The Presence of Arbuscular Mycorrhizal Fungi in the Rhizosphere of Transgenic Rapeseed Overexpressing a Trichoderma Thkel1 Gene Improves Plant Development and Yield
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. AMF Material
2.3. AMF Inoculation and Plant Growth Conditions
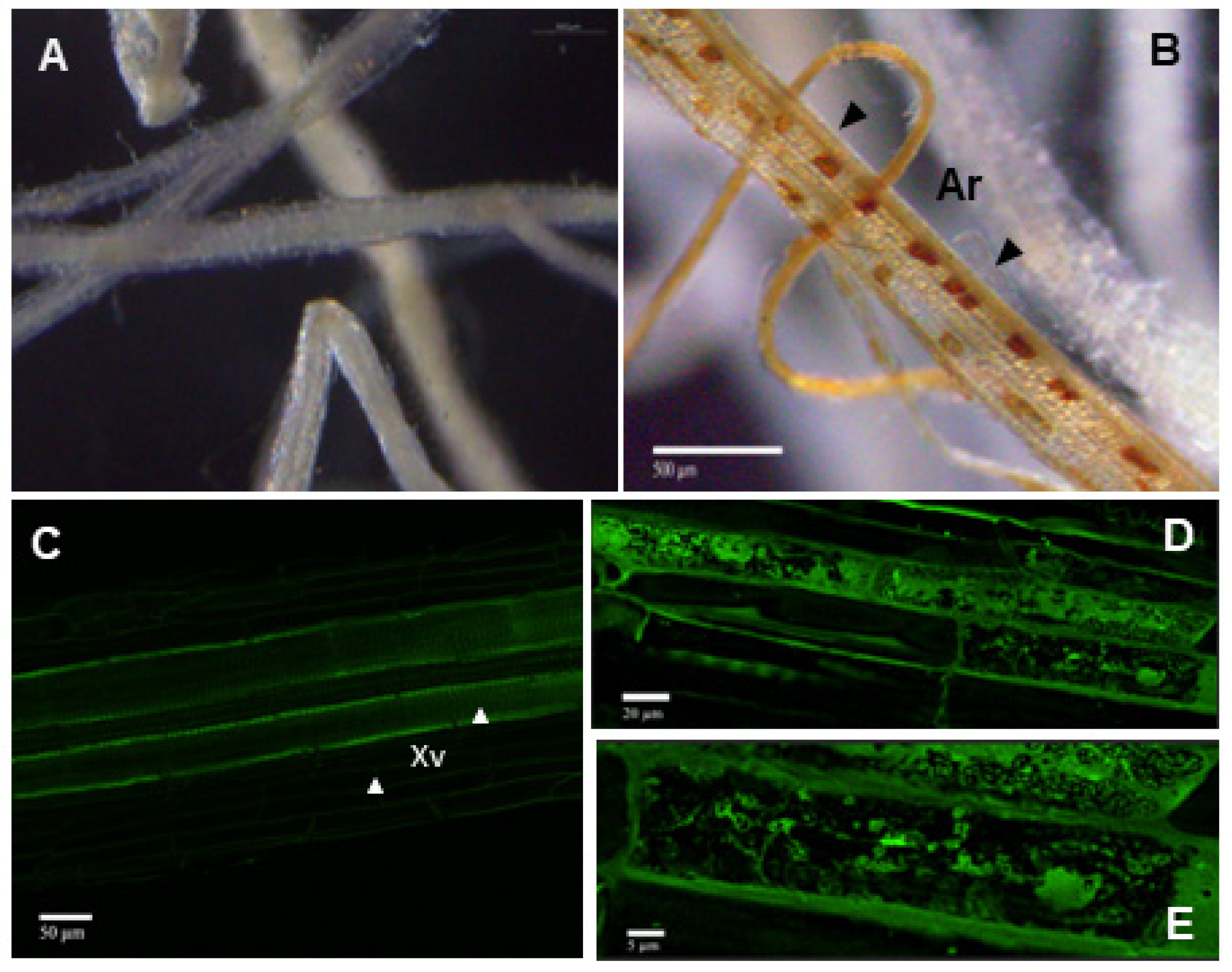
2.4. Visualization of the AMF Structures in Rapeseed Roots
2.5. DNA- Detection of AMF at the Roots
2.6. Phosphorus and Iron Measurement
2.7. GintPT and GintAMT2 Gene Expression Analysis by qPCR
2.8. GSL and Oil Profiles
2.8.1. Sample Extraction
2.8.2. LC/ESI-QqTOF-MS Analyses
2.9. Statistical Analysis
3. Results
3.1. Presence of AMF in Thkel1-Overexpressing Rapeseed Plants
3.2. Gene Expression Analysis of Phosphate and Ammonium Transporters of the AMF Rhizophagus Irregularis
3.3. Changes in P and Fe Content in AMF-BnKel Plants
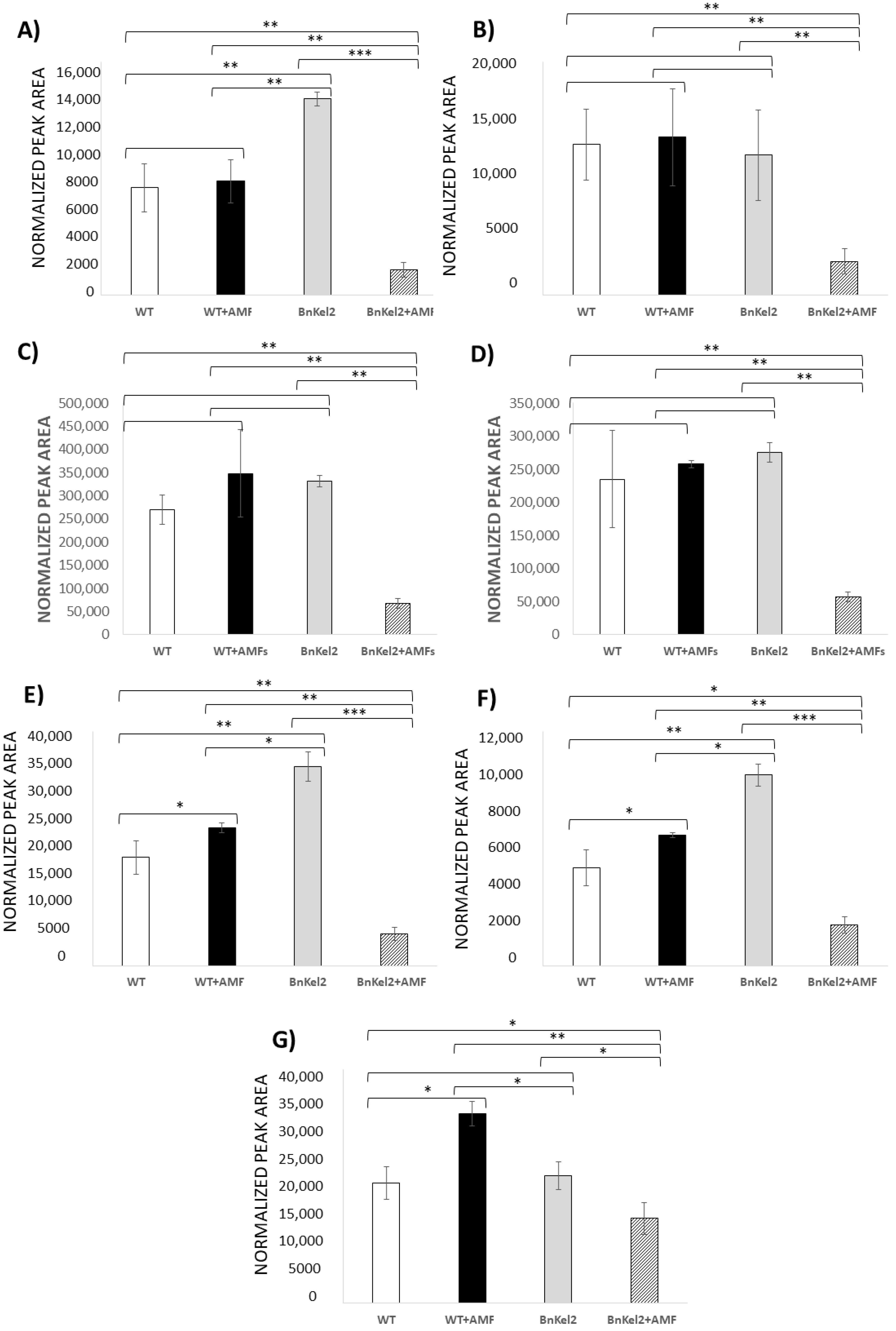
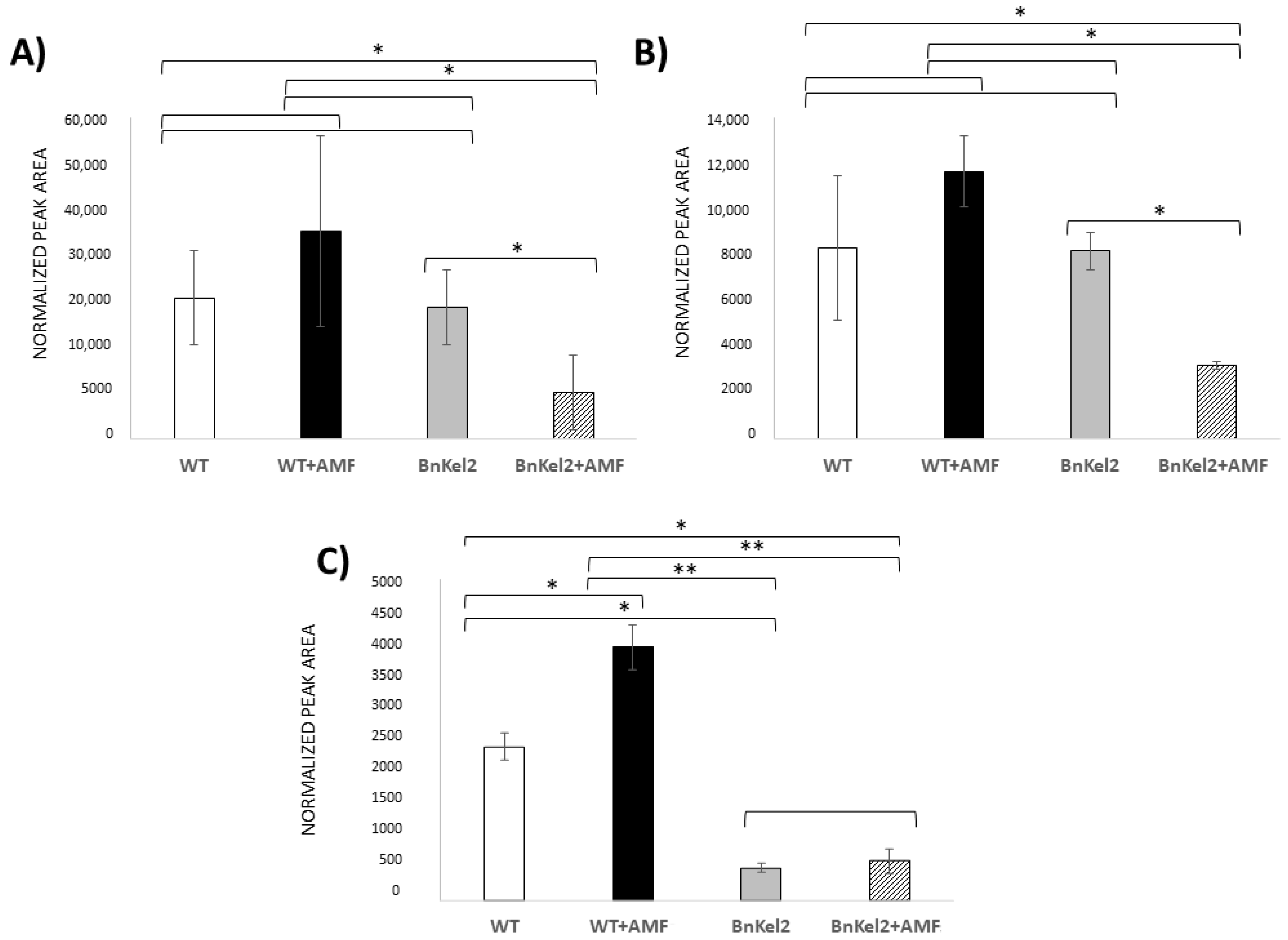
3.4. Reduction of GSL Content in BnKel Roots Inoculated with AMF
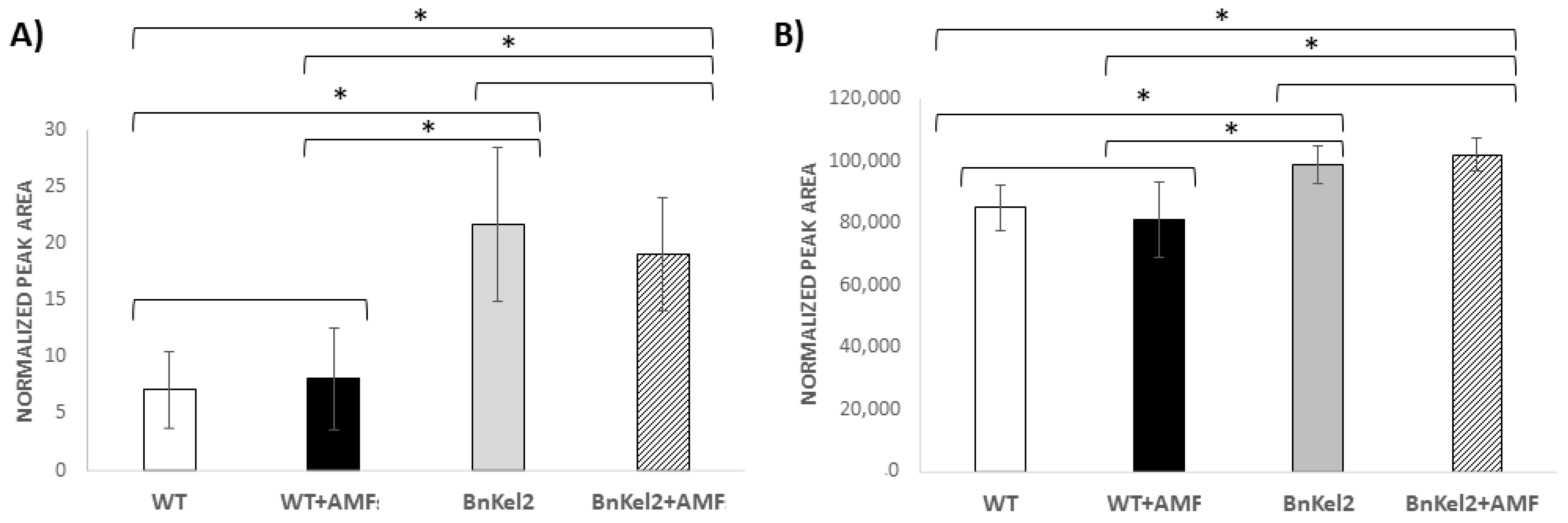
3.5. Reduction of GSL Content in BnKel Seeds and Seed Oil Content
3.6. Rapeseed Seed Yield
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, C.H.; Sun, R.; Hu, Y.; Zeng, L.; Zhang, N.; Cai, L.; Zhang, Q.; Koch, M.A.; Al-Shehbaz, I.; Edger, P.P.; et al. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol. 2015, 33, 394–412. [Google Scholar] [CrossRef] [PubMed]
- Cartea, M.E.; Francisco, M.; Soengas, P.; Velasco, P. Phenolic compounds in Brassica vegetables. Molecules 2011, 16, 251–280. [Google Scholar] [CrossRef] [PubMed]
- Angelino, D.; Jeffery, E. Glucosinolate hydrolysis and bioavailability of resulting isothiocyanates: Focus on glucoraphanin. J. Funct. Foods 2014, 7, 67–76. [Google Scholar] [CrossRef]
