Improved Real-Time Models for Object Detection and Instance Segmentation for Agaricus bisporus Segmentation and Localization System Using RGB-D Panoramic Stitching Images
Abstract
:1. Introduction
- To obtain panoramic information in low-visibility working environments, this paper introduces a cost-effective and high-precision image stitching method. The method utilizes multiple local color images and depth information with the fast-stitching algorithm to acquire a high-precision panoramic image through bimodal fusion.
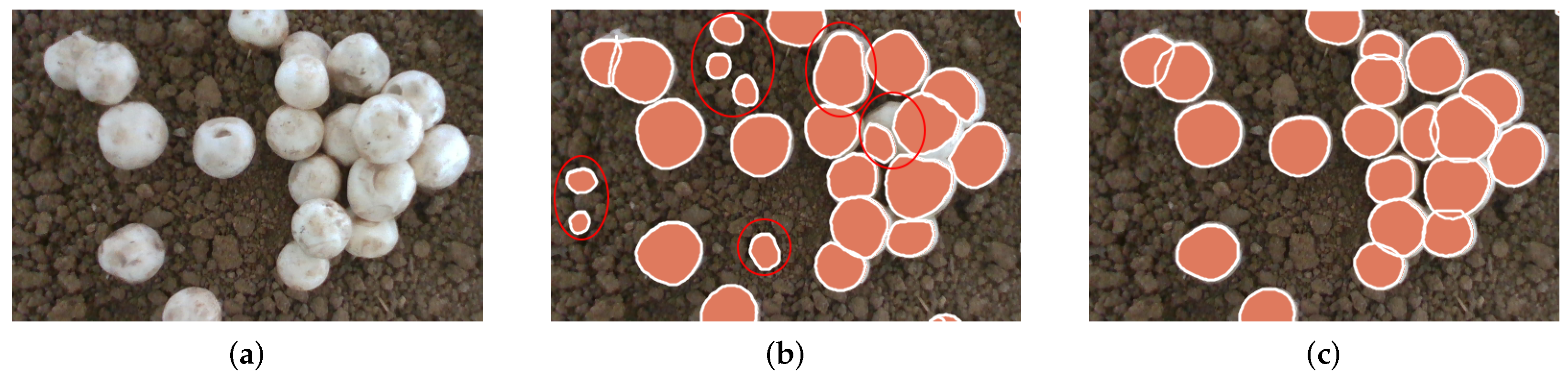
- This paper proposes using SimAM’s global attention mechanism and SPD-PAFPN’s lightweight feature fusion module in the RTMDet-Ins instance segmentation network model to address the extensive occlusion caused by A. bisporus’ clustered growth habits. This enhancement improves the handling capability of occluded A. bisporus and small-sized targets. Furthermore, the segmentation and detection capabilities of occluded areas for A. bisporus are further elevated by employing bimodal fusion images.
2. Materials and Methods
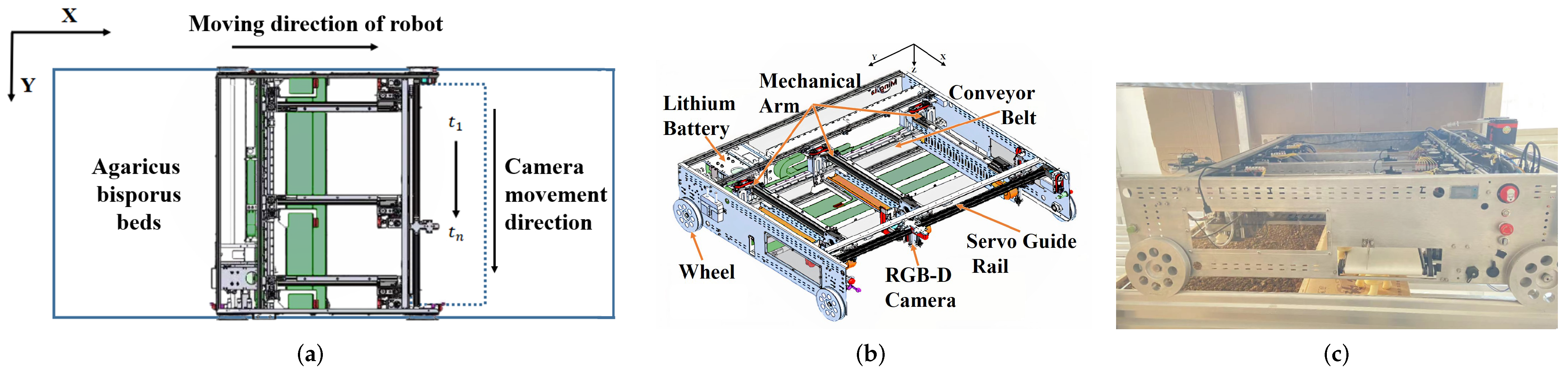
2.1. Harvest Robot Design
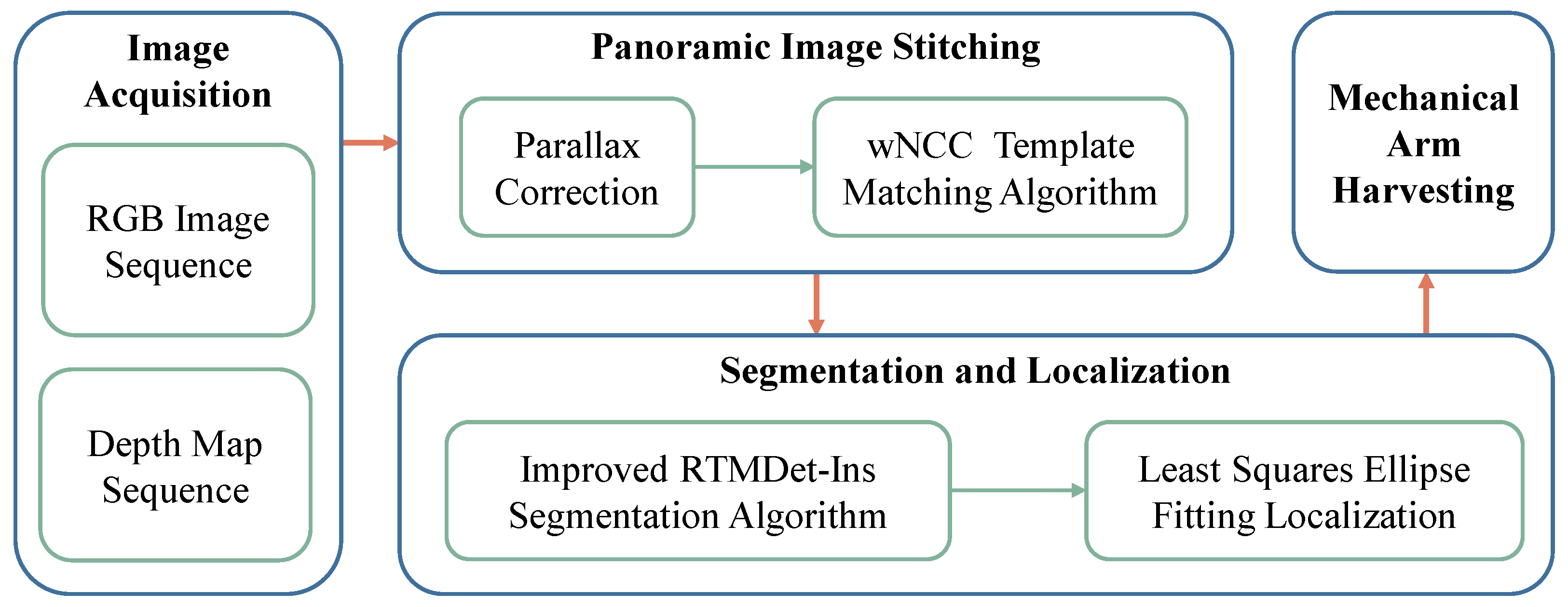
2.2. Visual Module Workflow Design
2.3. High-Precision Fusion Image Stitching Algorithm Based on Vision Correction
