Genome-Wide Association Study Reveals Loci and New Candidate Gene Controlling Seed Germination in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Evaluation of Seed Germination
2.3. Genome-Wide Association Study
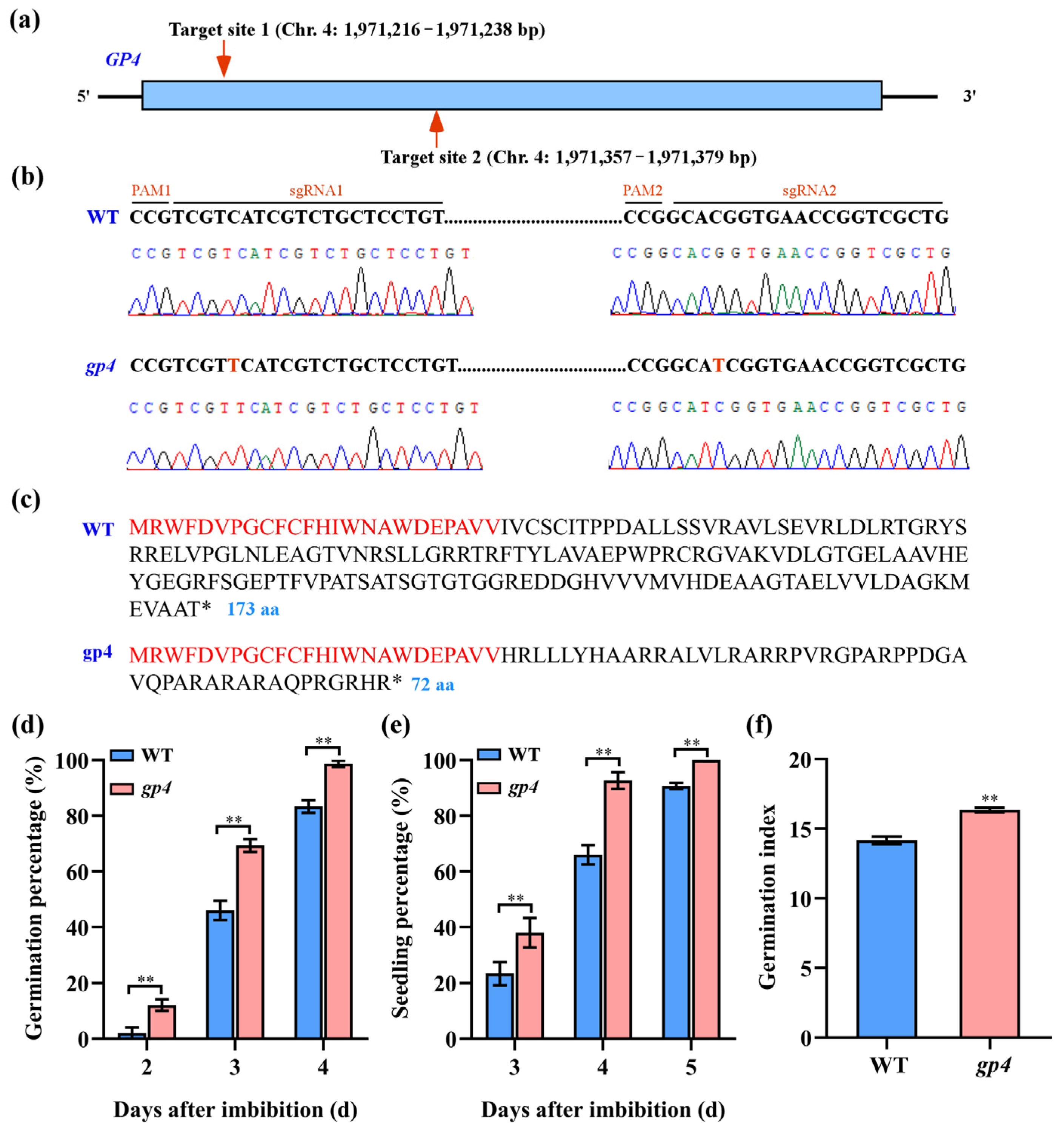
2.4. Generation and Identification of gp4 Mutant
2.5. Data Analysis
3. Results
3.1. Characterization of Seed Germination in INDICA Varietal Group
3.2. GWAS of Seed Germination in INDICA Varietal Group
3.3. Identification of GP4 as a Candidate Gene for Seed Germination
3.4. Functional Validation of GP4 through CRISPR-Cas9 System
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, K.; Lee, H.G.; Yoon, S.; Kim, H.U.; Seo, P.J. The Arabidopsis MYB96 transcription factor is a positive regulator of ABSCISIC ACID-INSENSITIVE4 in the control of seed germination. Plant Physiol. 2015, 168, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Hou, D.; Li, Y.; Chao, H.; Zhang, K.; Wang, H.; Xiang, J.; Raboanatahiry, N.; Wang, B.; Li, M. Integration of proteomic and genomic approaches to dissect seed germination vigor in Brassica napus seeds differing in oil content. BMC Plant Biol. 2019, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Mahender, A.; Anandan, A.; Pradhan, S.K. Early seedling vigour, an imperative trait for direct-seeded rice: An overview on physio-morphological parameters and molecular markers. Planta 2015, 241, 1027–1050. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Yang, R.; Wang, F.; Fu, J.; Yang, W.; Bai, T.; Wang, S.; Yin, H. Effects of gibberellin priming on seedling emergence and transcripts involved in mesocotyl elongation in rice under deep direct-seeding conditions. J. Zhejiang Univ. Sci. B 2021, 22, 1002–1021. [Google Scholar] [CrossRef]
- Miura, K.; Lin, S.; Yano, M.; Nagamine, T. Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor. Appl. Genet. 2002, 104, 981–986. [Google Scholar] [CrossRef]
- Fujino, K.; Sekiguchi, H.; Matsuda, Y.; Sugimoto, K.; Ono, K.; Yano, M. Molecular identification of a major quantitative trait locus, qLTG3–1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12623–12628. [Google Scholar] [CrossRef]
