Fungal Colonization of the Anatomical Parts of Soybean Seeds Supplied with Different Nitrogen Rates and Inoculated with Bradyrhizobium japonicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Experiment
2.2. Laboratory Analyses
2.2.1. Mycobiome Analysis
2.2.2. Molecular Analysis
2.3. Statistical Analysis
2.4. Weather Conditions
3. Results
3.1. Mycobiome Analysis
3.2. Molecular Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sudarić, A.; Kočar, M.M.; Duvnjak, T.; Zdunić, Z.; Kulundžić, A.M. Improving seed quality of soybean suitable for growing in Europe. In Soybean for Human Consumption and Animal Feed; IntechOpen: London, UK, 2019; Available online: https://www.intechopen.com/online-first/improving-seed-quality-of-soybean-suitable-for-growing-in-europe (accessed on 28 April 2024).
- Niwińska, B.; Witaszek, K.; Niedbała, G.; Pilarski, K. Seeds of n-GM Soybean Varieties Cultivated in Poland and Their Processing Products as High-Protein Feeds in Cattle Nutrition. Agriculture 2020, 10, 174. [Google Scholar] [CrossRef]
- Hosseini, B.; Voegele, R.T.; Link, T.I. Diagnosis of Soybean Diseases Caused by Fungal and Oomycete Pathogens: Existing Methods and New Developments. J. Fungi 2023, 9, 587. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Konishi, M.; Yanagisawa, S. Molecular basis of the nitrogen response in plants. Soil. Sci. Plant Nutr. 2017, 4, 329–341. [Google Scholar] [CrossRef]
- Horchani, F.; Hajri, R.; Aschi-Smiti, S. Effect of ammonium or nitrate nutrition on photosynthesis, growth, and nitrogen assimilation in tomato plants. J. Plant Nutr. Soil. Sci. 2010, 173, 610–617. [Google Scholar] [CrossRef]
- Sharma, S. Impacts of nitrogen on plant disease severity and plant defense mechanism. Fundam. Appl. Agric. 2020, 5, 303–314. [Google Scholar] [CrossRef]
- Wang, Q.; Li, S.; Li, J.; Huang, D. The Utilization and Roles of Nitrogen in Plants. Forests 2024, 15, 1191. [Google Scholar] [CrossRef]
- Szpunar-Krok, E.; Bobrecka-Jamro, D.; Pikuła, W.; Janńczak-Pieniążek, M. Effect of Nitrogen Fertilization and Inoculation with Bradyrhizobium japonicum on Nodulation and Yielding of Soybean. Agronomy 2023, 13, 1341. [Google Scholar] [CrossRef]
- Nandi, R.G.; Bara, J.K.; Shrivastava, P. Antimicrobial activity of Rhizobium japonicum and Bradyrhizobium japonicum on different plant pathogenic fungal strains. Microbiol. Commun. Biosci. Biotech. Res. Comm. 2019, 12, 435–439. [Google Scholar] [CrossRef]
- Mabrouk, Y.; Hemissi, I.; Salem, I.B.; Mejri, S.; Saidi, M.; Belhadj, O. Potential of Rhizobia in Improving Nitrogen Fixation and Yields of Legumes. In Symbiosis; Rigobelo, E.C., Ed.; IntechOpen: Rijeka, Croatia, 2018; Chapter 6. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Jabborova, D.; Wirth, S.J.; Alam, P.; Alyemeni, M.N.; Ahmad, P. Interactive Effects of Nutrients and Bradyrhizobium japonicum on the Growth and Root Architecture of Soybean (Glycine max L.). Front. Microbiol. 2018, 9, 1000. [Google Scholar] [CrossRef]
- Lin, F.; Chhapekar, S.S.; Vieira, C.C.; Da Silva, M.P.; Rojas, A.; Lee, D.; Liu, N.; Pardo, E.M.; Lee, Y.-C.; Dong, Z.; et al. Breeding for disease resistance in soybean: A global perspective. Theor. Appl. Genet. 2022, 135, 3773–3872. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, M.; Sun, Y.; Gu, Z.; Wang, R.; Saydin, A.; Shen, Q.; Guo, S. Nitrate increased cucumber tolerance to fusarium wilt by regulating fungal toxin production and distribution. Toxins 2017, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, P.; Thakur, S.; Nagre, S.P.; Anand, K.; Saraswat, S.; Mohare, S. Review of soybean breeding for disease and insect resistance. Front. Crop Improv. 2022, 10, 541–548. [Google Scholar]
