Identifying Cassava Genotypes Resistant to the African Cassava Whitefly, Bemisia tabaci (Gennadius)
Abstract
1. Introduction
2. Materials and Methods
2.1. Cassava Genotypes
2.2. Whitefly Colonies
2.3. Choice Tests
2.4. No-Choice Tests
2.5. Data Analysis
3. Results
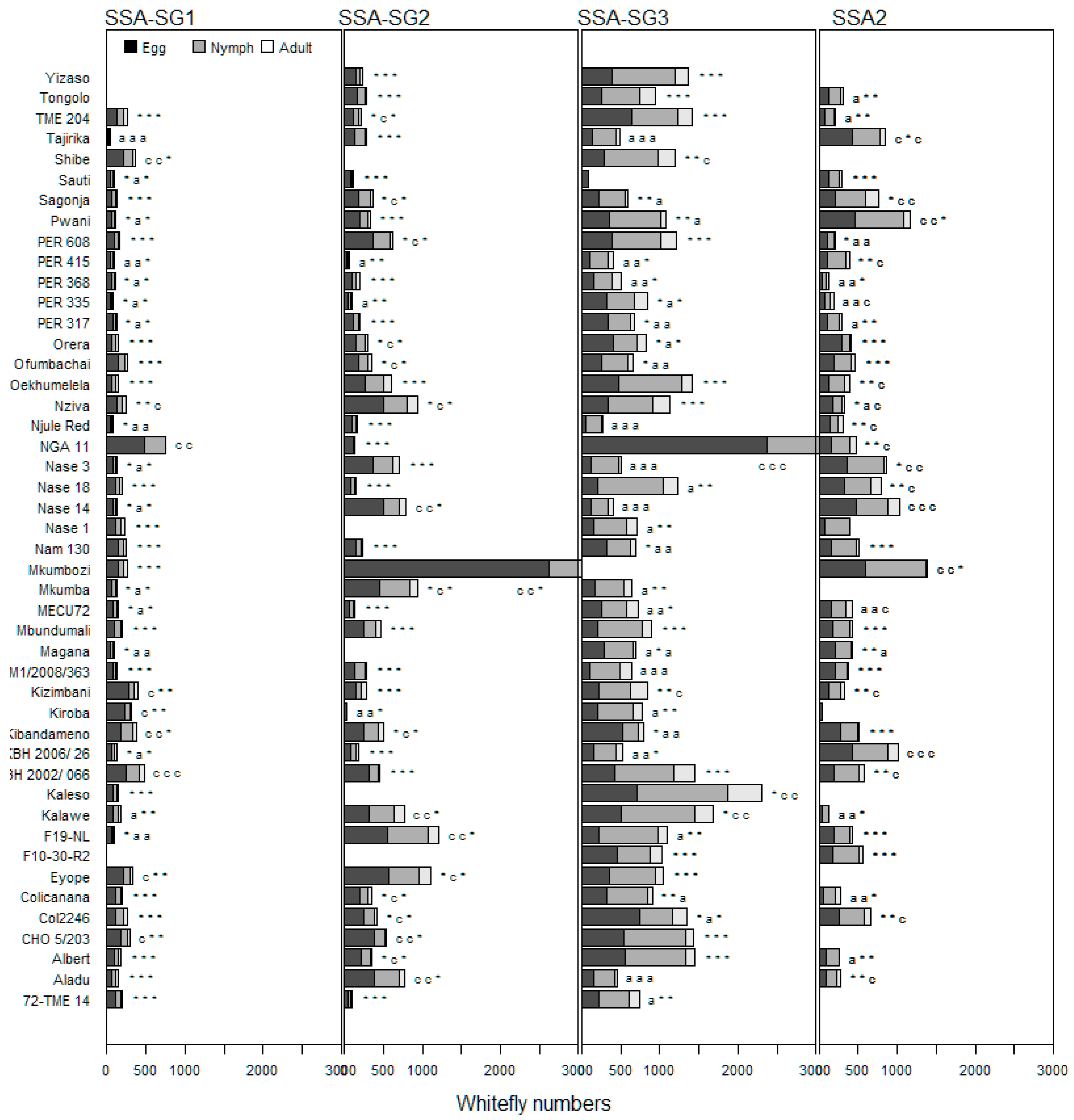
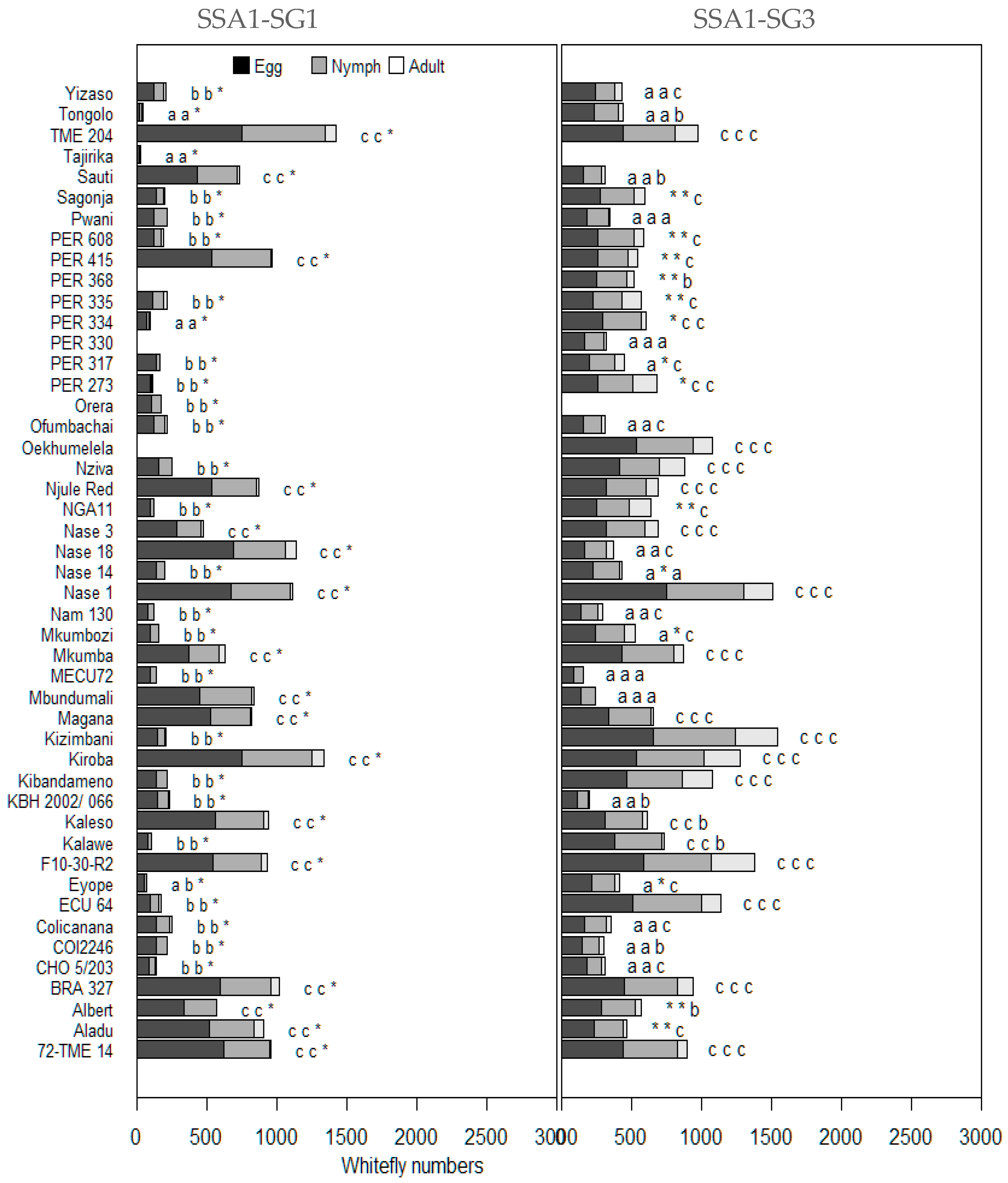
3.1. Performance of Cassava Genotypes in Choice Tests
3.2. Performance of Cassava Genotypes in No-Choice Tests
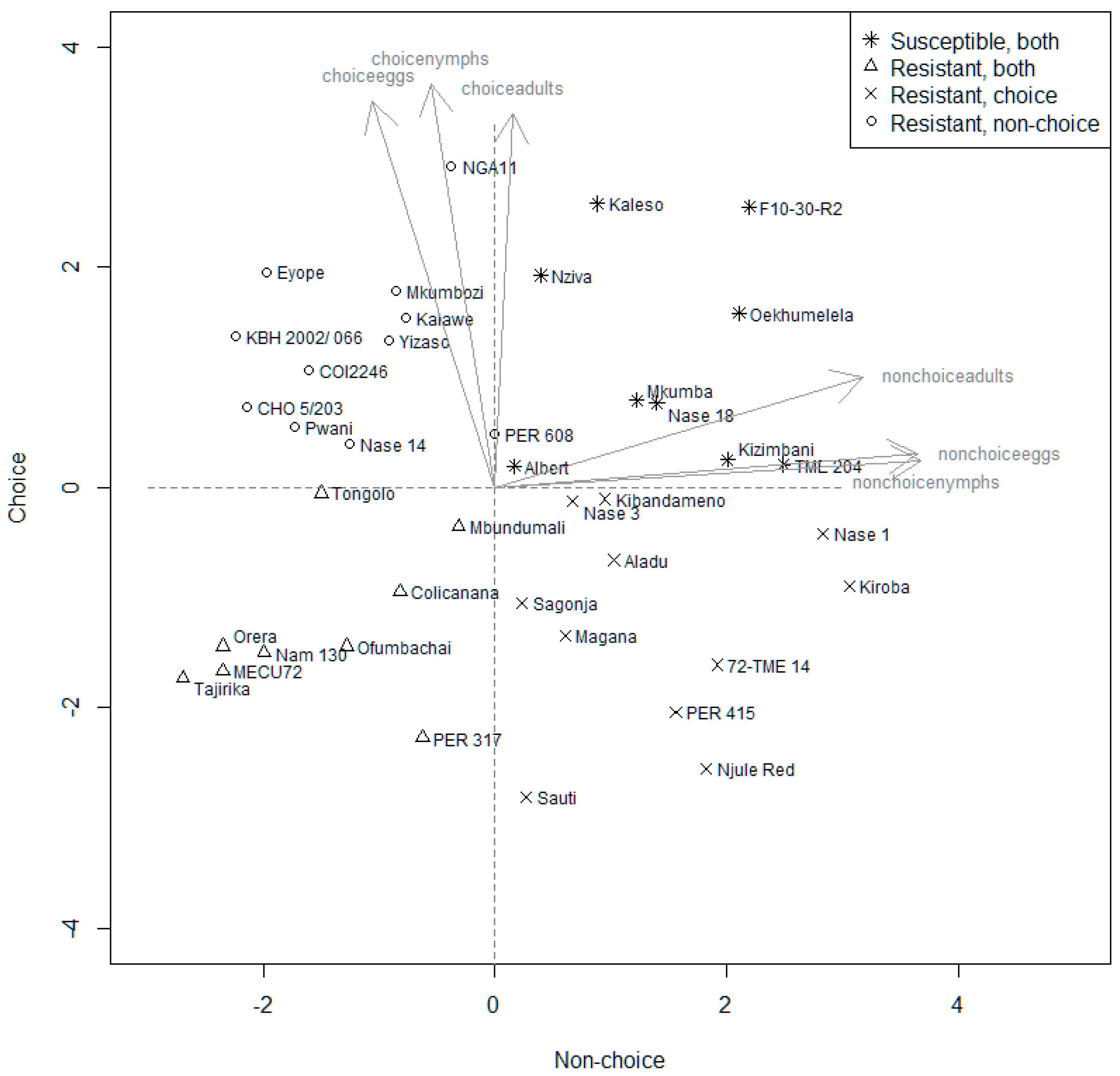
3.3. Principal Components Analysis—Genotypes Used in Both Choice and No-Choice Experiments
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliveira, M.R.; Henneberry, V.; Anderson, T.J. History, current status, and collaborative research projects for Bemisia tabaci. Crop Prot. 2001, 20, 709–723. [Google Scholar] [CrossRef]
- Lin, T.B.; Schwartz, A.; Saranga, Y. Photosynthesis and productivity of cotton under silverleaf whitefly stress. Crop Sci. 1999, 39, 174–184. [Google Scholar] [CrossRef]
- Omongo, C.A.; Opio, S.M.; Bayiyana, I.; Otim, M.H.; Omara, T.; Wamani, S.; Ocitti, P.; Bua, A.; Macfadyen, S.; Colvin, J. African cassava whitefly and viral disease management through timed application of imidacloprid. Crop Prot. 2022, 158, 106015. [Google Scholar] [CrossRef]
- Dubern, J. Transmission of African cassava mosaic geminivirus by the whitefly (Bemisia tabaci). Trop. Sci. 1994, 34, 82–91. [Google Scholar]
- Maruthi, M.N.; Hillocks, R.J.; Mtunda, K.; Raya, M.D.; Muhanna, M.; Kiozia, H.; Rekha, A.R.; Colvin, J.; Thresh, J.M. Transmission of Cassava brown streak virus by Bemisia tabaci (Gennadius). J. Phytopathol. 2005, 153, 307–312. [Google Scholar] [CrossRef]
- Jones, D.R. Plant viruses transmitted by whitefies. Eur. J. Plant Pathology 2003, 109, 195–219. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Ann. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef] [PubMed]