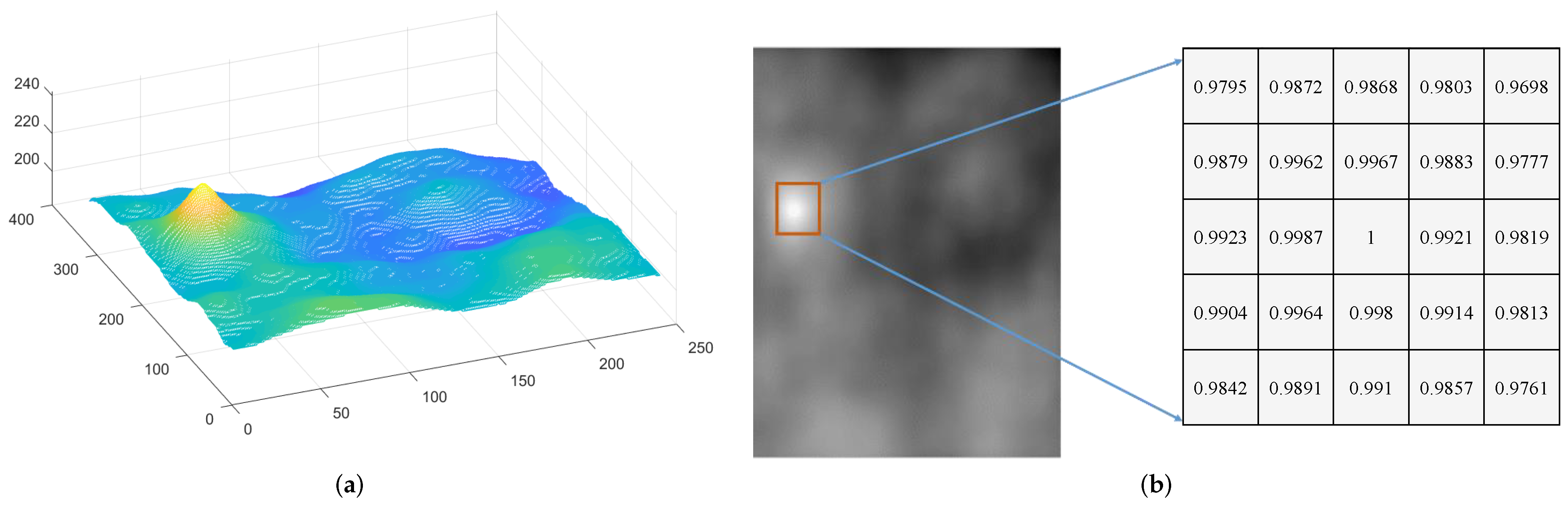
2.3.1. Parallax Correction
2.3.2. Panorama Stitching
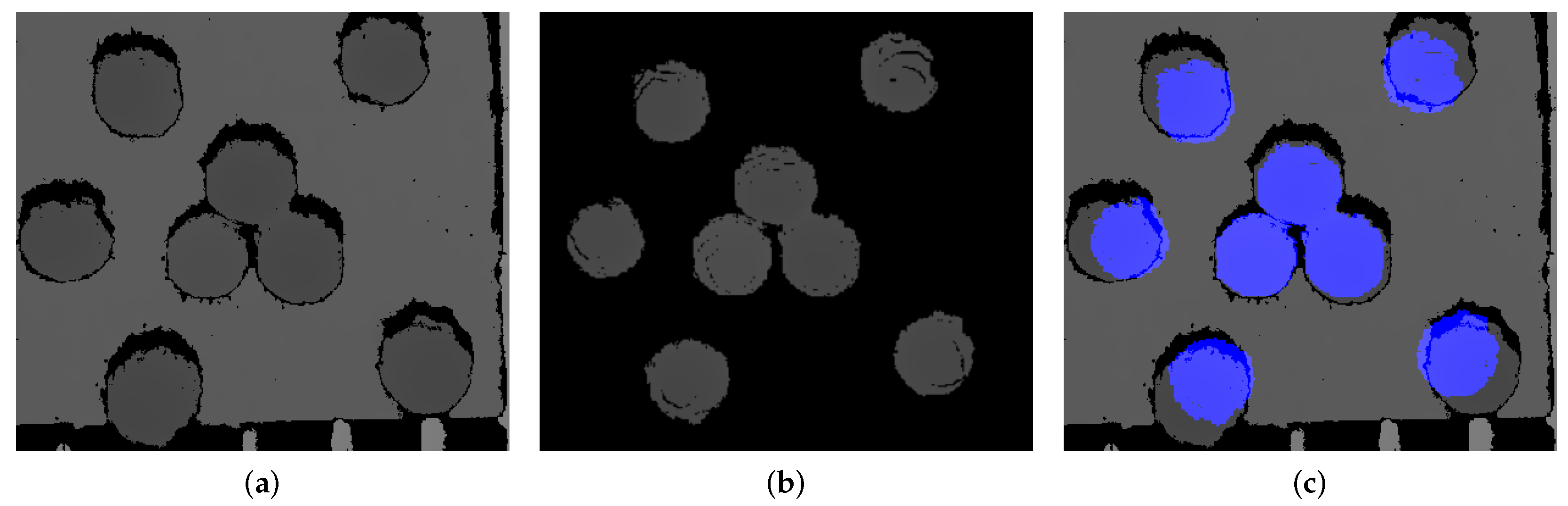
2.4. Improved RTMDet-Ins Fusion Image Experimental Segmentation and Localization Algorithm
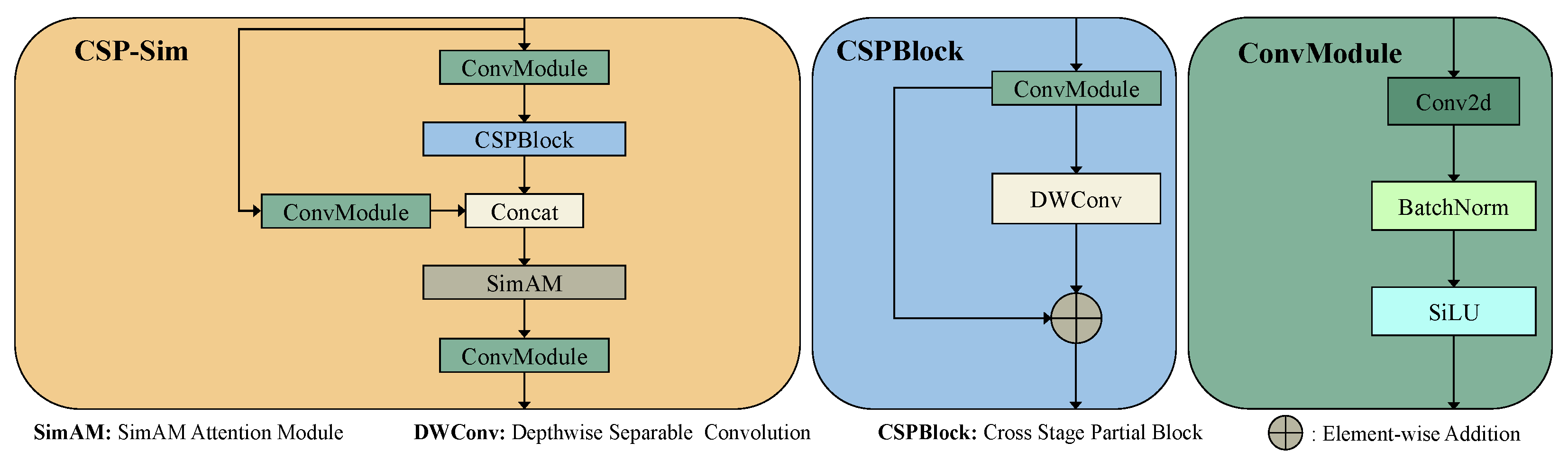
2.4.1. Improved to the Base Unit CSP-Sim
2.4.2. Improved Feature Fusion Module
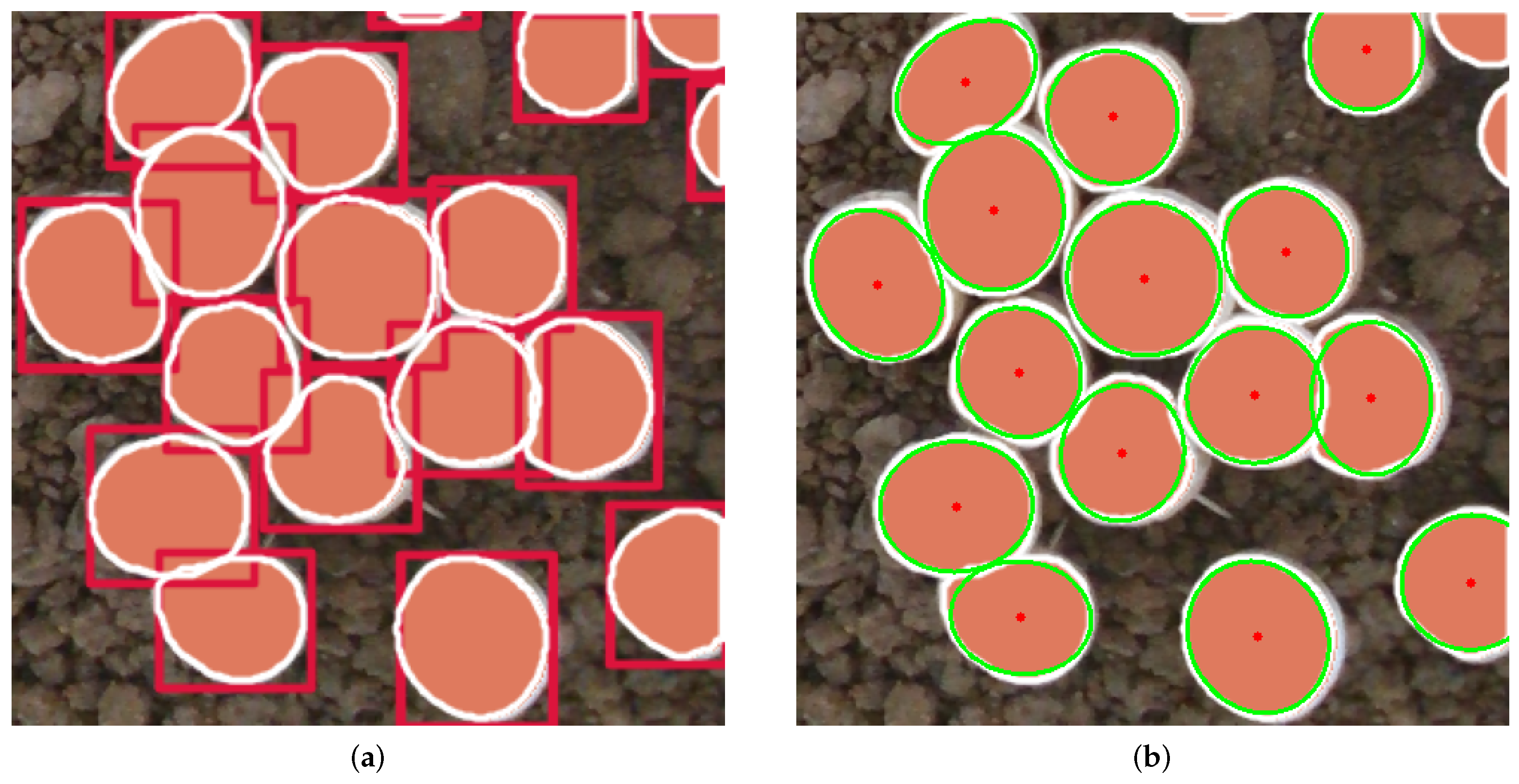
2.4.3. Localization Algorithm Based on Least Squares Ellipse Fitting
2.4.4. Performance Indicators
3. Results and Discussion
3.1. Experimental Environment and Training Strategy
3.2. Analysis of the Stitch Experiment Results
3.3. Ablation Study
3.4. Analysis of Detection Experiment Results
3.5. Analysis of Center Positioning Experiment Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Sławińska, A.; Jabłońska-Ryś, E.; Gustaw, W. Physico-Chemical, Sensory, and Nutritional Properties of Shortbread Cookies Enriched with Agaricus bisporus and Pleurotus ostreatus Powders. Appl. Sci. 2024, 14, 1938. [Google Scholar] [CrossRef]
- Iqbal, T.; Sohaib, M.; Iqbal, S.; Rehman, H. Exploring Therapeutic Potential of Pleurotus ostreatus and Agaricus bisporus Mushrooms against Hyperlipidemia and Oxidative Stress Using Animal Model. Foods 2024, 13, 709. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pan, B.; Wan, Y. Research on the application and future development of visual recognition in modern agriculture. In Proceedings of the 2023 International Conference on Data Science, Advanced Algorithm and Intelligent Computing (DAI 2023); Atlantis Press: Amsterdam, The Netherlands, 2024; pp. 25–35. [Google Scholar]
- Lu, C.P.; Liaw, J.J.; Wu, T.C.; Hung, T.F. Development of a mushroom growth measurement system applying deep learning for image recognition. Agronomy 2019, 9, 32. [Google Scholar] [CrossRef]
- Wee, B.S.; Chin, C.S.; Sharma, A. Survey of Mushroom Harvesting Agricultural Robots and Systems Design. IEEE Trans. AgriFood Electron. 2024, 2, 59–80. [Google Scholar] [CrossRef]
- Li, J.; Feng, Q.; Ru, M.; Sun, J.; Guo, X.; Zheng, W. Design of Shiitake Mushroom Robotic Picking Grasper: Considering Stipe Compressive Stress Relaxation. Machines 2024, 12, 241. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Z. The Vision-Based Target Recognition, Localization, and Control for Harvesting Robots: A Review. Int. J. Precis. Eng. Manuf. 2024, 25, 409–428. [Google Scholar] [CrossRef]
- Eastwood, D.C.; Herman, B.; Noble, R.; Dobrovin-Pennington, A.; Sreenivasaprasad, S.; Burton, K.S. Environmental regulation of reproductive phase change in Agaricus bisporus by 1-octen-3-ol, temperature and CO2. Fungal Genet. Biol. 2013, 55, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.; Miles, S.; Butler, J.; Baldwin, M.; Noble, R. AE—Automation and emerging technologies: Automatic mushroom harvester development. J. Agric. Eng. Res. 2001, 78, 15–23. [Google Scholar] [CrossRef]
- Yu, G.; Luo, J.; Zhao, Y. Region marking technique based on sequential scan and segmentation method of mushroom images. Trans. Chin. Soc. Agric. Eng. (Trans. CSAE) 2006, 22, 139–142. [Google Scholar]
- Zhou, R.; Chang, Z.; Sun, Y.; Fan, P.; Tan, C. A Novel Watershed Image Segmentation Algorithm Based on Quantum Inspired Morphology. J. Inf. Comput. Sci. 2015, 12, 4331–4338. [Google Scholar] [CrossRef]
- Chen, C.; Yi, S.; Mao, J.; Wang, F.; Zhang, B.; Du, F. A Novel Segmentation Recognition Algorithm of Agaricus bisporus Based on Morphology and Iterative Marker-Controlled Watershed Transform. Agronomy 2023, 13, 347. [Google Scholar] [CrossRef]
- Yang, S.; Ni, B.; Du, W.; Yu, T. Research on an improved segmentation recognition algorithm of overlapping Agaricus bisporus. Sensors 2022, 22, 3946. [Google Scholar] [CrossRef] [PubMed]
- Baisa, N.L.; Al-Diri, B. Mushrooms detection, localization and 3d pose estimation using rgb-d sensor for robotic-picking applications. arXiv 2022, arXiv:2201.02837. [Google Scholar]
