Effect of Introgression of Ty-1 and ty-5 Genes on Productivity, Quality, and Antioxidant Compounds in De la Pera Tomato Breeding Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.1.1. Genealogy
De la Pera UMH1203
De la Pera UMH1406
De la Pera UMH175 and UMH220
2.2. Field Test
2.3. Parameters Assessed
2.3.1. Yield and Average Fruit Weight
2.3.2. Total Soluble Solids and Titratable Acidity
2.3.3. Total Antioxidant Activity and Total Phenolic Compounds
2.4. Statistical Analysis
3. Results
3.1. Yield and Average Fruit Weight
3.2. Total Soluble Solids and Titratable Acidity
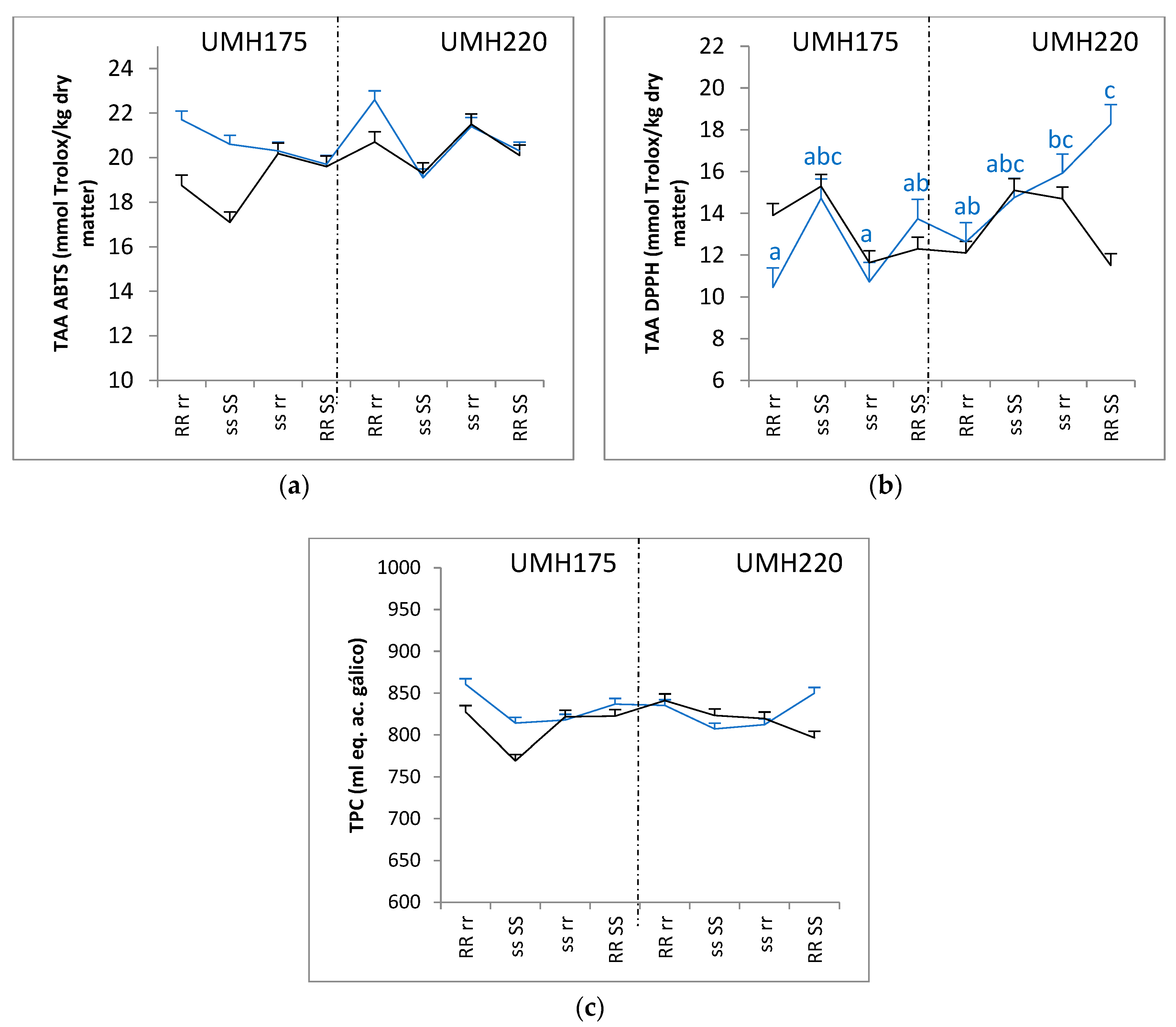
3.3. Total Antioxidant Activity and Total Phenolic Compounds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shahriari, Z.; Su, X.; Zheng, K.; Zhang, Z. Advances and Prospects of Virus-Resistant Breeding in Tomatoes. Int. J. Mol. Sci. 2023, 24, 15448. [Google Scholar] [CrossRef]
- Anbinder, I.; Reuveni, M.; Azari, R.; Paran, I.; Nahon, S.; Shlomo, H.; Chen, L.; Lapidot, M.; Levin, I. Molecular dissection of Tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 2009, 119, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Vasudeva, R.S.; Sam Raj, J. A leaf curl disease of tomato. Phytopatology 1948, 38, 364–369. [Google Scholar]
- Cohen, S.; Harpaz, I. Periodic, rather than continual adquisition of new tomato virus by its vector, the tobacco whitefly (Bemisia tabaci Gennadius). Entomol. Exp. Appl. 1964, 7, 155–166. [Google Scholar] [CrossRef]
- Picó, B.; Díez, M.J.; Nuez, F. Viral diseases causing the greatest economic losses to the tomato crop. II. The Tomato yellow leaf curl virus—A review. Sci. Hortic. 1996, 67, 151–196. [Google Scholar] [CrossRef]
- Moriones, E.; Arnó, J.; Accotto, G.P.; Noris, E.; Cavallarin, L. First report of Tomato yellow leaf curl virus in Spain. Plant Dis. 1993, 77, 953. [Google Scholar] [CrossRef]
- Reina, J.; Jiménez, J.; Bejarano, E.R.; Guerra, J.M.; Cuadrado, I.M.; García, C. El virus del rizado amarillo del tomate (TYLCV). Hortofruticultura 1994, 6, 36–40. [Google Scholar]
- Navas-Castillo, J.; Sanchez-Campos, S.; Díaz, J.A.; Sáez-Alonso, E.; Moriones, E. First report of Tomato yellow leaf curl virus-Is in Spain: Coexistence of two different geminiviruses in the same epidemic outbreak. Plant Dis. 1997, 81, 1461. [Google Scholar] [CrossRef]
- Pilowsky, M.; Cohen, S. Tolerance to Tomato yellow leaf curl virus derived from L. peruvianum. Plant Dis. 1990, 74, 248–250. [Google Scholar] [CrossRef]