- Legg, J.P.; Sseruwagi, P.; Boniface, S.; Okao-Okuja, G.; Shirima, R.; Bigirimana, S.; Gashaka, G.; Herrmann, H.W.; Jeremiah, S.; Obiero, H.; et al. Spatio-temporal patterns of genetic change amongst populations of cassava Bemisia tabaci whiteflies driving virus pandemics in East and Central Africa. Virus Res. 2014, 186, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Robson, F.; Hird, D.L.; Boa, E. Cassava brown streak: A deadly virus on the move. Plant Pathol. 2024, 73, 221–241. [Google Scholar] [CrossRef]
- Legg, J.P.; French, R.; Rogan, D.; Okao-Okuja, G.; Brown, J.K. A distinct Bemisia tabaci (Gennadius) (Hemiptera: Sternorrhyncha: Aleyrodidae) genotype cluster is associated with the epidemic of severe cassava mosaic virus disease in Uganda. Mol. Ecol. 2002, 11, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Esterhuizen, L.L.; Mabasa, K.G.; Van Heerden, S.W.; Czosnek, H.; Brown, J.K.; Van Heerden, H.; Rey ME, C. Genetic identification of members of the Bemisia tabaci cryptic species complex from South Africa reveals native and introduced haplotypes. J. Appl. Entomol. 2013, 137, 122–135. [Google Scholar] [CrossRef]
- Mugerwa, H.; Rey, M.E.C.; Alicai, T.; Ateka, E.; Atuncha, H.; Ndunguru, J.; Sseruwagi, P. Genetic diversity and geographic distribution of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) genotypes associated with cassava in East Africa. Ecol. Evol. 2012, 2, 2749–2762. [Google Scholar] [CrossRef]
- De Barro, P.J.; Liu, S.-S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A statement of species status. Annu. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef]
- Dinsdale, A.; Cook, L.; Riginos, C.; Buckley, Y.M.; De Barro, P. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Am. 2010, 103, 196–208. [Google Scholar] [CrossRef]
- Ghosh, S.; Bouvaine, S.; Maruthi, M.N. Prevalence and genetic diversity of endosymbiotic bacteria infecting cassava whiteflies in Africa. BMC Microbiol. 2015, 15, 93. [Google Scholar] [CrossRef] [PubMed]