- Hassan, M.N.; Zainal, Z.; Ismail, I. Plant Kelch containing F-box proteins: Structure, evolution and functions. RSC Adv. 2015, 5, 42808–42814. [Google Scholar] [CrossRef]
- Martínez-Ballesta, M.C.; Carvajal, M. Myrosinase in Brassicaceae: The most important issue for glucosinolate turnover and food quality. Phytochem. Rev. 2015, 14, 1045–1051. [Google Scholar] [CrossRef]
- Berruti, A.; Lumini, E.; Balestrini, R.; Bianciotto, V. Arbuscular mycorrhizal fungi as natural biofertilizers: Let’s benefit from past successes. Front. Microbiol. 2016, 6, 1559. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.L.; Hermosa, R.; Lorito, M.; Monte, E. Trichoderma: A multipurpose plant beneficial microorganism for eco-sustainable agriculture. Nat. Rev. Microbiol. 2023, 21, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Hermosa, R.; Viterbo, A.; Chet, I.; Monte, E. Plant-beneficial effects of Trichoderma and of its genes. Microbiology 2012, 158, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Mendoza, A.; Zaid, R.; Lawry, R.; Hermosa, R.; Monte, E.; Horwitz, B.A.; Mukherjee, P.K. Molecular dialogues between Trichoderma and roots: Role of the fungal secretome. Fungal Biol. Rev. 2018, 32, 62–85. [Google Scholar] [CrossRef]
- Nicolás, C.; Hermosa, R.; Rubio, B.; Mukherjee, P.K.; Monte, E. Trichoderma genes in plants for stress tolerance-status and prospects. Plant Sci. 2014, 228, 71–78. [Google Scholar] [CrossRef]
- MacLean, A.M.; Bravo, A.; Harrison, M.J. Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell 2017, 29, 2319–2335. [Google Scholar] [CrossRef]
- Roth, R.; Paszkowski, U. Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr. Opin. Plant Biol. 2017, 39, 50–56. [Google Scholar] [CrossRef]