- Chiotta, M.L.; Alaniz Zanon, M.S.; Palazzini, J.M.; Scandiani, M.M.; Formento, A.N.; Barros, G.G.; Chulze, S.N. Pathogenicity of Fusarium graminearum and F. meridionale on soybean pod blight and trichothecene accumulation. Plant Pathol. 2016, 65, 1492–1497. [Google Scholar] [CrossRef]
- Zhao, L.; Wei, X.; Zheng, T.; Guo, Y.-N.; Wang, J.; Deng, J.-X.; Li, M.-J. Evaluation of pathogenic Fusarium spp. associated with soybean seed (Glycine max) in Hubei Province, China. Plant Dis. 2022, 106, 3178–3186. [Google Scholar] [CrossRef]
- Vidić, M.; Đorđević, V.; Petrović, K.; Miladinović, J. Review of soybean resistance to pathogens. Ratar. Povrt. 2013, 50, 52–61. [Google Scholar] [CrossRef]
- Carvalho, E.R.; Reis, L.V.; Rocha, D.K.; Penido, A.C.; da Rosa Mavaieie, D.P.; de Fátima Ferreira, V.; Oliveira, J.A. Incidence of fungal species in stored soybean seeds in relation to cooling before packing and to packing material. SCAP 2021, 44, 193–202. [Google Scholar] [CrossRef]
- Bhattacharya, K.; Raha, S. Deteriorative changes of maize, groundnut and soybean seeds by fungi in storage. Mycopathologia 2002, 155, 135–141. [Google Scholar] [CrossRef]
- Broggi, L.E.; González, H.H.; Resnik, S.L.; Pacin, A. Alternaria alternata prevalence in cereal grains and soybean seeds from Entre Ríos, Argentina. Rev. Iberoam. Micol. 2007, 24, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Haikal, N.Z. Effect of filtrates of pathogenic fungi of soybean on seed germination and seedling parameters. J. Appl. Sci. Res. 2008, 4, 48–52. [Google Scholar]
- Medić-Pap, S.; Milošević, M.; Jasnić, S. Soybean seed-borne fungi in the Vojvodina province. Phytopathol. Pol. 2007, 45, 55–65. [Google Scholar]
- Nasir, N. Detecting seed borne fungi of soybean by different incubation methods. Plant Pathol. J. 2003, 2, 114–118. [Google Scholar] [CrossRef]
- Żelechowski, M.; Molcan, T.; Bilska, K.; Myszczyński, K.; Olszewski, J.; Karpiesiuk, K.; Wyrębek, J.; Kulik, T. Patterns of Diversity of Fusarium Fungi Contaminating Soybean Grains. Toxins 2021, 13, 884. [Google Scholar] [CrossRef] [PubMed]
- Olszak-Przybyś, H.; Korbecka-Glinka, G. The Diversity of Seed-Borne Fungi Associated with Soybean Grown in Southern Poland. Pathogens 2024, 13, 769. [Google Scholar] [CrossRef] [PubMed]
- Rao, T.V.; Rajeswari, B.; Keshavulu, K.; Varma, V.S. Studies on seedborne fungi of soybean. SSRG Int. J. Agric. Environ. Sci. (SSRG-IJAES) 2015, 2, 16–24. [Google Scholar]
- Sobczak, P.; Zawiślak, K.; Żukiewicz-Sobczak, W.; Mazur, J.; Nadulski, R.; Kozak, M. The assessment of microbiological purity of selected components of animal feeds and mixtures which underwent thermal processing. J. Cent. Eur. Agric. 2016, 17, 303–314. [Google Scholar] [CrossRef]
- Garcia, D.; Barros, G.; Chulze, S.; Ramos, A.J.; Sanchis, V.; Marín, S. Impact of cycling temperatures on Fusarium verticillioides and Fusarium graminnearum growth and mycotoxins production in soybean. J. Sci. Food Agric. 2012, 92, 2952–2959. [Google Scholar] [CrossRef]
- Hartman, G.L.; McCormick, S.P.; O’Donnell, K. Trichothecene-Producing Fusarium Species Isolated from Soybean Roots in Ethiopia and Ghana and their Pathogenicity of Soybean. Plant Dis. 2019, 103, 2070–2075. [Google Scholar] [CrossRef]