- Shi, C.; Nie, J.; Mo, Y.; Zhang, C.; Zhu, C.; Zang, X. High precision scene stitching and recognition of agaricus bisporus based on depth camera. In Proceedings of the Third International Computing Imaging Conference (CITA 2023), Sydney, Australia, 1–3 June 2023; SPIE: Bellingham, WA, USA, 2023; Volume 12921, pp. 1001–1006. [Google Scholar]
- Tian, Z.; Shen, C.; Chen, H. Conditional convolutions for instance segmentation. In Proceedings of the Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, 23–28 August 2020; Proceedings, Part I 16. Springer: Berlin/Heidelberg, Germany, 2020; pp. 282–298. [Google Scholar]
- Wang, X.; Zhang, R.; Kong, T.; Li, L.; Shen, C. Solov2: Dynamic and fast instance segmentation. Adv. Neural Inf. Process. Syst. 2020, 33, 17721–17732. [Google Scholar]
- Zhong, M.; Han, R.; Liu, Y.; Huang, B.; Chai, X.; Liu, Y. Development, integration, and field evaluation of an autonomous Agaricus bisporus picking robot. Comput. Electron. Agric. 2024, 220, 108871. [Google Scholar] [CrossRef]
- Briechle, K.; Hanebeck, U.D. Template matching using fast normalized cross correlation. In Proceedings of the Optical Pattern Recognition XII, Orlando, FL, USA, 16–20 April 2001; SPIE: Bellingham, WA, USA, 2001; Volume 4387, pp. 95–102. [Google Scholar]
- Lyu, C.; Zhang, W.; Huang, H.; Zhou, Y.; Wang, Y.; Liu, Y.; Zhang, S.; Chen, K. Rtmdet: An empirical study of designing real-time object detectors. arXiv 2022, arXiv:2212.07784. [Google Scholar]
- Yang, L.; Zhang, R.Y.; Li, L.; Xie, X. Simam: A simple, parameter-free attention module for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; PMLR: New York, NY, USA, 2021; pp. 11863–11874. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–16 July 2017; pp. 1251–1258. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–16 July 2017; pp. 2117–2125. [Google Scholar]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path aggregation network for instance segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8759–8768. [Google Scholar]
- Sunkara, R.; Luo, T. No more strided convolutions or pooling: A new CNN building block for low-resolution images and small objects. In Proceedings of the Joint European Conference on Machine Learning and Knowledge Discovery in Databases, Grenoble, France, 19–23 September 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 443–459. [Google Scholar]
- Fausto Milletari, N.; V-Net, A.S.A. Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. Available online: https://arxiv.org/abs/1606.04797 (accessed on 2 January 2024).