- Mugerwa, H.; Wang, H.L.; Sseruwagi, P.; Seal, S.; Colvin, J. Whole-genome single nucleotide polymorphism and mating compatibility studies reveal the presence of distinct species in sub-Saharan Africa Bemisia tabaci whiteflies. Insect Sci. 2021, 28, 1553–1566. [Google Scholar] [CrossRef] [PubMed]
- Omongo, C.A.; Kawuki, R.; Bellotti, A.C.; Alicai, T.; Baguma, Y.; Maruthi, M.N.; Bua, A.; Colvin, J. African Cassava Whitefly, Bemisia tabaci, Resistance in African and South American Cassava Genotypes. J. Integr. Agric. 2012, 11, 327–336. [Google Scholar] [CrossRef]
- Carabali, A.; Bellotti, A.C.; Montoya-Lerma, J.; Fregene, M. Resistance to the whitefly, Aleurotrachelus socialis, in wild populations of cassava, Manihot tristis. J. Insect Sci. 2010, 10, 170. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carabalí, A.; Montoya-Lerma, J.; Belloti, A.C.; Fregene, M.; Gallego, G. Resistance to the Whitefly Aleurotrachelus socialis (Hemiptera: Aleyrodidae) and SSR Marker Identification in Advanced Populations of the Hybrid Manihot esculenta subsp. Manihot flabellifolia. J. Integr. Agric. 2013, 12, 2217–2228. [Google Scholar]
- Bellotti, A.C.; Arias, B. Host plant resistance to whiteflies with emphasis on cassava as a case study. Crop Prot. 2001, 20, 813–823. [Google Scholar] [CrossRef]
- Parsa, S.; Medina, C.; Rodríguez, V. Sources of pest resistance in cassava. Crop Prot. 2015, 68, 79–84. [Google Scholar] [CrossRef]
- Painter, R.H. Insect Resistance Incrop Plants; The Macmillan company: New York, NY, USA, 1951; Soil Science; Volume 72, p. 481. [Google Scholar]
- Kogan, M.; Ortman, E.F. Antixenosis–a new term proposed to define Painter’s “nonpreference” modality of resistance. Bull. ESA 1978, 24, 175–176. [Google Scholar] [CrossRef]
- Smith, C.M. Plant Resistance to Arthropods: Molecular and Conventional Approaches; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Sulistyo, A.; Inayati, A. Mechanisms of antixenosis, antibiosis, and tolerance of fourteen soybean genotypes in response to whiteflies (Bemisia tabaci). Biodiversitas J. Biol. Divers. 2016, 17, 447–453. [Google Scholar] [CrossRef]