- Okorski, A.; Polak-Śliwińska, M.; Karpiesiuk, K.; Pszczółkowska, A.; Kozera, W. Real time pcr: A good tool to estimate mycotoxin contamination in pig diets. World Mycotoxin J. 2017, 10, 219–228. [Google Scholar] [CrossRef]
- Jedziniak, P.; Panasiuk, Ł.; Pietruszka, K.; Posyniak, A. Multiple mycotoxins analysis in animal feed with LC-MS/MS: Comparison of extract dilution and immunoaffinity clean-up. J. Sep. Sci. 2019, 42, 1240–1247. [Google Scholar] [CrossRef]
- Alshannaq, A.; Yu, J.-H. Occurrence, Toxicity, and Analysis of Major Mycotoxins in Food. Int. J. Environ. Res. Public. Health. 2017, 14, 632. [Google Scholar] [CrossRef]
- Zhang, C.; Qu, Z.; Hou, J.; Yao, Y. Contamination and Control of Mycotoxins in Grain and Oil Crops. Microorganisms 2024, 12, 567. [Google Scholar] [CrossRef] [PubMed]
- Pleadin, J.; Frece, J.; Markov, K. Mycotoxins in food and feed. In Advances in Food and Nutrition Research; Toldra, F., Ed.; Elsevier: Cambridge, UK, 2019; pp. 297–345. [Google Scholar]
- El-Sayed, R.A.; Jebur, A.B.; Kang, W.; El-Demerdash, F.M. An overview on the major mycotoxins in food products: Characteristics, toxicity, and analysis. J. Future Foods. 2022, 2, 91–102. [Google Scholar] [CrossRef]
- Qu, Z.; Ren, X.; Du, Z.; Hou, J.; Li, Y.; Yao, Y.; Yi, A. Fusarium mycotoxins: The major food contaminants. mLIfe 2024, 3, 176–206. [Google Scholar] [CrossRef] [PubMed]
- Cegielska–Radziejewska, R.; Szablewski, T.; Karolczak, K.; Kaczmarek, A.; Kijowski, J. An immunoenzymatic method for the determination of mycotoxins contents in cereals and feeds. Nauka Przyr. Technol. 2009, 3, 114. [Google Scholar]
- Costa, J.H.; Saraiva, K.D.C.; Morais, V.D.; Oliveira, J.T.A.; Sousa, D.O.B.; Fernandes de Melo, D.; Morais, J.K.S.; Vasconcelos, I.M. Reference gene identification for real-time PCR analyses in soybean leaves under fungus (Cercospora kikuchii) infection and treatments with salicylic and jasmonic acids. Australas. Plant Pathol. 2016, 45, 191–199. [Google Scholar] [CrossRef]
- Chang, X.; Li, H.; Naeem, M.; Wu, X.; Yong, T.; Song, C.; Liu, T.; Chen, W.; Yang, W. Diversity of the Seedborne Fungi and Pathogenicity of Fusarium Species Associated with Intercropped Soybean. Pathogens 2020, 9, 531. [Google Scholar] [CrossRef]
- Gilman, J.C. A Manual of Soil Fungi; Oxford And Ibh Publishing, Co.: New Delhi, India, 1957. [Google Scholar]
- Gerlach, W.; Nirenberg, H. The Genus Fusarium—A Pictorial Atlas; Biologische Bundesanstalt für Land- und Forstwirtschaft: Berlin, Germany, 1982. [Google Scholar]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; Blackwell Publishing: Hoboken, NY, USA, 2006; pp. 81–159. [Google Scholar]
- Watanabe, I.; Kakishima, M.; Adachi, Y.; Nakajima, H. Potential mycotoxin productivity of Alternaria alternata isolated from garden trees. Mycotoxins 2007, 57, 34–45. [Google Scholar] [CrossRef]
- Kulik, T.; Pszczółkowska, A.; Fordoński, G.; Olszewski, J. PCR approach based on the esyn1 gene for the detection of potential enniatin–producing Fusarium species. Int. J. Food Microbiol. 2007, 116, 319–324. [Google Scholar] [CrossRef]
- Hue, F.X.; Huerre, M.; Rouffault, M.A.; Bievre, C. Specific Detection of Fusarium Species in Blood and Tissues by PCR Technique. J. Clin. Microbiol. 1999, 37, 2434–2438. [Google Scholar] [CrossRef]
