, 4957–4962. [Google Scholar] [CrossRef]
- León, L.; Rallo, L. Análisis de aceituna intacta mediante espectroscopía en el infrarrojo cercano (NIRS): Una herramienta de utilidad en programas de mejora de olivo. Grasas y Aceites 2003, 54, 41–47. [Google Scholar] [CrossRef]
- Barros, A.S.; Nunes, A.; Martins, J.; Delgadillo, I. Determination of Oil and Water in Olive and Olive Pomace by NIR and Multivariate Analysis. Sens. Instrum. Food Qual. 2009, 3, 180–186. [Google Scholar] [CrossRef]
- Bendini, A.; Cerretani, L.; Carrasco-Pancorbo, A.; Gómez-Caravaca, A.M.; Segura-Carretero, A.; Fernández-Gutiérrez, A.; Lercker, G. Phenolic Molecules in Virgin Olive Oils: A Survey of Their Sensory Properties, Health Effects, Antioxidant Activity and Analytical Methods. An Overview of the Last Decade Alessandra. Molecules 2007, 12, 1679–1719. [Google Scholar] [CrossRef] [PubMed]
- Cayuela, J.A.; García, J.M.; García, J.M.; Caliani, N.; Caliani, N. NIR Prediction of Fruit Moisture, Free Acidity and Oil Content in Intact Olives. Grasas y Aceites 2009, 60, 194–202. [Google Scholar] [CrossRef]
- Cayuela, J.A.; Camino, M.D.C.P. Prediction of Quality of Intact Olives by near Infrared Spectroscopy. Eur. J. Lipid Sci. Technol. 2010, 112, 1209–1217. [Google Scholar] [CrossRef]
- Allouche, Y.; López, E.F.; Maza, G.B.; Márquez, A.J. Near Infrared Spectroscopy and Artificial Neural Network to Characterise Olive Fruit and Oil Online for Process Optimisation. J. Near Infrared Spectrosc. 2015, 23, 111–121. [Google Scholar] [CrossRef]
- Christy, A.A.; Kasemsumran, S.; Du, Y.; Ozaki, Y. The Detection and Quantification of Adulteration in Olive Oil by Near-Infrared Spectroscopy and Chemometrics. Anal. Sci. 2004, 20, 935–940. [Google Scholar] [CrossRef]
- Bellincontro, A.; Caruso, G.; Mencarelli, F.; Gucci, R. Oil Accumulation in Intact Olive Fruits Measured by near Infrared Spectroscopy–Acousto-optically Tunable Filter. J. Sci. Food Agric. 2013, 93, 1259–1265. [Google Scholar] [CrossRef]
- Bellincontro, A.; Taticchi, A.; Servili, M.; Esposto, S.; Farinelli, D.; Mencarelli, F. Feasible Application of a Portable NIR-AOTF Tool for On-Field Prediction of Phenolic Compounds during the Ripening of Olives for Oil Production. J. Agric. Food Chem. 2012, 60, 2665–2673. [Google Scholar] [CrossRef]
- Cayuela Sánchez, J.A.; Moreda, W.; García, J.M. Rapid Determination of Olive Oil Oxidative Stability and Its Major Quality Parameters Using Vis/NIR Transmittance Spectroscopy. J. Agric. Food Chem. 2013, 61, 8056–8062. [Google Scholar] [CrossRef]
- Cayuela, J.A.; Yousfi, K.; Carmen Martínez, M.; García, J.M. Rapid Determination of Olive Oil Chlorophylls and Carotenoids by Using Visible Spectroscopy. J. Am. Oil Chem. Soc. 2014, 91, 1677–1684. [Google Scholar] [CrossRef]
- Cayuela, J.A. Rapid NIR Determination of Alkyl Esters in Virgin Olive Oil. Grasas y Aceites 2017, 68, 195. [Google Scholar] [CrossRef]
- Garrido-Varo, A.; Sánchez, M.-T.; De La Haba, M.-J.; Torres, I.; Pérez-Marín, D. Fast, Low-Cost and Non-Destructive Physico-Chemical Analysis of Virgin Olive Oils Using Near-Infrared Reflectance Spectroscopy. Sensors 2017, 17, 2642. [Google Scholar] [CrossRef] [PubMed]
- Cayuela, J.A.; García, J.F. Sorting Olive Oil Based on Alpha-Tocopherol and Total Tocopherol Content Using near-Infra-Red Spectroscopy (NIRS) Analysis. J. Food Eng. 2017, 202, 79–88. [Google Scholar] [CrossRef]