- Friedmann, M.; Lapidot, M.; Cohen, S.; Pilowsky, M. A novel source of resistance to Tomato yellow leaf curl virus exhibiting a symptomless reaction to viral infection. J. Am. Soc. Hortic. Sci. 1998, 123, 1004–1007. [Google Scholar] [CrossRef]
- Kalloo, G.; Banerjee, M.K. Transfer of Tomato leaf curl virus resistance from Lycopersicon hirsutum f. glabratum to L. esculentum. Plant Breed. 1990, 105, 156–159. [Google Scholar] [CrossRef]
- Kasrawi, M.A.; Suwwan, M.A.; Mansour, A. Sources of resistance to Tomato-yellow-leaf-curl-virus (TYLCV) in Lycopersicon species. Euphytica 1988, 37, 61–64. [Google Scholar] [CrossRef]
- Laterrot, H. Resistance genitors to Tomato yellow leaf curl virus (TYLCV). Tomato Leaf Curl. Nswl. 1992, 1, 2–4. [Google Scholar]
- Scott, J.W.; Stevens, M.R.; Barten, J.H.M.; Thome, C.R.; Polston, J.E.; Schuster, D.J.; Serra, C.A. Introgression of resistance to whitefly-transmitted geminiviruses from Lycopersicon chilense to tomato. In Bemisia: 1995 Taxonomy, Biology, Damate, Control and Management; Gerling, D., Mayer, R.T., Eds.; Intercept Ltd.: Andover, UK, 1995; pp. 357–367. [Google Scholar]
- Vidavsky, F.; Czosnek, H. Tomato breeding lines resistant and tolerant to tomato yellow leaf curl virus issued from Lycopersicon hirsutum. Phytopathology 1998, 88, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Zamir, D.; Michelson, I.E.; Zakay, Y.; Navot, N.; Zeidan, M.; Sarfatti, M.; Eshed, Y.; Harel, E.; Pleban, T.; van Oss, H.; et al. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, Ty-1. Theor. Appl. Genet. 1994, 88, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Hanson, P.M.; Bernacchi, D.; Green, S.; Tanksley, S.D.; Muniyappa, V.; Padmaja, A.S.; Chen, H.; Kuo, G.; Fang, D.; Chen, J. Mapping a Wild Tomato Introgression Asociated with Tomato Yellow Leaf Curl Virus Resistance in a Cultivated Tomato Line. J. Am. Soc. Hortic. Sci. 2000, 125, 15–20. [Google Scholar] [CrossRef]
- Dueñas, F.; Martínez, Y.; Álvarez, M.; Moya, C.; Peteira, B.; Arias, Y.; Diez, M.J.; Hanson, P.; Shagarodsky, T. Caracterización agromorfológica y evaluación de la resistencia al TYLCV en nuevos genotipos de tomate (Solanum lycopersicum L.) como apoyo al programa de mejoramiento genético de la hortaliza para la enfermedad. Cultiv. Trop. 2008, 29, 53–60. [Google Scholar]
- Hanson, P.M.; Green, S.K.; Kuo, G. Ty-2, a gene on chromosome 11 conditioning geminivirus resistance in tomato. In Report of the Tomato Genetics Cooperative; Scott, J.W., Ed.; University of Florida: Wimauma, FL, USA, 2006; Volume 56, pp. 17–18. [Google Scholar]