- Vegi, A.; Wolf-Hall, C. Multiplex real-time pcr marhod for detection and quantification of mycotoxigenic fungi belong to three different genera. J. Food Sci. 2013, 78, 70–76. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Podleśny, J.; Podleśna, A.; Nędzi, M. Occurrence of fungal diseases caused by fungi on faba bean (Vicia faba L. var. minor Harz.) plants in different regions of Poland. Prog. Plant Prot. 2017, 57, 190–195. [Google Scholar] [CrossRef]
- Rocha, L.F.; Srour, A.Y.; Pimentel, M.; Subedi, A.; Bond, J.P.; Fakhoury, A.; Ammar, H.A. A panel of qPCR assays to detect and quantify soybean soil-borne pathogens. Lett. Appl. Microbiol. 2022, 76, ovac023. [Google Scholar] [CrossRef] [PubMed]
- Rojas, J.A.; Miles, T.D.; Coffey, M.D.; Martin, F.N.; Chilvers, M.I. Development and application of qPCR and RPA genus- and species-specific detection of Phytophthora sojae and P. sansomeana root rot pathogens of soybean. Plant Dis. 2017, 101, 1171–1181. [Google Scholar] [CrossRef]
- Wang, J.; Jacobs, J.L.; Byrne, J.M.; Chilvers, M.I. Improved diagnoses and quantification of Fusarium virguliforme, causal agent of soybean sudden death syndrome. Phytopathology 2015, 105, 378–387. [Google Scholar] [CrossRef]
- Kim, T.G.; Knudsen, G.R. Quantitative real-time PCR effectively detects and quantifies colonization of sclerotia of Sclerotinia sclerotiorum by Trichoderma spp. Appl. Soil. Ecol. 2008, 40, 100–108. [Google Scholar] [CrossRef]
- Wu, L.; Hwang, S.-F.; Strelkov, S.E.; Fredua-Agyeman, R.; Oh, S.-H.; Bélanger, R.R.; Wally, O.; Kim, Y.-M. Pathogenicity, Host Resistance, and Genetic Diversity of Fusarium Species under Controlled Conditions from Soybean in Canada. J. Fungi 2024, 10, 303. [Google Scholar] [CrossRef]
- Ciampi-Guillardi, M.; Ramiro, J.; de Mores, M.H.D.; Barbieri, C.G.; Massola, N.S., Jr. Multiplex qPCR assay for direct detection and quantification of Colletotrichum, Corynespora cassiicola and Sclerotninia sclerotiorum in soybean seeds. Plant Dis. 2020, 104, 3002–3009. [Google Scholar] [CrossRef]
- Janda, K.; Wolska, J. Study on the quantitative and qualitative of fungi colonizing soybeans (Glycine max L.). Pomeranian J. Life Sci. 2015, 61, 426–432. [Google Scholar] [PubMed]
- Kinnikar, A.; Desai, P.; Jahagirdar, S. Identification and Detection of Seed Borne Diseases of Soybean Using Image Processing—A Survey. Int. J. Emerg. Technol. Comput. Sci. Electron. 2015, 14, 363. [Google Scholar]
- Olszak-Przybyś, H.; Korbecka-Glinka, G.; Patkowska, E. Identification and Pathogenicity of Fusarium Isolated from Soybean in Poland. Pathogens 2023, 12, 1162. [Google Scholar] [CrossRef] [PubMed]
- Escamilla, D.; Rosso, M.L.; Zhang, B. Identification of fungi associated with soybeans and effective seed disinfection treatments. Food Sci. Nutr. 2019, 7, 3194–3205. [Google Scholar] [CrossRef]
- Pszczółkowska, A.; Okorski, A.; Fordoński, G.; Kotecki, A.; Kozak, M.; Dzienis, G. Effect of weather conditions on yield and health status of Faba bean seeds in Poland. Agronomy 2020, 10, 48. [Google Scholar] [CrossRef]
- Marcinkowska, J. Fungi occurrence on seeds of field pea. Acta Mycol. 2008, 43, 77–89. [Google Scholar] [CrossRef]
- Mengistu, A.; Sinclair, J. Seed borne microorganisms of Ethiopian-grown soybean and chickpea seeds. Plant Diesease. Rep. 1979, 63, 616–619. [Google Scholar]
- Jędryczka, M.; Kaczmarek, J. Infestation of commercial seed lots of narrow-leafed lupin by pathogenic and saprotrophic fungi. Fragm. Agron. 2012, 29, 63–69. [Google Scholar]