- Delaux, P.M. Comparative phylogenomics of symbiotic associations. New Phytol. 2017, 213, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Bruisson, S.; Bapaume, L.; Darbon, G.; Glauser, G.; Schorderet, M.; Reinhardt, D. VAPYRIN attenuates defence by repressing PR gene induction and localized lignin accumulation during arbuscular mycorrhizal symbiosis of Petunia hybrida. New Phytol. 2021, 229, 3481–3496. [Google Scholar] [CrossRef] [PubMed]
- Fernández, I.; Cosme, M.; Stringlis, I.A.; Yu, K.; de Jonge, R.; van Wees, S.M.; Pozo, M.J.; Pieterse, C.M.J.; van der Heijden, M.G.A. Molecular dialogue between arbuscular mycorrhizal fungi and the nonhost plant Arabidopsis thaliana switches from initial detection to antagonism. New Phytol. 2019, 223, 867–881. [Google Scholar] [CrossRef]
- Cosme, M.; Fernández, I.; Van der Heijden, M.G.A.; Pieterse, C.M.J. Non-mycorrhizal plants: The exceptions that prove the rule. Trends Plant Sci. 2018, 23, 577–587. [Google Scholar] [CrossRef]
- Cosme, M.; Fernández, I.; Declerck, S.; Van der Heijden, M.G.A.; Pieterse, C.M.J. A coumarin exudation pathway mitigates arbuscular mycorrhizal incompatibility in Arabidopsis thaliana. Plant Mol. Biol. 2021, 106, 319–334. [Google Scholar] [CrossRef]
- Hermosa, R.; Botella, L.; Keck, E.; Jiménez, J.Á.; Montero-Barrientos, M.; Arbona, V.; Gómez-Cadenas, A.; Monte, E.; Nicolás, C. The overexpression in Arabidopsis thaliana of a Trichoderma harzianum gene that modulates glucosidase activity, and enhances tolerance to salt and osmotic stresses. J. Plant Physiol. 2011, 168, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Gumz, F.; Krausze, J.; Eisenschmidt, D.; Backenköhler, A.; Barleben, L.; Brandt, W.; Wittstock, U. The crystal structure of the thiocyanate-forming protein from Thlaspi arvense, a kelch protein involved in glucosinolate breakdown. Plant Mol. Biol. 2015, 89, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J.; Hermosa, R.; Monte, E.; Nicolás, C. Trichoderma harzianum favours the access of arbuscular mycorrhizal fungi to non-host Brassicaceae roots and increases plant productivity. Sci. Rep. 2019, 9, 11650. [Google Scholar] [CrossRef]