- Romanow, L.R.; Ponti, O.M.B.; Mollema, C. Resistance in tomato to the greenhouse whitefly: Analysis of population dynamics. Entomol. Exp. Appl. 1991, 60, 247–259. [Google Scholar] [CrossRef]
- Erb, W.A.; Lindquist, R.K.; Flickinger, N.J.; Casey, M.L. Resistance of selected interspecific lycopersicon hybrids to greenhouse whitefly (Homoptera: Aleurodidae). Fla. Entomol. 1994, 77, 104. [Google Scholar] [CrossRef]
- Baldin, E.L.L.; Beneduzzi, R.A. Characterization of antibiosis and antixenosis to the whitefly silverleaf Bemisia tabaci B biotype (Hemiptera: Aleyrodidae) in several squash varieties. J. Pest Sci. 2010, 83, 223–229. [Google Scholar] [CrossRef]
- Maruthi, M.N.; Whitfield, E.C.; Otti, G.; Tumwegamire, S.; Kanju, E.; Legg, J.P.; Mkamilo, G.; Kawuki, R.; Benesi, I.; Zacarias, A.; et al. A method for generating virus-free cassava plants to combat viral disease epidemics in Africa. Physiol. Mol. Plant Pathol. 2019, 105, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Perez-Fons, L.; Ovalle, T.M.; Maruthi, M.N.; Colvin, J.; Lopez-Lavalle, L.A.B.; Fraser, P.D. The metabotyping of an East African cassava diversity panel: A core collection for developing biotic stress tolerance in cassava. PLoS ONE 2020, 15, e0242245. [Google Scholar] [CrossRef] [PubMed]
- Mugerwa, H.; Seal, S.; Wang, H.L.; Patel, M.V.; Kabaalu, R.; Omongo, C.A.; Alicai, T.; Tairo, F.; Ndunguru, J.; Sseruwagi, P.; et al. African ancestry of New World, Bemisia tabaci-whitefly species. Sci. Rep. 2018, 8, 2734. [Google Scholar] [CrossRef] [PubMed]
- Byrne, D.N.; Bellows, T.S., Jr. Whitefly biology. Annu. Rev. Entomol. 1991, 36, 431–457. [Google Scholar] [CrossRef]
- de Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research R Package Version 1.3-5. 2021. Available online: https://CRAN.R-project.org/package=agricolae (accessed on 30 March 2022).