- Wang, C.Y.; Liao, H.Y.M.; Wu, Y.H.; Chen, P.Y.; Hsieh, J.W.; Yeh, I.H. CSPNet: A new backbone that can enhance learning capability of CNN. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 390–391. [Google Scholar]
- Sajjadi, M.S.; Vemulapalli, R.; Brown, M. Frame-recurrent video super-resolution. In Proceedings of the IEEE Conference on Computer Vision and pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6626–6634. [Google Scholar]
- Fitzgibbon, A.; Pilu, M.; Fisher, R.B. Direct least square fitting of ellipses. IEEE Trans. Pattern Anal. Mach. Intell. 1999, 21, 476–480. [Google Scholar] [CrossRef]


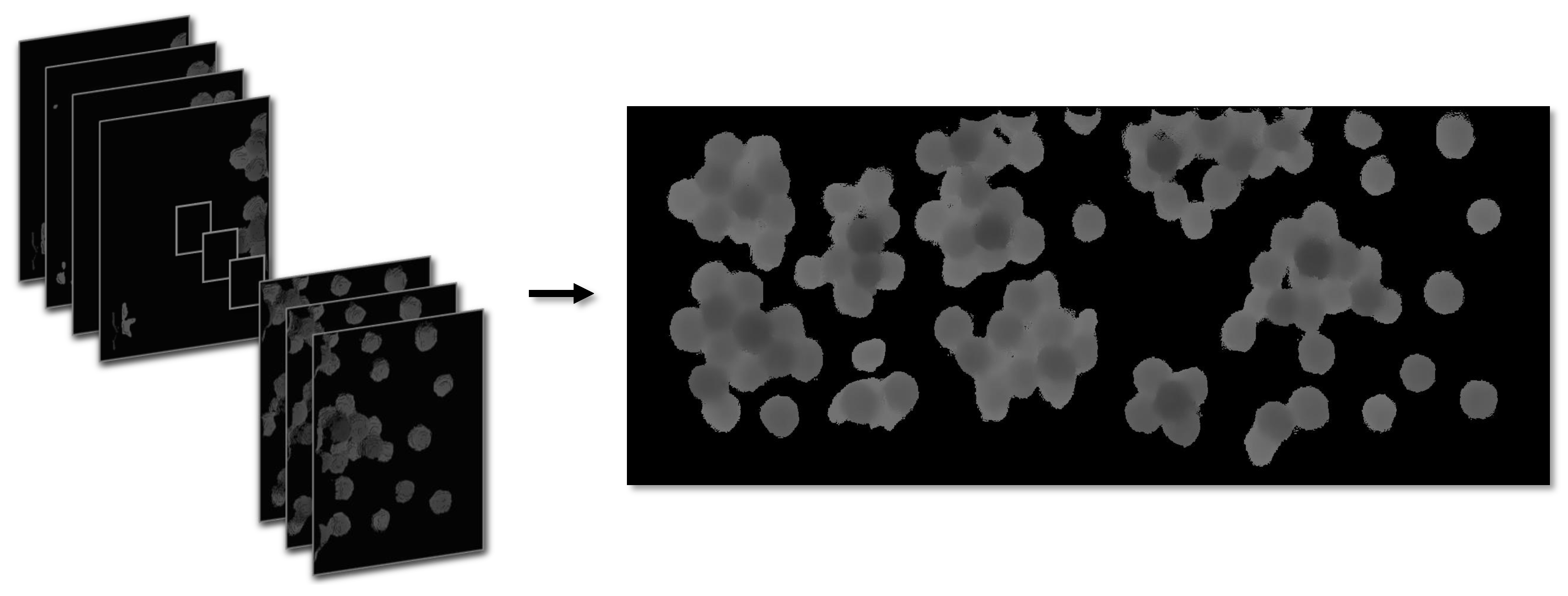


| Sample ID | No. 1 | No. 2 | No. 3 | No. 4 | No. 5 | No. 6 | No. 7 | No. 8 | No. 9 | No. 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Panoramic Stitching Errors after Disparity Correction/mm | 1.855 | 1.932 | 2.021 | 1.903 | 2.029 | 1.892 | 1.923 | 1.929 | 2.004 | 1.969 |
| Panoramic Stitching Errors without Correction/mm | 15.236 | 17.768 | 18.234 | 16.246 | 18.675 | 19.023 | 15.912 | 16.824 | 18.897 | 17.932 |
| Methods | SimAM | SPD-Conv | Params (M) | FLOPs (G) | Running Time (ms) | AP50 (%) | AP75 (%) |
|---|---|---|---|---|---|---|---|
| RTMDet-Ins | 5.61 | 11.87 | 26.26 | 96.50 | 93.48 | ||
| RTMDet-Ins + SimAM | ✔ | 5.46 | 11.87 | 25.73 | 97.91 | 95.73 | |
| RTMDet-Ins + SPD-Conv | ✔ | 5.53 | 10.74 | 26.15 | 96.93 | 94.20 | |
| Improved RTMDet-Ins | ✔ | ✔ | 5.35 | 10.74 | 25.38 | 98.50 | 96.10 |
| Sample ID | Number | Improved RTMDet-Ins | SOLOv2 | CondInst | Watershed | ||||
|---|---|---|---|---|---|---|---|---|---|
| Correct Number | Correct Rate (%) | Correct Number | Correct Rate (%) | Correct Number | Correct Rate (%) | Correct Number | Correct Rate (%) | ||
| No. 1 | 31 | 31 | 100.00 | 31 | 100.00 | 31 | 100.00 | 29 | 93.55 |
| No. 2 | 41 | 41 | 100.00 | 39 | 95.12 | 41 | 100.00 | 38 | 92.68 |
| No. 3 | 38 | 37 | 97.37 | 35 | 92.11 | 36 | 97.74 | 34 | 89.47 |
| No. 4 | 33 | 33 | 100.00 | 33 | 100.00 | 33 | 100.00 | 30 | 90.91 |
| No. 5 | 33 | 32 | 96.97 | 32 | 96.97 | 32 | 96.97 | 30 | 90.91 |
| No. 6 | 44 | 43 | 97.73 | 41 | 93.18 | 41 | 93.18 | 39 | 88.64 |
| No. 7 | 38 | 38 | 100.00 | 37 | 97.37 | 36 | 94.74 | 36 | 94.74 |
| No. 8 | 29 | 29 | 100.00 | 28 | 96.55 | 28 | 96.55 | 28 | 96.55 |
| No. 9 | 52 | 51 | 98.08 | 51 | 98.08 | 50 | 96.15 | 48 | 92.31 |
| No. 10 | 53 | 51 | 96.23 | 50 | 94.34 | 51 | 96.23 | 49 | 92.45 |
| Average Correct Rate | 98.64% | 96.37% | 96.86% | 92.22% | |||||
| Manual Positiomng | Least-Squares Ellipse Fitting | Hough Transform Circle Fitting | |
|---|---|---|---|
| Positioning example |  |  |  |