- Sugimoto, K.; Takeuchi, Y.; Ebana, K.; Miyao, A.; Hirochika, H.; Hara, N.; Ishiyama, K.; Kobayashi, M.; Ban, Y.; Hattori, T.; et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 2010, 107, 5792–5797. [Google Scholar] [CrossRef]
- Ye, H.; Feng, J.; Zhang, L.; Zhang, J.; Mispan, M.S.; Cao, Z.; Beighley, D.H.; Yang, J.; Gu, X.Y. Map-Based Cloning of Seed Dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 2015, 169, 2152–2165. [Google Scholar]
- Lai, Y.; Cheng, J.; He, Y.; Yang, B.; Wang, Z.; Zhang, H. Identification of QTLs with additive, epistatic, and QTL × seed maturity interaction effects for seed vigor in rice. Plant Mol. Biol. Rep. 2015, 34, 160–171. [Google Scholar] [CrossRef]
- Yang, J.; Yang, G.; Yang, M.; Su, L.; Xia, A.; Li, D.; Huang, C.; Zhou, D.; Liu, Y.; Wang, H.; et al. Quantitative trait locus analysis of seed germination and early seedling growth in rice. Front. Plant Sci. 2019, 10, 1582. [Google Scholar] [CrossRef]
- Riedelsheimer, C.; Lisec, J.; Czedik-Eysenberg, A.; Sulpice, R.; Flis, A.; Grieder, C.; Altmann, T.; Stitt, M.; Willmitzer, L.; Melchinger, A.E. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc. Natl. Acad. Sci. USA 2012, 109, 8872–8877. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Velasco-Punzalan, M.; Pacleb, M.; Valdez, R.; Kretzschmar, T.; McNally, K.L.; Ismail, A.M.; Cruz, P.C.S.; Sackville Hamilton, N.R.; Hay, F.R. Variation in seed longevity among diverse Indica rice varieties. Ann. Bot. 2019, 124, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Han, B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, B.; Xu, J.; Peng, L.; Sun, S.; Huang, Z.; Jiang, X.; He, Y.; Wang, Z. A genome-wide association study reveals that the 2-oxoglutarate/malate translocator mediates seed vigor in rice. Plant J. 2021, 108, 478–491. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Fan, K.; Wang, Y.; Tian, L.; Zhang, C.; Sun, W.; He, H.; Yu, S. OsGRETCHENHAGEN3-2 modulates rice seed storability via accumulation of abscisic acid and protective substances. Plant Physiol. 2021, 186, 469–482. [Google Scholar] [CrossRef]
- Castano-Duque, L.; Ghosal, S.; Quilloy, F.A.; Mitchell-Olds, T.; Dixit, S. An epigenetic pathway in rice connects genetic variation to anaerobic germination and seedling establishment. Plant Physiol. 2021, 186, 1042–1059. [Google Scholar] [CrossRef]