- Di Francesco, A.; Di Foggia, M.; Corbetta, M.; Baldo, D.; Ratti, C.; Baraldi, E. Biocontrol Activity and Plant Growth Promotion Exerted by Aureobasidium pullulans Strains. J. Plant Growth Regul. 2021, 40, 1233–1244. [Google Scholar] [CrossRef]
- Hafez, M.; Abdelmagid, A.; Aboukhaddour, R.; Adam, L.R.; Daayf, F. Fusarium Root Rot Complex in Soybean: Molecular Characterization, Trichothecene Formation, and Cross-Pathogenicity. Phytopathology 2021, 111, 2287–2302. [Google Scholar] [CrossRef]
- Diaz-Arias, M.M.; Leandro, L.F.S.; Munkvold, G. Aggressiveness of Fusarium species and impact of root infection on growth and yield of soybean. Phytopathology 2013, 103, 822–832. [Google Scholar] [CrossRef]
- Petrović, K.; Orzali, L.; Krsmanović, S.; Valente, M.T.; Tolimir, M.; Pavlov, J.; Riccioni, L. Genetic Diversity and Pathogenicity of the Fusarium Species Complex on Soybean in Serbia. Plant Dis. 2024, 108, 1851–1860. [Google Scholar] [CrossRef] [PubMed]
- Pszczółkowska, A.; Okorski, A.; Fordoński, G.; Prusiński, J.; Faligowska, A.; Borowska, M. Fungal colonization of seeds of three lupine species in different regions of Poland. Acta Agrobot. 2017, 70, 1714. [Google Scholar] [CrossRef]
- Pszczółkowska, A.; Okorski, A.; Fordoński, G.; Faligowska, A.; Kaszkowiak, E.; Olszewski, J.; Chareńska, A. The frequency of occurrence of pathogenic and saprotrophic fungi in pea seeds in different regions of Poland. Legume Res.-Int. J. 2019, 42, 270–276, 118:2357149008. [Google Scholar] [CrossRef]
- Ozgonen, H.; Gulcu, M. Determination of mycoflora of pea (Pisum sativum) seeds and the effects of Rhizobium leguminosorum on fungal pathogens of peas. Afr. J. Biotechnol. 2011, 10, 6235–6240. [Google Scholar] [CrossRef]
- Alomran, M.M.; Lupien, S.L.; Coyne, C.J.; Dugan, F.M. Mycobiota of Lupinus albus seed from a public germplasm collection. N. Am. Fungi 2013, 8, 1–15. [Google Scholar] [CrossRef]
- Ahmed, O.; Balogun, O.S.; Fawole, O.B.; Fabiyi, O.A.; Aliyu, T.H.; Kassoum, K.O. Seed-borne fungi of soybeans (Glycine max [L.] Merr) in the Guinea savannah agroecology of Nigeria. J. Agric. Sci. 2016, 61, 57–68. [Google Scholar] [CrossRef]
- Ahmed, O.; Fawole, O.B.; Balogun, O.S. Component analysis for seed mycoflora in four cultivars of cowpea (Vigna unguiculata [L.] Walp). Cent. Point (Sci. Ed.) 2006, 14, 71–77. [Google Scholar]
- Aiswarya, N.; Bhattiprolu, S.L.; Bayyapu Reddy, K.; Rama Rao, G. Location of seedborne fungi in farmers saved samples of groundnut variety tag 24. Int. J. Chem. Stud. 2019, 7, 3103–3105. [Google Scholar]
- Martín, I.; Gálvez, L.; Guasch, L.; Palmero, D. Fungal Pathogens and Seed Storage in the Dry State. Plants 2022, 11, 3167. [Google Scholar] [CrossRef]
- Tayyab, M.; Ali, Q.; Sultana, M.; Iqbal, A.; Ali Khan, R.M.; Hiza Noor, H.; Ahmad, K.; Aleem, M.; Anmol, S.; Faisal, M.; et al. Detection of Seed Borne Fungi Associated with Glycine max and its Management Through Physio–Chemical Treatments. J. Xi’an Shiyou Univ. Nat. Sci. Ed. 2024, 20, 543–603. [Google Scholar]
- Klaedtke, S.; Jacques, M.A.; Raggi, L.; Préveaux, A.; Bonneau, S.; Negri, V.; Chablem, V.; Barret, M. Terroir is a key driver of seed-associated microbial assemblages. Environ. Microbiol. 2016, 18, 1792–1804. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Deng, J.C.; Yang, C.Q.; Huang, N.; Chang, X.L.; Zhang, J.; Yang, F.; Liu, W.G.; Wang, X.C.; Yong, T.W.; et al. Fungal Diversity in Field Mold-Damaged Soybean Fruits and Pathogenicity Identification Based on High-Throughput rDNA Sequencing. Front. Microbiol. 2017, 8, 779. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-B.; Zhai, H.-C.; Huang, S.-X.; Cai, J.-P. A site-directed CO2 detection method for monitoring the spoilage of stored grains by insects and fungi in Chinese horizontal warehouses. J. Stored Prod. Res. 2014, 59, 146–151. [Google Scholar] [CrossRef]