| Coordinate | (523, 235) | (523, 230) | (522, 228) |
| LER | 0 | 0.78% | 1.15% |
| Sample ID | Manual | Least-Squares Ellipse Fitting | Hough Transform Circle Fitting | ||
|---|---|---|---|---|---|
| Algorithm Location | LER (%) | Algorithm Location | LER (%) | ||
| No. 1 | (210, 71) | (211, 72) | 0.21% | (208, 69) | 0.42% |
| No. 2 | (378, 261) | (380, 261) | 0.11% | (380, 257) | 0.73% |
| No. 3 | (104, 338) | (102, 335) | 0.58% | (102, 340) | 0.42% |
| No. 4 | (300, 420) | (303, 424) | 0.79% | (303, 423) | 0.63% |
| No. 5 | (233, 178) | (235, 178) | 0.11% | (233, 174) | 0.63% |
| No. 6 | (157, 409) | (152, 406) | 0.74% | (155, 412) | 0.58% |
| No. 7 | (215, 295) | (218, 298) | 0.63% | (213, 292) | 0.58% |
| No. 8 | (303, 255) | (308, 254) | 0.43% | (306, 251) | 0.79% |
| No. 9 | (61, 183) | (61, 182) | 0.16% | (57, 183) | 0.22% |
| No. 10 | (129, 133) | (129, 132) | 0.16% | (128, 130) | 0.52% |
| No. 11 | (328, 163) | (329, 164) | 0.21% | (325, 159) | 0.79% |
| No. 12 | (146, 243) | (146, 242) | 0.16% | (148, 240) | 0.58% |
| No. 13 | (445, 384) | (446, 383) | 0.21% | (441, 387) | 0.68% |
| No. 14 | (117, 44) | (118, 44) | 0.05% | (118, 45) | 0.21% |
| No. 15 | (381, 29) | (380, 29) | 0.05% | (379, 28) | 0.26% |
| mean LER | 0.31% | 0.52% | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, C.; Mo, Y.; Ren, X.; Nie, J.; Zhang, C.; Yuan, J.; Zhu, C. Improved Real-Time Models for Object Detection and Instance Segmentation for Agaricus bisporus Segmentation and Localization System Using RGB-D Panoramic Stitching Images. Agriculture 2024, 14, 735. https://doi.org/10.3390/agriculture14050735
Shi C, Mo Y, Ren X, Nie J, Zhang C, Yuan J, Zhu C. Improved Real-Time Models for Object Detection and Instance Segmentation for Agaricus bisporus Segmentation and Localization System Using RGB-D Panoramic Stitching Images. Agriculture. 2024; 14(5):735. https://doi.org/10.3390/agriculture14050735
Chicago/Turabian StyleShi, Chenbo, Yuanzheng Mo, Xiangqun Ren, Jiahao Nie, Chun Zhang, Jin Yuan, and Changsheng Zhu. 2024. "Improved Real-Time Models for Object Detection and Instance Segmentation for Agaricus bisporus Segmentation and Localization System Using RGB-D Panoramic Stitching Images" Agriculture 14, no. 5: 735. https://doi.org/10.3390/agriculture14050735
APA StyleShi, C., Mo, Y., Ren, X., Nie, J., Zhang, C., Yuan, J., & Zhu, C. (2024). Improved Real-Time Models for Object Detection and Instance Segmentation for Agaricus bisporus Segmentation and Localization System Using RGB-D Panoramic Stitching Images. Agriculture, 14(5), 735. https://doi.org/10.3390/agriculture14050735