- Yang, B.; Chen, M.; Zhan, C.; Liu, K.; Cheng, Y.; Xie, T.; Zhu, P.; He, Y.; Zeng, P.; Tang, H.; et al. Identification of OsPK5 involved in rice glycolytic metabolism and GA/ABA balance for improving seed germination via genome-wide association study. J. Exp. Bot. 2022, 73, 3446–3461. [Google Scholar] [CrossRef]
- Yoshida, H.; Hirano, K.; Yano, K.; Wang, F.; Mori, M.; Kawamura, M.; Koketsu, E.; Hattori, M.; Ordonio, R.L.; Huang, P.; et al. Genome-wide association study identifies a gene responsible for temperature-dependent rice germination. Nat. Commun. 2022, 13, 5665. [Google Scholar] [CrossRef]
- Dai, L.; Lu, X.; Shen, L.; Guo, L.; Zhang, G.; Gao, Z.; Zhu, L.; Hu, J.; Dong, G.; Ren, D.; et al. Genome-wide association study reveals novel QTLs and candidate genes for seed vigor in rice. Front. Plant Sci. 2022, 13, 1005203. [Google Scholar] [CrossRef]
- Yang, B.; Zeng, J.; Chen, S.; Li, S.; Wu, L.; Wan, X. Genome-wide association study reveals the genetic basis of seed germination in Japonica rice. Agriculture 2022, 13, 118. [Google Scholar] [CrossRef]
- Yang, B.; Chen, S.; Zheng, Z.; Zeng, J.; Liu, J.; Zhao, H.; Zheng, Y. Genome-wide association studies for rice seed germination under drought stress using 3VmrMLM. Food Energy Secur. 2024, 13, e529. [Google Scholar] [CrossRef]
- Prasad, C.T.M.; Kodde, J.; Angenent, G.C.; Hay, F.R.; McNally, K.L.; Groot, S.P.C. Identification of the rice Rc Gene as a main regulator of seed survival under dry storage conditions. Plant Cell Environ. 2023, 46, 1962–1980. [Google Scholar] [CrossRef] [PubMed]
- Zhan, C.; Zhu, P.; Chen, Y.; Chen, X.; Liu, K.; Chen, S.; Hu, J.; He, Y.; Xie, T.; Luo, S.; et al. Identification of a key locus, qNL3.1, associated with seed germination under salt stress via a genome-wide association study in rice. Theor. Appl. Genet. 2023, 136, 58. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ma, Y.; Zhao, H.; Guo, L.; Guo, Y.; Liu, C. Genome-wide association studies identified OsTMF as a gene regulating rice seed germination under salt stress. Front. Plant Sci. 2024, 15, 1384246. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Tuấn, P.A.; Izydorczyk, M.S.; Ayele, B.T. Ethylene regulates post-germination seedling growth in wheat through spatial and temporal modulation of ABA/GA balance. J. Exp. Bot. 2019, 71, 1985–2004. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, Z.; Ren, Z.; Zhi, L.; Yao, B.; Su, C.; Liu, L.; Li, X. SCFAtPP2-B11 Modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. PLoS Genet. 2017, 13, e1006947. [Google Scholar] [CrossRef]
- Cutler, A.J.; Krochko, J.E. Formation and breakdown of ABA. Trends Plant Sci. 1999, 4, 472–478. [Google Scholar] [CrossRef]
- Yan, J.; Zhao, C.; Zhou, J.; Ye, Y.; Wang, P.; Zhu, X.; Tang, G.; Bressan, R.A.; Zhu, J. The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006416. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Kim, D.H.; Hwang, I. ABA homeostasis and signaling involving multiple subcellular compartments and multiple receptors. Plant Cell Rep. 2013, 32, 807–813. [Google Scholar] [CrossRef]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A.D. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003, 131, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.D.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.D.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–12240. [Google Scholar] [CrossRef] [PubMed]
- Burbidge, A.; Grieve, T.M.; Jackson, A.; Thompson, A.; McCarty, D.R.; Taylor, I.B. Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 1999, 17, 427–431. [Google Scholar] [CrossRef]
- Chernys, J.T.; Zeevaart, J.A.D. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 2000, 124, 343–354. [Google Scholar] [CrossRef]
- Qin, X.; Zeevaart, J.A.D. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002, 128, 544–551. [Google Scholar] [CrossRef]
- Zhao, K.; Tung, C.-W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, J.; Bao, Y.; Wang, F.; Zhang, H. Quantitative trait loci analysis for rice seed vigor during the germination stage. J. Zhejiang Univ. Sci. B 2010, 11, 958–964. [Google Scholar] [CrossRef]