- Ji, Y.; Scott, J.W.; Hanson, P.; Graham, E.; Maxwell, D.P. Sources of resistance, inheritance, and location of genetic loci conferring resistance to members of the tomatoinfecting begomoviruses. In Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Henryk, C., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 343–362. [Google Scholar]
- Ji, Y.; Schuster, D.J.; Scott, J.W. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol. Breed. 2007, 20, 271–284. [Google Scholar] [CrossRef]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.; Scott, J.W.; Edwards, J.D.; Bai, Y. The Tomato Yellow Leaf Curl Virus Resistance Genes Ty-1 and Ty-3 are Allelic and Code for DFDGD-Class RNA-Dependent RNA Polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef]
- Ji, Y.; Scott, J.W.; Schuster, D.J. Toward Fine Mapping of the Tomato Yellow Leaf Curl Virus Resistance Gene Ty-2 on Chromosome 11 of Tomato. HortScience 2009, 44, 614–618. [Google Scholar] [CrossRef]
- Hutton, S.F.; Scott, J.W.; Schuster, D.J. Recessive Resistance to Tomato yellow leaf curl virus from the Tomato Cultivar Tyking Is Located in the Same Region as Ty-5 on Chromosome 4. HortScience 2012, 47, 324–327. [Google Scholar] [CrossRef]
- Lapidot, M.; Karniel, U.; Gelbart, D.; Fogel, D.; Evenor, D.; Kutsher, Y.; Makhbash, Z.; Nahon, S.; Shlomo, H.; Chen, L.; et al. A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota. PLoS Genet. 2015, 11, e1005538. [Google Scholar] [CrossRef] [PubMed]
- Hutton, S.F.; Scott, J.W. Ty-6, a major begomovirus resistance gene located on chromosome 10. In Report of the Tomato Genetics Cooperative; Scott, J.W., Ed.; University of Florida: Wimauma, FL, USA, 2014; Volume 64, pp. 14–18. [Google Scholar]
- Scott, J.W.; Hutton, S.F.; Freeman, J.H. Fla. 8638B and Fla. 8624 Tomato Breeding Lines with Begomovirus Resistance Genes ty-5 Plus Ty-6 and Ty-6, respectively. HortScience 2015, 50, 1405–1407. [Google Scholar] [CrossRef]
- Prasanna, H.C.; Sinha, D.P.; Rai, G.K.; Krishna, R.; Kashyap, S.P.; Singh, N.K.; Singh, M.; Malathi, V.G. Pyramiding Ty-2 and Ty-3 genes for resistance to monopartite and bipartite tomato leaf curl viruses of India. Plant Pathol. 2014, 64, 256–264. [Google Scholar] [CrossRef]
- Elbaz, M.; Hanson, P.; Fgaier, S.; Laarif, A. Evaluation of tomato entries with different combinations of resistance genes to tomato yellow leaf curl disease in Tunisia. Plant Breed. 2016, 135, 525–530. [Google Scholar] [CrossRef]