- Vierheilig, H.; Schweiger, P.; Brundrett, M. An overview of methods for the detection and observation of arbuscular mycorrhizal fungi in roots. Physiol. Plant. 2005, 125, 393–404. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Young, J.P.W. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 2008, 65, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, J.; Huang, S.; Guo, T.; Deng, L.; Hua, W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 2014, 538, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J.; Hermosa, R.; Monte, E.; Nicolás, C. The Trichoderma harzianum Kelch protein ThKEL1 plays a key role in root colonization and the induction of systemic defense in Brassicaceae plants. Front. Plant Sci. 2019, 10, 1478. [Google Scholar] [CrossRef] [PubMed]
- Illescas, M.; Rubio, M.B.; Hernández-Ruiz, V.; Morán-Diez, M.E.; Martínez de Alba, A.E.; Nicolás, C.; Monte, E.; Hermosa, R. Effect of inorganic top dressing and Trichoderma harzianum seed-inoculation on crop yield and the shaping of root microbial communities of wheat plants cultivated under high basal N fertilization. Front. Plant Sci. 2020, 11, 575861. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- De Ollas, C.; González-Guzmán, M.; Pitarch, Z.; Matus, J.T.; Candela, H.; Rambla, J.L.; Granell, A.; Gómez-Cadenas, A.; Arbona, V. Identification of ABA-mediated genetic and metabolic responses to soil flooding in tomato (Solanum lycopersicum L. Mill). Front. Plant Sci. 2021, 12, 613059. [Google Scholar] [CrossRef] [PubMed]