- Wamani, S.; Opio, S.M.; Omara, T.; Ocitti, P.; Colvin, J.; Omongo, C.A. Resistance to Cassava Whitefly (Bemisia tabaci) among Eastern and Southern African Elite Cassava Genotypes. Insects 2024, 15, 258. [Google Scholar]
- Omondi, A.B.; Obeng-Ofori, D.; Kyerematen, R.A.; Danquah, E.Y. Host preference and suitability of some selected crops for two biotypes of Bemisia tabaci in Ghana. Entomol. Exp. Appl. 2005, 115, 393–400. [Google Scholar] [CrossRef]
- Katono, K.; Macfadyen, S.; Omongo, C.A.; Odong, T.L.; Colvin, J.; Karungi, J.; Otim, M.H. Influence of cassava morphological traits and environmental conditions on field populations of Bemisia tabaci. Insects 2021, 12, 604. [Google Scholar] [CrossRef] [PubMed]
- Charles, A.L.; Sriroth, K.; Huang, T.C. Proximate composition, mineral contents, hydrogen cyanide and phytic acid of 5 cassava genotypes. Food Chem. 2005, 92, 615–620. [Google Scholar] [CrossRef]
- Manano, J.; Ogwok, P.; Byarugaba-Bazirake, G.W. Chemical composition of major cassava varieties in Uganda, targeted for industrialisation. J. Food Res. 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Mweine, P. Effect of Bemisia Tabasco (Whitefly) on Photosynthetic Capacity and Secondary Metabolite Composition of Selected Cassava Varieties Grown in Uganda. Ph.D. Dissertation, Makerere University, Kampala, Uganda, 2022. [Google Scholar]
- Mwila, N.; Nuwamanya, E.; Odong, T.L.; Badji, A.; Agbahoungba, S.; Ibanda, P.A.; Mwala, M.; Sohati, P.; Kyamanywa, S.; Rubaihayo, P.R. Genotype by environment interaction unravels influence on secondary metabolite quality in cassava infested by Bemisia tabaci. J. Agric. Sci. 2018, 10, 192–209. [Google Scholar] [CrossRef][Green Version]
- Martinez, M.; Diaz, I. Plant Cyanogenic-Derived Metabolites and Herbivore Counter-Defences. Plants 2024, 1, 1239. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, J.A.; Muola, A. How should plant resistance to herbivores be measured? Front. Plant Sci. 2017, 8, 663. [Google Scholar] [CrossRef]
- Mudereri, B.T.; Kimathi, E.; Chitata, T.; Moshobane, M.C.; Abdel-Rahman, E.M. Landscape-scale biogeographic distribution analysis of the whitefly, Bemisia tabaci (Gennadius, 1889) in Kenya. Int. J. Trop. Insect Sci. 2021, 41, 1585–1599. [Google Scholar] [CrossRef]
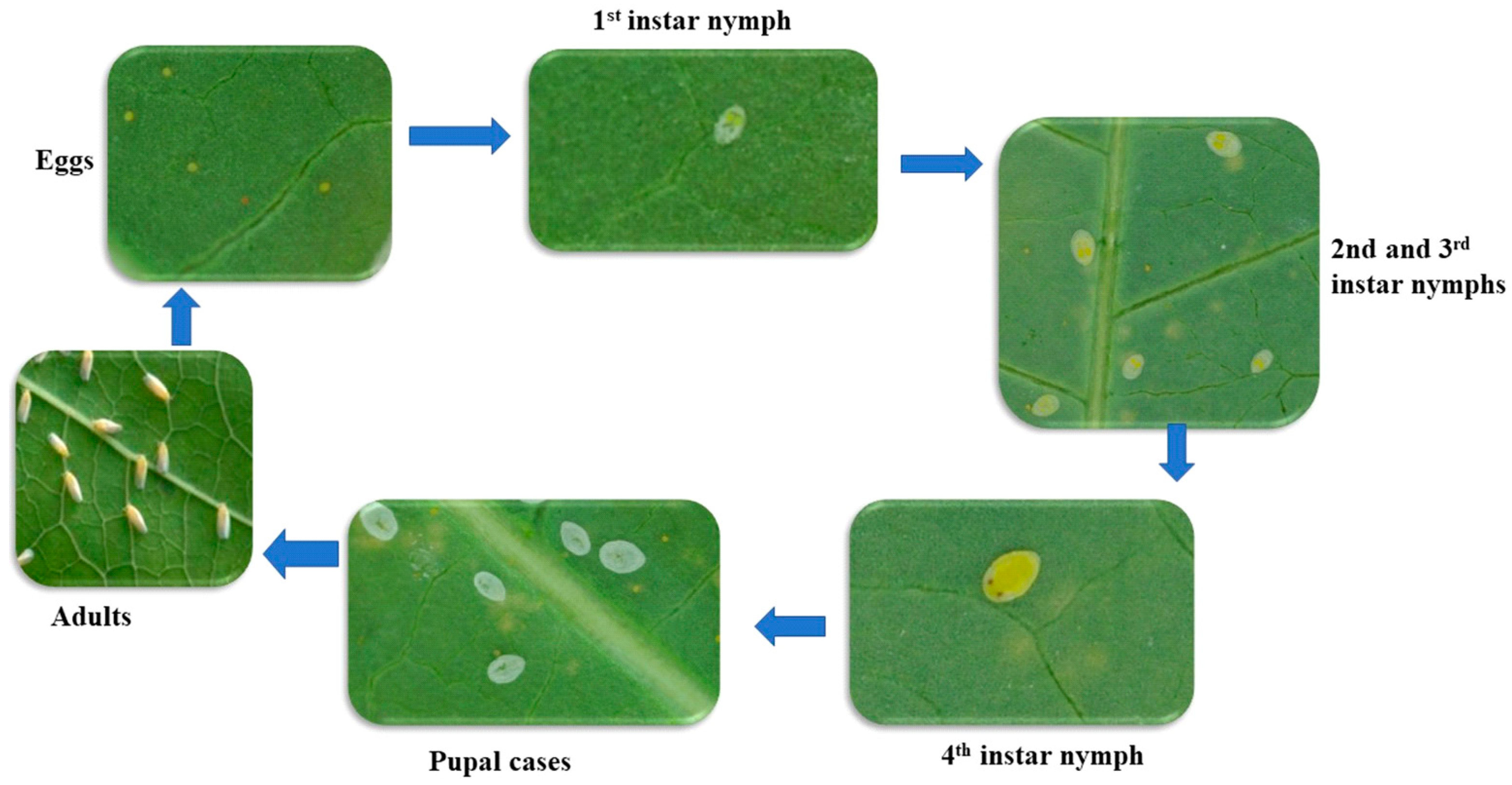
| S/No | Variety | Country of Origin | Characteristics * |
|---|---|---|---|
| 1 | BRA 327 | Brazil | Whitefly-susceptible (Aleurotracheus socialis) |
| 2 | ECU 64 | Ecuador | Whitefly-resistant (Aleurotracheus socialis) |
| 3 | Col2246 | Colombia | Whitefly-susceptible (Aleurotracheus socialis) |
| 4 | MECU72 (ECU72) | Ecuador | Whitefly-resistant (Aleurotracheus socialis) |
| 5 | PER 273 | Peru | Whitefly-resistant (Aleurotracheus socialis) |
| 6 | PER 330 | Peru | Whitefly-resistant (Aleurotracheus socialis) |
| 7 | PER 334 | Peru | Whitefly-resistant (Aleurotracheus socialis) |
| 8 | PER 317 | Peru | Whitefly-resistant (Aleurotracheus socialis) |
| 9 | PER 335 | Peru | Whitefly-resistant (Aleurotracheus socialis) |