- Al-Shihi, A.A.; Peter, H.; Al-Sadi, A.M.; Al-Yahyai, R.A.; Briddon, R.W.; Deadman, M.; Shahid, M.S. Evaluation of tomato inbred lines for resistance to the tomato yellow leaf curl disease complex in Oman. Crop Prot. 2018, 110, 91–98. [Google Scholar] [CrossRef]
- Rubio, F.; Alonso, A.; García-Martínez, S.; Ruiz, J.J. Introgresion of virus-resistance genes into traditional tomato varieties (Solanum lycopersicum L.): Effects on yield and quality. Sci. Hortic. 2016, 198, 183–190. [Google Scholar] [CrossRef]
- Verlaan, M.G.; Szinay, D.; Hutton, S.F.; de Jong, H.; Visser, R.G.F.; Scott, J.W.; Bai, Y. Chromosomal rearrangements between tomato and Solanum chilense hamper mapping and breeding of the TYLCV resistance gene Ty-1. Plant J. 2011, 68, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.A.; Carbonell, P.; Salinas, J.F.; Alonso, A.; Grau, A.; Ruiz, J.J.; García-Martínez, S. Diseño de marcadores moleculares ligados al gen Ty-1 para la búsqueda de individuos recombinantes que hayan perdido parte de la carga de ligamiento asociada al gen Ty-1. In Libro de Resúmenes del III Congreso Universitario en Innovación y Sostenibilidad Agroalimentaria; LIMENCOP: Orihuela, Spain, 2022. [Google Scholar]
- García-Martínez, S.; Grau, A.; Alonso, A.; Rubio, F.; Valero, M.; Ruiz, J. UMH 1203, a Multiple Virus-resistant Fresh-market Tomato Breeding Line for Open-field Conditions. HortScience 2012, 47, 124–125. [Google Scholar] [CrossRef]
- Perez de Castro, A.; Blanca, J.M.; Díez, M.J.; Nuez Viñals, F. Identification of a CAPS marker tightly linked to the Tomato yellow leaf curl disease resistance gene Ty-1 in tomato. Eur. J. Plant Pathol. 2007, 117, 347–356. [Google Scholar] [CrossRef]
- García-Martínez, S.; Sánchez, C.; Castelló, J.; Grau, A.; Valero, M.; Ferrández, A.; Ruiz, J.J. Empleo de marcadores moleculares para la introducción múltiple de genes de resistencia a virosis (ToMV, TSWV y TYLCV) en variedades tradicionales de tomate alicantinas. Agrícola Vergel. 2003, 255, 140–143. [Google Scholar]
- Robles-Sánchez, R.M.; Rojas-Graü, M.A.; Odriozola-Serrano, I.; González-Aguilar, G.A.; Martín-Belloso, O. Effect of minimal processing on bioactive compounds and antioxidant activity of fresh-cut ‘Kent’mango (Mangifera indica L.). Postharvest Biol. Technol. 2009, 51, 384–390. [Google Scholar] [CrossRef]