- McCouch, S.R.; Wright, M.H.; Tung, C.W.; Maron, L.G.; McNally, K.L.; Fitzgerald, M.A.; Singh, N.; DeClerck, G.; Agosto-Perez, F.; Korniliev, P.; et al. Open access resources for genome-wide association mapping in rice. Nat. Commun. 2016, 7, 10532. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacteriu allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Wang, Y.; Zhang, C.; He, H.; Yu, S. Genetic dissection of seed dormancy using chromosome segment substitution lines in rice (Oryza sativa L.). Int. J. Mol. Sci. 2020, 21, 1344. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Shi, J.; Gui, J.; Zhou, H.; Yan, Y.; Zhu, X.; Xie, B.; Liu, X.; He, J. Rice seed protrusion quantitative trait loci mapping through genome-wide association study. Plants 2024, 13, 134. [Google Scholar] [CrossRef]
- Lin, Q.; Wu, F.; Sheng, P.; Zhang, Z.; Zhang, X.; Guo, X.; Wang, J.; Cheng, Z.; Wang, J.; Wang, H.; et al. The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 2015, 2, 7981. [Google Scholar] [CrossRef]
- Islam, M.R.; Naveed, S.A.; Zhang, Y.; Li, Z.; Zhao, X.; Fiaz, S.; Zhang, F.; Wu, Z.; Hu, Z.; Fu, B.; et al. Identification of candidate genes for salinity and anaerobic tolerance at the germination stage in rice by genome-wide association analyses. Front. Genet. 2022, 13, 822516. [Google Scholar] [CrossRef]
- Zhou, Y.; Xie, Y.; Cai, J.; Liu, C.; Zhu, H.; Jiang, R.; Zhong, Y.; Zhang, G.; Tan, B.; Liu, G.; et al. Substitution mapping of qtls controlling seed dormancy using single segment substitution lines derived from multiple cultivated rice donors in seven cropping seasons. Theor. Appl. Genet. 2017, 130, 1191–1205. [Google Scholar] [CrossRef]
- Martínez-Andújar, C.; Ordiz, M.I.; Huang, Z.; Nonogaki, M.; Beachy, R.N.; Nonogaki, H. Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis Thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 2011, 108, 17225–17229. [Google Scholar] [CrossRef]
- Huo, H.; Dahal, P.; Kunusoth, K.; McCallum, C.M.; Bradford, K.J. Expression of 9-cis-EPOXYCAROTENOID DIOXYGENASE4 is essential for thermoinhibition of lettuce seed germination but not for seed development or stress tolerance. Plant Cell 2013, 25, 884–900. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, J.; Yang, T.; Dong, J.; Yang, W.; Chen, L.; Zhou, L.; Chen, J.; Liu, B.; Zhang, S.; et al. Genome-wide association mapping and gene expression analysis identify OsCPS1 as a new candidate gene controlling early seedling length in rice. Front. Plant Sci. 2022, 13, 976669. [Google Scholar] [CrossRef]