- Belaj, A.; Dominguez-García, M.D.C.; Atienza, S.G.; Martín Urdíroz, N.; De la Rosa, R.; Satovic, Z.; Martín, A.; Kilian, A.; Trujillo, I.; Valpuesta, V.; et al. Developing a Core Collection of Olive (Olea Europaea L.) Based on Molecular Markers (DArTs, SSRs, SNPs) and Agronomic Traits. Tree Genet. Genomes 2012, 8, 365–378. [Google Scholar] [CrossRef]
- Williams, P.C.; Sobering, D.C. Comparison of Commercial near Infrared Transmittance and Reflectance Instruments for Analysis of Whole Grains and Seeds. J. Near Infrared Spectrosc. 1993, 1, 25–32. [Google Scholar] [CrossRef]
- León, L.; Velasco, L.; de la Rosa, R. Initial Selection Steps in Olive Breeding Programs. Euphytica 2015, 201, 453–462. [Google Scholar] [CrossRef]
- Armenta, S.; Garrigues, S.; De La Guardia, M. Determination of Edible Oil Parameters by near Infrared Spectrometry. Anal. Chim. Acta 2007, 596, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Nenadis, N.; Tsikouras, I.; Xenikakis, P.; Tsimidou, M.Z. Fourier Transform Mid-infrared Spectroscopy Evaluation of Early Stages of Virgin Olive Oil Autoxidation. Eur. J. Lipid Sci. Technol. 2013, 115, 526–534. [Google Scholar] [CrossRef]
- Sikorska, E.; Khmelinskii, I.; Sikorski, M. Vibrational and Electronic Spectroscopy and Chemometrics in Analysis of Edible Oils. In Methods in Food Analysis; Cruz, R.M.S., Khmelinskii, I., Vieira, M., Eds.; CRC Press: Boca Raton, FL, USA, 2016; pp. 211–244. ISBN 978-0-429-06983-3. [Google Scholar]
- García Martín, J.F. Potential of Near-Infrared Spectroscopy for the Determination of Olive Oil Quality. Sensors 2022, 22, 2831. [Google Scholar] [CrossRef]
- Mailer, R.J. Rapid Evaluation of Olive Oil Quality by NIR Reflectance Spectroscopy. J. Am. Oil Chem. Soc. 2004, 81, 823–827. [Google Scholar] [CrossRef]
- Manley, M.; Eberle, K. Comparison of Fourier Transform near Infrared Spectroscopy Partial Least Square Regression Models for South African Extra Virgin Olive Oil Using Spectra Collected on Two Spectrophotometers at Different Resolutions and Path Lengths. J. Near Infrared Spectrosc. 2006, 14, 111–126. [Google Scholar] [CrossRef]
- Uncu, O.; Ozen, B. Prediction of Various Chemical Parameters of Olive Oils with Fourier Transform Infrared Spectroscopy. LWT–Food Sci. Technol. 2015, 63, 978–984. [Google Scholar] [CrossRef]
- Eldin, A.B. Near Infra Red Spectroscopy. In Wide Spectra of Quality Control; IntechOpen: London, UK, 2011. [Google Scholar] [CrossRef]
- Nicolaï, B.M.; Beullens, K.; Bobelyn, E.; Peirs, A.; Saeys, W.; Theron, K.I.; Lammertyn, J. Nondestructive Measurement of Fruit and Vegetable Quality by Means of NIR Spectroscopy: A Review. Postharvest Biol. Technol. 2007, 46, 99–118. [Google Scholar] [CrossRef]
- Sim, J.; McGoverin, C.; Oey, I.; Frew, R.; Kebede, B. Near-Infrared Reflectance Spectroscopy Accurately Predicted Isotope and Elemental Compositions for Origin Traceability of Coffee. Food Chem. 2023, 427, 136695. [Google Scholar] [CrossRef] [PubMed]
- Fearn, T. Flat or natural? A note on the choice of calibration samples. In Near Infra-Red Spectroscopy: Bridging the Gap between Data Analysis and NIR Applications; Hildrum, K.I., Isaksson, T., Naes, T., Tandberg, A., Eds.; Ellis Horwood Ltd.: Chichester, UK, 1992; pp. 61–66. [Google Scholar]
- Esbensen, K.H.; Geladi, P.; Larsen, A. Mythbusters in Chemometrics. NIR News 2012, 23, 19–21. [Google Scholar] [CrossRef]