- Hart, M.M.; Trevors, J.T. Microbe management: Application of mycorrhyzal fungi in sustainable agriculture. Front. Ecol. Environ. 2005, 3, 533–539. [Google Scholar] [CrossRef]
- Xu, M.J.; Wang, H.Z. Proteomic analysis reveals the mechanisms of Mycena dendrobii promoting transplantation survival and growth of tissue culture seedlings of Dendrobium officinale. J. Appl. Microbiol. 2015, 118, 1444–1455. [Google Scholar] [CrossRef]
- Sugimura, Y.; Saito, K. Comparative transcriptome analysis between Solanum lycopersicum L. and Lotus japonicus L. during arbuscular mycorrhizal development. J. Soil Sci. Plant Nutr. 2017, 63, 127–136. [Google Scholar] [CrossRef]
- Takahara, M.; Magori, S.; Soyano, T.; Okamoto, S.; Yoshida, C.; Yano, K.; Sato, S.; Tabata, S.; Yamaguchi, K.; Shigenobu, S.; et al. Too much love, a novel Kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume-Rhizobium symbiosis. Plant Cell Physiol. 2013, 54, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, J.A. Vesicular-arbuscular mycorrhizal infection of “host” and “non-host” plants: Effect on the growth responses of the plants and competition between them. Soil Biol. Biochem. 1986, 18, 607–610. [Google Scholar] [CrossRef]
- Kabir, Z.; O’Halloran, I.P.; Hamel, C. The proliferation of fungal hyphae in soils supporting mycorrhizal and non-mycorrhizal plants. Mycorrhiza 1997, 6, 477–480. [Google Scholar] [CrossRef]
- Schreiner, R.; Koide, R.T. Antifungal compounds from the roots of mycotrophic and non-mycotrophic plant species. New Phytol. 1993, 123, 99–105. [Google Scholar] [CrossRef]
- Aguilar, R.; Carreón-Abud, Y.; López-Carmona, D.; Larsen, J. Organic fertilizers alter the composition of pathogens and arbuscular mycorrhizal fungi in maize roots. J. Phytopathol. 2017, 165, 448–454. [Google Scholar] [CrossRef]