- Kadam, N.N.; Jagadish, K.; Struik, P.C.; van der Linden, C.G.; Yin, X. Incorporating genome-wide association into eco-physiological simulation to identify markers for improving rice yields. J. Exp. Bot. 2019, 70, 2575–2586. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xiang, Z.; Liu, M.; Wang, S.; Zhang, L.; Cai, D.; Huang, Y.; Mao, D.; Fu, J.; Chen, L. ABA biosynthesis gene OsNCED3 contributes to preharvest sprouting resistance and grain development in rice. Plant Cell Environ. 2022, 46, 1384–1401. [Google Scholar] [CrossRef]

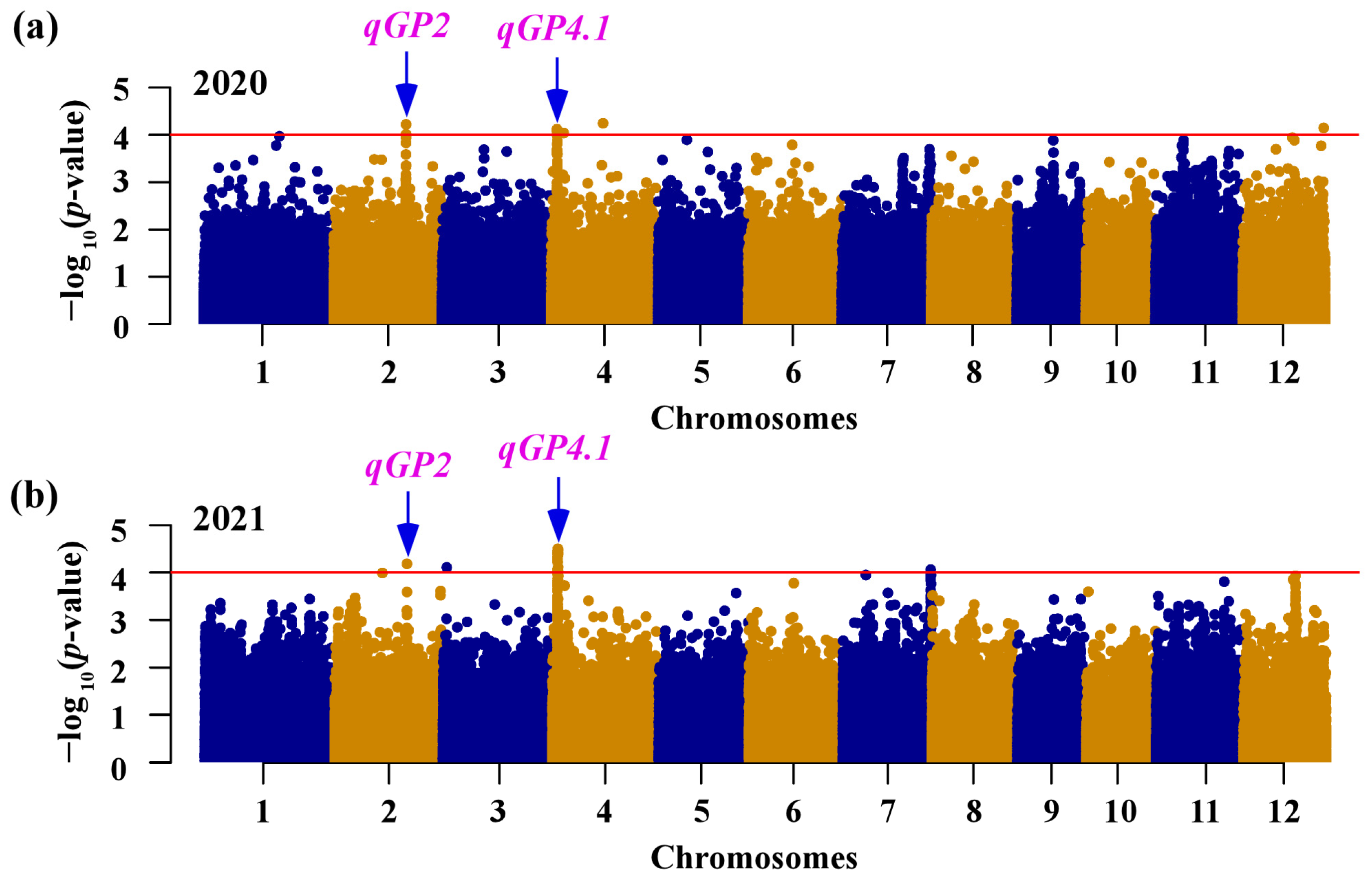

| Years | Range (%) | Mean (%) | SD (%) | CV | Heritability (%) |
|---|---|---|---|---|---|
| 2020 | 0.00~99.33 | 56.02 | 28.75 | 0.51 | 94.19 |
| 2021 | 0.00~100.00 | 58.04 | 27.74 | 0.48 |
| Loci | Years | Chr. | Lead SNP Position (bp) | p Value | Known QTLs/Genes | Reference |
|---|---|---|---|---|---|---|
| qGP2 | 2020/2021 | 2 | 24,056,673 | 6.01 × 10−5 | qDOM2.3/qPP2-1 | [43,44] |
| qGP3 | 2021 | 3 | 1,350,796 | 7.78 × 10−5 | TE | [45] |
| qGP4.1 | 2020/2021 | 4 | 1,966,105 | 3.14 × 10−5 | qPP4/qRI4a | [44,46] |
| qGP4.2 | 2020 | 4 | 4,155,560 | 9.14 × 10−5 | ||
| qGP4.3 | 2020 | 4 | 17,194,678 | 5.68 × 10−5 | ||
| qGP7 | 2021 | 7 | 29,274,866 | 8.68 × 10−5 | OsCLSY1 | [16] |
| qGP12 | 2020 | 12 | 26,981,655 | 7.12 × 10−5 | qSD12-1 | [47] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Chen, G.; Peng, Z.; Liu, J.; Zheng, Y.; Yang, B. Genome-Wide Association Study Reveals Loci and New Candidate Gene Controlling Seed Germination in Rice. Agriculture 2024, 14, 1613. https://doi.org/10.3390/agriculture14091613
Chen S, Chen G, Peng Z, Liu J, Zheng Y, Yang B. Genome-Wide Association Study Reveals Loci and New Candidate Gene Controlling Seed Germination in Rice. Agriculture. 2024; 14(9):1613. https://doi.org/10.3390/agriculture14091613
Chicago/Turabian StyleChen, Shaona, Guanlong Chen, Zepeng Peng, Jiping Liu, Yixiong Zheng, and Bin Yang. 2024. "Genome-Wide Association Study Reveals Loci and New Candidate Gene Controlling Seed Germination in Rice" Agriculture 14, no. 9: 1613. https://doi.org/10.3390/agriculture14091613