| 10 | PER 368 | Peru | Whitefly-resistant (Aleurotracheus socialis) |
| 11 | PER 415 | Peru | Whitefly-resistant (Aleurotracheus socialis) |
| 12 | PER 608 | Peru | Whitefly-resistant (Aleurotracheus socialis) |
| 13 | Aladu (Alado alado) | Uganda (Landraces) | Whitefly-resistant (Bemisia tabaci) |
| 14 | Magana | Uganda (Landrace) | Whitefly-resistant (Bemisia tabaci) |
| 15 | Nam 130 (NaroCass1/UG3/Tz30) | Uganda (cultivar) | Whitefly-resistant (Bemisia tabaci) |
| 16 | Njule Red | Uganda (cultivar) | Whitefly-resistant (Bemisia tabaci) |
| 17 | Ofumbachai | Uganda (Landrace) | Whitefly-resistant (Bemisia tabaci) |
| 18 | Tongolo | Uganda (Landrace) | Whitefly-resistant (Bemisia tabaci) |
| 19 | LM1/2008/363 | Kenya | Moderate resistance to CMD and CBSD |
| 20 | F19-NL | Kenya | Moderate resistance to CMD and CBSD |
| 21 | Tajirika | Kenya | Moderate resistance to CMD and CBSD |
| 22 | Shibe | Kenya | Moderate resistance to CMD and CBSD |
| 23 | F10-30-R2 | Kenya | Moderate resistance to CMD and CBSD |
| 24 | Kibandameno | Kenya | Susceptible to CMD and CBSD |
| 25 | Mkumbozi | Tanzania | Moderate resistance to CMD and CBSD |
| 26 | Kaleso | Kenya | Resistant to CBSD and susceptible to CMD |
| 27 | Yizaso | Malawi | Moderate resistance to CMD and CBSD |
| 28 | Mbundumali | Malawi | Susceptible to CMD and CBSD |
| 29 | Sauti | Malawi | Moderate resistance to CMD and CBSD |
| 30 | CHO 5/203 | Malawi | Moderate resistance to CMD and CBSD |
| 31 | Sagonja | Malawi | Moderate resistance to CMD and CBSD |
| 32 | Kalawe | Malawi | Moderate resistance to CMD and CBSD |
| 33 | Oekhumelela | Mozambique | Moderate resistance to CMD and CBSD |
| 34 | Eyope | Mozambique | Moderate resistance to CMD and CBSD |
| 35 | Nziva | Mozambique | Susceptible to CMD, moderate to CBSD |
| 36 | Colicanana | Mozambique | Susceptible to CMD, moderate to CBSD |
| 37 | Orera | Mozambique | Susceptible to CMD, moderate to CBSD |
| 38 | KBH 2002/066 | Tanzania | Moderate resistance to CMD and CBSD |
| 39 | Pwani | Tanzania | Moderate resistance to CMD, strong to CBSD |
| 40 | Mkumba | Tanzania | Moderate resistance to CMD, strong to CBSD |
| 41 | Kizimbani | Tanzania | Moderate resistance to CMD and CBSD |
| 42 | KBH 2006/26 | Tanzania | Moderate resistance to CMD and CBSD |
| 43 | Albert | Tanzania | Susceptible to CMD and CBSD |
| 44 | Kiroba | Tanzania | Moderate resistance to CMD and CBSD |
| 45 | Nase 3 (Migyera) | Uganda | Moderate resistance to CMD and CBSD |
| 46 | TME 204 | Uganda | Strong resistance to CMD, susceptible CBSD |
| 47 | Tz 130 (NaroCass1) | Tanzania | Strong resistance to CMD, moderate CBSD |
| 48 | Nase 18 | Uganda | Strong resistance to CMD, moderate CBSD |
| 49 | 72-TME 14 | Uganda | Strong resistance to CMD, moderate CBSD |
| 50 | Nase 14 | Uganda | Strong resistance to CMD, moderate CBSD |
| 51 | Nase 1 | Uganda | Strong resistance to CMD, moderate CBSD |
| Interaction | Degree of Freedom | Deviance Chi-Square Units | p-Value |
|---|---|---|---|
| genotype | 45 | 560.61 | <0.00001 |
| Life stage | 2 | 783.07 | <0.00001 |
| Populations | 3 | 928.08 | <0.00001 |