- Glenn, M.G.; Chew, F.S.; Williams, P.H. Influence of glucosinolate content of Brassica (Cruciferae) roots on growth of vesicular–arbuscular mycorrhizal fungi. New Phytol. 1998, 110, 217–225. [Google Scholar] [CrossRef]
- DeMars, B.G.; Boerner, R.E. Arbuscular mycorrhizal development in three crucifers. Mycorrhiza 1995, 5, 405–408. [Google Scholar] [CrossRef]
- Liu, Y.; Xiong, Z.; Wu, W.; Ling, H.Q.; Kong, D. Iron in the symbiosis of plants and microorganisms. Plants 2023, 12, 1958. [Google Scholar] [CrossRef] [PubMed]
- Kabir, A.H.; Debnath, T.; Das, U.; Prity, S.A.; Haque, A.; Rahman, M.M.; Parvez, M.S. Arbuscular mycorrhizal fungi alleviate Fe-deficiency symptoms in sunflower by increasing iron uptake and its availability along with antioxidant defense. Plant Physiol. Biochem. 2020, 150, 254–262. [Google Scholar] [CrossRef]
- Pongrac, P.; Vogel-Mikuš, K.; Poschenrieder, C.; Barcelo, J.; Tolra, R.; Regvar, M. Arbuscular mycorrhiza in glucosinolate-containing plants: The story of the metal hyperaccumulator Noccaea (Thlaspi) praecox (Brassicaceae). In Molecular Microbial Ecology of the Rhizosphere; de Brujin, F.J., Ed.; Wiley: Hoboken, NJ, USA, 2013; pp. 1023–1032. [Google Scholar]
- Vierheilig, H.; Bennett, R.; Kiddle, G.; Kaldorf, M.; Ludwig-Müller, J. Differences in glucosinolate patterns and arbuscular mycorrhizal status of glucosinolate-containing plant species. New Phytol. 2000, 146, 343–352. [Google Scholar] [CrossRef]
- Cuong, D.M.; Kim, J.K.; Bong, S.J.; Baek, S.A.; Jeon, J.; Park, J.S.; Park, S.U. Comparative analysis of glucosinolates and metabolite profiling of green and red mustard (Brassica juncea) hairy roots. 3 Biotech. 2018, 8, 382. [Google Scholar] [CrossRef] [PubMed]