- Serrano, A.; De La Rosa, R.; Sánchez-Ortiz, A.; Cano, J.; Pérez, A.G.; Sanz, C.; Arias-Calderón, R.; Velasco, L.; León, L. Chemical Components Influencing Oxidative Stability and Sensorial Properties of Extra Virgin Olive Oil and Effect of Genotype and Location on Their Expression. LWT 2021, 136, 110257. [Google Scholar] [CrossRef]
- Navas-López, J.F.; Cano, J.; de la Rosa, R.; Velasco, L.; León, L. Genotype by Environment Interaction for Oil Quality Components in Olive Tree. Eur. J. Agron. 2020, 119, 126115. [Google Scholar] [CrossRef]


| Mean | SD 1 | Minimum | Maximum | Skewness | Kurtosis | |
|---|---|---|---|---|---|---|
| OSI | 14.68 | 6.46 | 0.40 | 38.73 | 0.73 | 0.80 |
| C160 | 15.46 | 2.82 | 8.73 | 23.65 | 0.16 | 0.08 |
| C161 | 1.50 | 0.62 | 0.03 | 4.20 | 0.98 | 1.56 |
| C180 | 2.54 | 0.56 | 0.00 | 4.70 | 0.35 | 1.27 |
| C181 | 68.34 | 6.65 | 41.04 | 84.57 | −1.31 | 2.62 |
| C182 | 10.31 | 4.72 | 2.52 | 28.94 | 1.36 | 2.31 |
| C183 | 0.81 | 0.23 | 0.00 | 1.86 | 0.31 | 2.21 |
| C181/182 | 8.27 | 4.35 | 1.42 | 33.56 | 1.58 | 4.88 |
| NPLS | RMSECV | R2CV | Slope | RPD | RER | |
|---|---|---|---|---|---|---|
| MPA | ||||||
| OSI | 9 | 2.99 | 0.79 | 0.80 | 2.16 | 12.81 |
| C160 | 9 | 1.25 | 0.80 | 0.81 | 2.26 | 11.92 |
| C161 | 13 | 0.30 | 0.77 | 0.80 | 2.07 | 13.92 |
| C180 | 14 | 0.33 | 0.67 | 0.69 | 1.70 | 14.41 |
| C181 | 4 | 1.91 | 0.92 | 0.91 | 3.48 | 22.82 |
| C182 | 3 | 0.83 | 0.97 | 0.97 | 5.69 | 31.91 |
| C183 | 14 | 0.15 | 0.60 | 0.63 | 1.53 | 12.63 |
| C181/182 | 3 | 2.43 | 0.69 | 0.70 | 1.79 | 13.23 |
| MicroNIR | ||||||
| OSI | 14 | 3.14 | 0.76 | 0.79 | 2.06 | 12.20 |
| C160 | 14 | 1.19 | 0.82 | 0.83 | 2.37 | 12.49 |
| C161 | 20 | 0.35 | 0.69 | 0.74 | 1.77 | 12.02 |
| C180 | 18 | 0.39 | 0.52 | 0.62 | 1.44 | 11.99 |
| C181 | 10 | 1.60 | 0.94 | 0.94 | 4.16 | 27.28 |
| C182 | 9 | 0.74 | 0.98 | 0.98 | 6.38 | 35.49 |
| C183 | 16 | 0.18 | 0.39 | 0.50 | 1.28 | 10.24 |
| C181/182 | 9 | 2.52 | 0.66 | 0.71 | 1.73 | 12.73 |
| Trait | Instrument | Genotype | Environment | GxE | Error | H2 |
|---|---|---|---|---|---|---|
| OSI | Reference | 61.00 | 26.19 | 2.66 | 10.14 | 0.78 |
| MPA | 71.20 | 22.94 | 0.23 | 5.63 | 0.86 | |
| MicroNIR | 23.67 | 53.72 | 5.81 | 16.80 | 0.43 | |
| C181 | Reference | 54.90 | 38.31 | 1.83 | 4.96 | 0.79 |
| MPA | 35.73 | 58.81 | 0.00 | 5.45 | 0.64 | |
| MicroNIR | 77.38 | 16.22 | 3.31 | 3.08 | 0.91 |
| Trait | Instrument | Genotype | Error | H2 | r |
|---|---|---|---|---|---|
| OSI | Reference | 90.39 | 9.61 | 0.90 | |
| MPA | 87.67 | 12.33 | 0.88 | 0.87 *** | |
| MicroNIR | 89.29 | 10.71 | 0.89 | 0.89 *** | |
| C181 | Reference | 96.50 | 3.50 | 0.97 | |
| MPA | 95.88 | 4.12 | 0.96 | 0.98 *** | |
| MicroNIR | 94.37 | 5.63 | 0.94 | 0.98 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yılmaz-Düzyaman, H.; de la Rosa, R.; Velasco, L.; Núñez-Sánchez, N.; León, L. Oil Quality Prediction in Olive Oil by Near-Infrared Spectroscopy: Applications in Olive Breeding. Agriculture 2024, 14, 721. https://doi.org/10.3390/agriculture14050721
Yılmaz-Düzyaman H, de la Rosa R, Velasco L, Núñez-Sánchez N, León L. Oil Quality Prediction in Olive Oil by Near-Infrared Spectroscopy: Applications in Olive Breeding. Agriculture. 2024; 14(5):721. https://doi.org/10.3390/agriculture14050721
Chicago/Turabian StyleYılmaz-Düzyaman, Hande, Raúl de la Rosa, Leonardo Velasco, Nieves Núñez-Sánchez, and Lorenzo León. 2024. "Oil Quality Prediction in Olive Oil by Near-Infrared Spectroscopy: Applications in Olive Breeding" Agriculture 14, no. 5: 721. https://doi.org/10.3390/agriculture14050721