| Genotype × life stage | 90 | 103.77 | 0.15213 |
| Genotype × populations | 120 | 617.86 | <0.00001 |
| Life stage × populations | 6 | 116.03 | <0.00001 |
| Genotype × life stage × populations | 233 | 172.37 | 0.99892 |
| Residual | 1797 | 2568.8 |
| Interaction | Degree of Freedom | Deviance Chi-Square Units | p-Value |
|---|---|---|---|
| Origin | 1 | 37.07 | <0.00001 |
| Life stage | 2 | 544.43 | <0.00001 |
| Populations | 3 | 670.8 | <0.00001 |
| Origin × life stage | 2 | 4.03 | 0.13332 |
| Origin × populations | 3 | 10.43 | 0.01524 |
| life stage × populations | 6 | 75.27 | <0.00001 |
| Origin × life stage × populations | 6 | 6.27 | 0.39363 |
| Residual | 2273 | 2674.11 |
| Genotype | Mean ± Standard Error | |
|---|---|---|
| NGA11 (Susceptible control) | 613.5 ± 120.6 | d |
| Mkumbozi | 531.4 ± 100.7 | cd |
| Kaleso | 319.2 ± 81.4 | cd |
| Eyope | 271.3 ± 52.1 | bcd |
| F10-30-R2 | 253.2 ± 56.7 | bcd |
| CHO 5/203 | 251.2 ± 46.6 | bcd |
| KBH 2002/066 | 243 ± 41.1 | bcd |
| F19-NL | 236.5 ± 44.4 | abcd |
| Yizaso | 232.3 ± 56.9 | abcd |
| Nziva | 231.5 ± 39.1 | abcd |
| Kalawe | 229.2 ± 36.7 | abcd |
| Col2246 (Susceptible control) | 227.3 ± 29.9 | abcd |
| Pwani | 225 ± 37.7 | abcd |
| Oekhumelela | 222.9 ± 37.7 | abcd |
| Shibe | 208.2 ± 49.1 | abcd |
| Albert | 202.3 ± 34.9 | abcd |
| Nase 3 | 196.3 ± 32.9 | abcd |
| Mkumba | 191.7 ± 35.9 | abcd |
| Nase 14 | 190.2 ± 32.2 | abcd |
| PER 608 | 188.9 ± 32.6 | abcd |
| Nase 18 | 186 ± 31.5 | abcd |
| Kibandameno | 176.4 ± 27.4 | abcd |
| Tongolo | 174 ± 33.4 | abcd |
| Mbundumali | 166.8 ± 27 | abcd |
| TME 204 | 162.3 ± 27.2 | abcd |
| KBH 2006/26 | 157.3 ± 25.7 | abcd |
| Aladu | 154.8 ± 25.7 | abcd |
| Kizimbani | 152.4 ± 30 | abcd |
| Colicanana | 144.9 ± 23.5 | abcd |
| Magana | 143.6 ± 29.4 | abcd |
| Nam 130 | 143.2 ± 23.4 | abcd |
| Tajirika | 143.1 ± 23.4 | abcd |
| Sagonja | 141.1 ± 25.1 | abc |
| Ofumbachai | 140.5 ± 24 | abc |
| Nase 1 | 133.4 ± 32.7 | abc |
| Orera | 132.6 ± 23.8 | abc |
| Kiroba | 131.3 ± 24.9 | abc |
| LM1/2008/363 | 120.1 ± 20.1 | abc |
| MECU72 (Resistant control) | 119.1 ± 15.2 | abc |
| 72-TME 14 | 109.2 ± 24.5 | abc |
| PER 317 | 108.4 ± 18 | abc |
| PER 335 | 99.4 ± 14.7 | abc |
| PER 415 | 91.1 ± 16.2 | ab |
| PER 368 | 76.1 ± 14.1 | ab |
| Njule Red | 61.4 ± 11.7 | a |
| Sauti | 55.3 ± 10.1 | a |
| Interaction | Degree of Freedom | Deviance Chi-Square Units | p-Value |
|---|---|---|---|
| Genotype | 46 | 983.02 | <0.00001 |
| Species | 1 | 167.48 | <0.00001 |
| Life stage | 2 | 1534.24 | <0.00001 |
| Genotype × populations | 41 | 579.21 | <0.00001 |
| Genotype × life stage | 92 | 272.42 | <0.00001 |
| Populations × life stage | 2 | 213.65 | <0.00001 |
| Genotype × populations × life stage | 82 | 207.46 | <0.00001 |
| Residual | 520 | 916.89 |
| Interaction | Degree of Freedom | Deviance Chi-Square Units | p-Value |
|---|---|---|---|
| origin | 1 | 8.62 | 0.00332 |
| Species | 1 | 24.14 | <0.00001 |
| Life stage | 2 | 372.08 | <0.00001 |
| Origin × populations | 1 | 2.84 | 0.09194 |
| Origin × life stage | 2 | 0.31 | 0.85642 |