- Tabart, J.; Kevers, C.; Pincemail, J.; Defraigne, J.O.; Dommes, J. Comparative antioxidant capacities of phenolic compounds measured by various tests. Food Chem. 2009, 113, 1226–1233. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventos, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Bernachi, D.; Emmatty, D.; Eshed, Y.; Inai, S.; Lopez, J.; Petiard, V.; Sayama, H.; Uhlig, J.; Zamir, D. Yield and quality evaluationson a pair of processing tomato lines nearly isogenic for the Tm2a gene forresistance to the tobacco mosaic virus. Euphytica 1998, 99, 77–83. [Google Scholar] [CrossRef]
- Lewis, R.S.; Linger, L.R.; Wolff, M.F.; Wernsman, E.A. The negative influence of N-mediated TMV resistance on yield in tobacco: Linkage drag versus pleiotropy. Theor. Appl. Genet. 2007, 115, 169–178. [Google Scholar] [CrossRef]
- Brouwer, D.J.; St Clair, D.A. Fine mapping of three quantitative trait loci for late blight resistance in tomato using near isogenic lines (NILs) and sub-NILs. Theor. Appl. Genet. 2004, 108, 628–638. [Google Scholar] [CrossRef]
- Cabrera, J.A.; Carbonell, P.; Salinas, J.F.; Alonso, A.; Grau, A.; García-Martínez, S.; Pérez de Castro, A.; Díez, M.J.; Ruiz, J.J. Estudio preliminar de la resistencia al virus del rizado amarillo del tomate en líneas de mejora de tomate De la pera. In Libro de Resúmenes del IX Congreso Ibérico y XVII Congreso Nacional de Ciencias Hortícolas; SECH: Mérida, Spain, 2023. [Google Scholar]
- Lipan, L.; Issa-Issa, H.; Moriana, A.; Zurita, N.M.; Galindo, A.; Martín-Palomo, M.J.; Andreu, L.; Carbonell-Barrachina, Á.A.; Hernández, F.; Corell, M. Scheduling Regulated Deficit Irrigation with Leaf Water Potential of Cherry Tomato in Greenhouse and its Effect on Fruit Quality. Agriculture 2021, 11, 669. [Google Scholar] [CrossRef]


| Gen | Accession | Species | Chromosome | Reference |
|---|---|---|---|---|
| Ty-1 | LA1969 | S. chilense | 6 | [16,22] |
| Ty-2 | B6013 | S. habrochaites | 11 | [11,17,19] |
| Ty-3 | LA1932, LA2779 | S. chilense | 6 | [20,21,22] |
| Ty-4 | LA1932 | S. chilense | 3 | [23] |
| ty-5 | TY172 | S. peruvianum | 4 | [2,10,24,25] |
| Ty-6 | LA1938 | S. chilense | 10 | [26] |
| Dates | 2022 | 2023 |
|---|---|---|
| Sowing | 11 February | 7 February |
| Planting | 29 April * | 4 April |
| Beginning of harvest | 13 July | 10 July |
| End of harvest | 03 August | 26 July |
| Phase | Fertilizer units | Overall Fertilization |
| 1 Vegetative development | 1 N–2 P2O5–1 K2O–1 CaO | 375 N–225 P2O5–550 K2O–190 CaO |
| 2 Flowering and fruit development | 1 N–1 P2O5–1 K2O–1 CaO | |
| 3 Ripening of the fruit | 1 N–0.3 P2O5–2 K2O–1 CaO |
| Year | Yield | Fruit Weight | TSS | TA | TAA | TPCs | ||
|---|---|---|---|---|---|---|---|---|
| ABTS | DPPH | |||||||
| GML Test | ||||||||
| 2022 | p-value | *** | *** | *** | *** | ns | * | ns |