- Anthony, M.A.; Celenza, J.L.; Armstrong, A.; Frey, S.D. Indolic glucosinolate pathway provides resistance to mychorrhizal fungal colonization in a non-host Brassicaceae. Ecosphere 2020, 11, e03100. [Google Scholar] [CrossRef]
- Sharma, A.; Sinharoy, S.; Bisht, N.C. The mysterious non-arbuscular mycorrhizal status of Brassicaceae species. Environ. Microbiol. 2023, 25, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J.; Díaz-González, S.; Díaz-Urbano, M.; Velasco, P.; Sacristán, S. Fungal endophytes of Brassicaceae: Molecular interactions and crop benefits. Front. Plant Sci. 2022, 13, 932288. [Google Scholar] [CrossRef] [PubMed]
- Tayo, T.; Dutta, N.; Sharma, K. Effect of feeding canola-quality rapeseed mustard meal on animal production—A review. Agri. Rev. 2012, 33, 114–121. [Google Scholar]
- Nour-Eldin, H.H.; Madsen, S.R.; Engelen, S.; Jørgensen, M.E.; Olsen, C.E.; Andersen, J.S.; Seynnaeve, D.; Verhoye, T.; Fulawka, R.; Denolf, P.; et al. Reduction of antinutritional glucosinolates in Brassica oilseeds by mutation of genes encoding transporters. Nat. Biotechnol. 2017, 35, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Egert, S.; Baxheinrich, A.; Lee-Barkey, Y.H.; Tschoepe, D.; Stehle, P.; Stratmann, B.; Wahrburg, U. Effects of a hypoenergetic diet rich in α-linolenic acid on fatty acid composition of serum phospholipids in overweight and obese patients with metabolic syndrome. Nutrition 2018, 49, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Scapicchio, P.L. Revisiting choline alphoscerate profile: A new, perspective, role in dementia? Int. J. Neurosci. 2013, 123, 444–449. [Google Scholar] [CrossRef]
- Baum, C.; El-Tohamy, W.; Gruda, N. Increasing the productivity and product quality of vegetable crops using arbuscular mycorrhizal fungi: A review. Sci. Hortic. 2015, 187, 131–141. [Google Scholar] [CrossRef]

| AMF Inoculated Plants | Plant | AMF | Ratio 3 | ||||