| Species × life stage | 2 | 69.55 | <0.00001 |
| Origin × populations × life stage | 2 | 0.14 | 0.93239 |
| Residual | 775 | 940.5 |
| Genotype | Mean ± Standard Error | CLD Code |
|---|---|---|
| 72-TME 14 | 307.7 ± 86.1 | h…p |
| Aladu | 227.9 ± 63.8 | k…p |
| Albert | 188.9 ± 52.9 | c…n |
| BRA 327 | 325.3 ± 112.2 | mnop |
| CHO 5/203 | 74.4 ± 20.9 | b…h |
| Col2246(Susceptible control) | 86.2 ± 24.2 | abc |
| Colicanana | 100.1 ± 28.1 | f…p |
| ECU 64 | 218.7 ± 61.2 | h….p |
| Eyope | 80.9 ± 22.7 | bcde |
| F10-30-R2 | 404.2 ± 116.4 | op |
| Kalawe | 138.8 ± 38.9 | bcdef |
| Kaleso | 258.1 ± 72.2 | i…p |
| KBH 2002/066 | 70.6 ± 19.8 | c…n |
| Kibandameno | 215.2 ± 60.2 | c…n |
| Kiroba | 435.4 ± 121.8 | p |
| Kizimbani | 292 ± 81.7 | g…p |
| Magana | 245.3 ± 68.7 | g…p |
| Mbundumali | 178.4 ± 50 | b…k |
| MECU72(Resistant control) | 47.6 ± 13.4 | ab |
| Mkumba | 249.9 ± 69.9 | lmnop |
| Mkumbozi | 112.4 ± 31.5 | b…j |
| Nam 130 | 68.6 ± 19.3 | bcdef |
| Nase 1 | 436.1 ± 122 | Mnop |
| Nase 14 | 103.9 ± 29.1 | abc |
| Nase 18 | 250.6 ± 70.1 | lmnop |
| Nase 3 | 194.1 ± 54.3 | j…p |
| NGA11(Susceptible control) | 125.6 ± 35.2 | c…n |
| Njule Red | 260.3 ± 72.8 | i…p |
| Nziva | 187.6 ± 52.5 | d…o |
| Oekhumelela | 358.6 ± 143.6 | nop |
| Ofumbachai | 91.5 ± 26.4 | e…p |
| Orera | 57.9 ± 23.3 | b…i |
| PER 273 | 130.2 ± 36.5 | c…m |
| PER 317 | 100.6 ± 28.2 | b…i |
| PER 330 | 105.7 ± 52.6 | b…h |
| PER 334 | 115.5 ± 32.4 | bcdefg |
| PER 335 | 129.7 ± 36.3 | g…p |
| PER 368 | 173.1 ± 69.4 | c…o |
| PER 415 | 250 ± 70 | j…p |
| PER 608 | 141.8 ± 43.6 | c…n |
| Pwani | 93.2 ± 26.2 | abc |
| Sagonja | 132 ± 37 | f…p |
| Sauti | 172.8 ± 48.4 | c…n |
| Tajirika | 8.4 ± 3.5 | a |
| TME 204 | 399.1 ± 111.6 | nop |
| Tongolo | 79.1 ± 22.2 | bcd |
| Yizaso | 106.2 ± 29.8 | c…l |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atim, J.; Kalyebi, A.; Bohorquez-Chaux, A.; Lopez-Lavalle, L.A.B.; Omongo, C.A.; Colvin, J.; Maruthi, M.N. Identifying Cassava Genotypes Resistant to the African Cassava Whitefly, Bemisia tabaci (Gennadius). Agriculture 2024, 14, 1016. https://doi.org/10.3390/agriculture14071016
Atim J, Kalyebi A, Bohorquez-Chaux A, Lopez-Lavalle LAB, Omongo CA, Colvin J, Maruthi MN. Identifying Cassava Genotypes Resistant to the African Cassava Whitefly, Bemisia tabaci (Gennadius). Agriculture. 2024; 14(7):1016. https://doi.org/10.3390/agriculture14071016
Chicago/Turabian StyleAtim, Jackie, Andrew Kalyebi, Adriana Bohorquez-Chaux, Luis Augusto Becerra Lopez-Lavalle, Christopher Abu Omongo, John Colvin, and M. N. Maruthi. 2024. "Identifying Cassava Genotypes Resistant to the African Cassava Whitefly, Bemisia tabaci (Gennadius)" Agriculture 14, no. 7: 1016. https://doi.org/10.3390/agriculture14071016
APA StyleAtim, J., Kalyebi, A., Bohorquez-Chaux, A., Lopez-Lavalle, L. A. B., Omongo, C. A., Colvin, J., & Maruthi, M. N. (2024). Identifying Cassava Genotypes Resistant to the African Cassava Whitefly, Bemisia tabaci (Gennadius). Agriculture, 14(7), 1016. https://doi.org/10.3390/agriculture14071016