| 2023 | p-value | *** | *** | * | *** | ns | ns | ns |
| Fisher’s Multiple Range Test | ||||||||
| 2022 | Genotype | |||||||
| 175 RR rr | 0.80 a | 37.69 abc | 6.05 d | 0.37 b | 21.70 | 10.46 a | 860.4 | |
| 175 ss SS | 2.05 c | 45.06 de | 5.92 cd | 0.43 d | 20.60 | 14.72 abc | 814.1 | |
| 175 ss rr | 1.36 b | 33.50 a | 5.73 bc | 0.45 e | 20.30 | 10.71 a | 817.8 | |
| 175 RR SS | 1.20 ab | 36.50 ab | 5.74 bc | 0.32 a | 19.70 | 13.73 ab | 836.8 | |
| 220 RR rr | 1.48 b | 39.81 bcd | 5.59 ab | 0.30 a | 22.60 | 12.63 ab | 835.3 | |
| 220 ss SS | 2.40 c | 50.76 f | 5.47 a | 0.40 c | 19.10 | 14.75 abc | 807.2 | |
| 220 ss rr | 2.02 c | 42.85 cde | 5.77 bc | 0.40 c | 21.40 | 15.91 bc | 812.2 | |
| 220 RR SS | 2.09 c | 45.56 ef | 5.47 a | 0.31 a | 20.30 | 18.27 c | 850.0 | |
| 2023 | Genotype | |||||||
| 175 RR rr | 1.43 a | 46.45 ab | 5.09 abc | 0.32 b | 18.75 | 13.90 | 827.3 | |
| 175 ss SS | 2.46 b | 57.13 c | 5.16 bc | 0.40 d | 17.10 | 15.30 | 769.2 | |
| 175 ss rr | 1.63 a | 48.08 ab | 5.13 bc | 0.29 a | 20.18 | 11.64 | 821.6 | |
| 175 RR SS | 1.53 a | 43.59 a | 4.99 ab | 0.29 a | 19.60 | 12.30 | 822.4 | |
| 220 RR rr | 1.65 a | 44.38 a | 5.01 ab | 0.28 a | 20.70 | 12.10 | 841.1 | |
| 220 ss SS | 3.08 c | 59.00 c | 5.11 bc | 0.36 c | 19.30 | 15.10 | 823.2 | |
| 220 ss rr | 2.58 b | 50.90 b | 5.23 c | 0.36 c | 21.5 | 14.70 | 819.5 | |
| 220 RR SS | 1.82 a | 47.81 ab | 4.90 a | 0.30 a | 20.10 | 11.50 | 796.4 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabrera, J.Á.; Carbonell, P.; Salinas, J.F.; Grau, A.; Alonso, A.; Hernández, F.; Ruiz, J.J.; García-Martínez, S. Effect of Introgression of Ty-1 and ty-5 Genes on Productivity, Quality, and Antioxidant Compounds in De la Pera Tomato Breeding Lines. Agriculture 2024, 14, 1192. https://doi.org/10.3390/agriculture14071192
Cabrera JÁ, Carbonell P, Salinas JF, Grau A, Alonso A, Hernández F, Ruiz JJ, García-Martínez S. Effect of Introgression of Ty-1 and ty-5 Genes on Productivity, Quality, and Antioxidant Compounds in De la Pera Tomato Breeding Lines. Agriculture. 2024; 14(7):1192. https://doi.org/10.3390/agriculture14071192
Chicago/Turabian StyleCabrera, José Ángel, Pedro Carbonell, Juan Francisco Salinas, Adrian Grau, Aranzazu Alonso, Francisca Hernández, Juan José Ruiz, and Santiago García-Martínez. 2024. "Effect of Introgression of Ty-1 and ty-5 Genes on Productivity, Quality, and Antioxidant Compounds in De la Pera Tomato Breeding Lines" Agriculture 14, no. 7: 1192. https://doi.org/10.3390/agriculture14071192