|---|---|---|---|---|---|---|---|
| Ct | SD | Qty 1 | Ct | SD | Qty 2 | ||
| WT | 21.28 | 0.05 | 0.84 | - | 0.0 | 0.0 | 0 c |
| BnKel1 | 20.75 | 0.02 | 1.19 | 27.53 | 0.12 | 0.78 | 0.65 ± 0.03 b |
| BnKel2 | 20.98 | 0.03 | 0.90 | 26.38 | 0.22 | 0.67 | 0.74 ± 0.04 a |
| Plants | P 1 | Fe 2 |
|---|---|---|
| WT | 2.990 ± 0.063 c | 82.075 ± 5.655 bc |
| WT + AMF | 2.592 ± 0.242 d | 71.715 ± 4.815 c |
| BnKel2 | 3.715 ± 0.017 b | 86.218 ± 7.765 b |
| BnKel2 + AMF | 4.994 ± 0.496 a | 138.379 ± 18.993 a |
| Plants | Seed Weight (g) |
|---|---|
| WT | 61.835 ± 6.226 c |
| WT + AMF | 53.653 ± 7.665 c |
| BnKel2 | 99.526 ± 9.678 b |
| BnKel2 + AMF | 155.596 ± 12.966 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicolás, C.; Calvo-Polanco, M.; Poveda, J.; Alonso-Ramírez, A.; Ascaso, J.; Arbona, V.; Hermosa, R. The Presence of Arbuscular Mycorrhizal Fungi in the Rhizosphere of Transgenic Rapeseed Overexpressing a Trichoderma Thkel1 Gene Improves Plant Development and Yield. Agriculture 2024, 14, 851. https://doi.org/10.3390/agriculture14060851
Nicolás C, Calvo-Polanco M, Poveda J, Alonso-Ramírez A, Ascaso J, Arbona V, Hermosa R. The Presence of Arbuscular Mycorrhizal Fungi in the Rhizosphere of Transgenic Rapeseed Overexpressing a Trichoderma Thkel1 Gene Improves Plant Development and Yield. Agriculture. 2024; 14(6):851. https://doi.org/10.3390/agriculture14060851
Chicago/Turabian StyleNicolás, Carlos, Mónica Calvo-Polanco, Jorge Poveda, Ana Alonso-Ramírez, Julio Ascaso, Vicent Arbona, and Rosa Hermosa. 2024. "The Presence of Arbuscular Mycorrhizal Fungi in the Rhizosphere of Transgenic Rapeseed Overexpressing a Trichoderma Thkel1 Gene Improves Plant Development and Yield" Agriculture 14, no. 6: 851. https://doi.org/10.3390/agriculture14060851