- Majewska-Sawka, A.; Nakashima, H. Endophyte transmission via seeds of Lolium perenne L. immunodetection of fungal antigens. Fungal Genet. Biol. 2004, 41, 534–541. [Google Scholar] [CrossRef]
- Ngugi, H.K.; Scherm, H. Biology of flower-infecting fungi. Annu. Rev. Phytopathol. 2006, 44, 261–282. [Google Scholar] [CrossRef]
- Wang, M.; Sun, Y.; Gu, Z.; Wang, R.; Sun, G.; Zhu, C.; Guo, S.; Shen, Q. Nitrate Protects Cucumber Plants Against Fusarium oxysporum by Regulating Citrate Exudation. Plant Cell Physiol. 2016, 57, 2001–2012. [Google Scholar] [CrossRef]
- Arora, N.K.; Kang, S.C.; Maheshwari, D.K. Isolation of siderophore-producing strains of Rhizobium meliloti and their biocontrol potential against Macrophomina phaseolina that causes charcoal rot of groundnut. Curr. Sci. 2001, 81, 673–677. [Google Scholar]
- Deshwal, V.K.; Dubey, R.C.; Maheshwari, D.K. Isolation of plant growth-promoting strains of Bradyrhizobium (Arachis) spp. with biocontrol potential against Macrophomina phaseolina causing charcoal rot of peanut. Curr. Sci. 2003, 84, 443. [Google Scholar]
- Mariastuti, H.D.; Listiyowati, S.; Wahyudi, A.T. Antifungal activity of soybean rhizosphere actinomycetes producing bioactive compounds against Fusarium oxysporum. Biodiversitas 2018, 19, 2127–2133. [Google Scholar] [CrossRef]
- Mishra, P.K.; Fox, R.T.; Culham, A. Development of a PCR-based assay for rapid and reliable identification of pathogenic Fusaria. FEMS Microbiol. Lett. 2003, 218, 329–332. [Google Scholar] [CrossRef]
- Nicholson, P.; Chandler, E.; Draeger, R.C.; Gosman, N.E.; Simpson, D.R.; Thomsett, M.; Wilson, A.H. Molecular tools to study epidemiology and toxicology of Fusarium head blight of cereals. Eur. J. Plant Pathol. 2003, 109, 691–703. [Google Scholar] [CrossRef]
- van Diepeningen, A.D.; Brankovics, B.; Iltes, J.; van der Lee, T.A.J.; Waalwijk, C. Diagnosis of Fusarium Infections: Approaches to Identification by the Clinical Mycology Laboratory. Curr. Fungal Infect. Rep. 2015, 9, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, M.J.; Dobson, D.W. Mycotoxin production by Aspergillus, Fusarium and Penicillium species. Int. J. Food Microbiol. 1998, 43, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef]
- Duarte, S.C.; Pena, A.; Lino, C.M. A review on ochratoxin A occurrence and effects of processing of cereal and cereal derived food products. Food Microbiol. 2010, 27, 187–198. [Google Scholar] [CrossRef]
- Perkowski, J.; Stuper, K.; Buśko, M.; Góral, T.; Kaczmarek, A.; Jeleń, H. Differences in metabolomic profiles of the naturally contaminated grain of barley, oats and rye. J. Cereal Sci. 2012, 56, 544–551. [Google Scholar] [CrossRef]
- Dawidziuk, A.; Koczyk, G.; Popiel, D.; Kaczmarek, J.; Buśko, M. Molecular diagnostics on the toxigenic potential of Fusarium spp. plant pathogens. J. Appl. Microbiol. 2014, 116, 1607–1620. [Google Scholar] [CrossRef]
- Perincherry, L.; Lalak-Kańczugowska, J.; Stępień, Ł. Fusarium-produced mycotoxins in plant-pathogen interactions. Toxins 2019, 11, 664. [Google Scholar] [CrossRef]
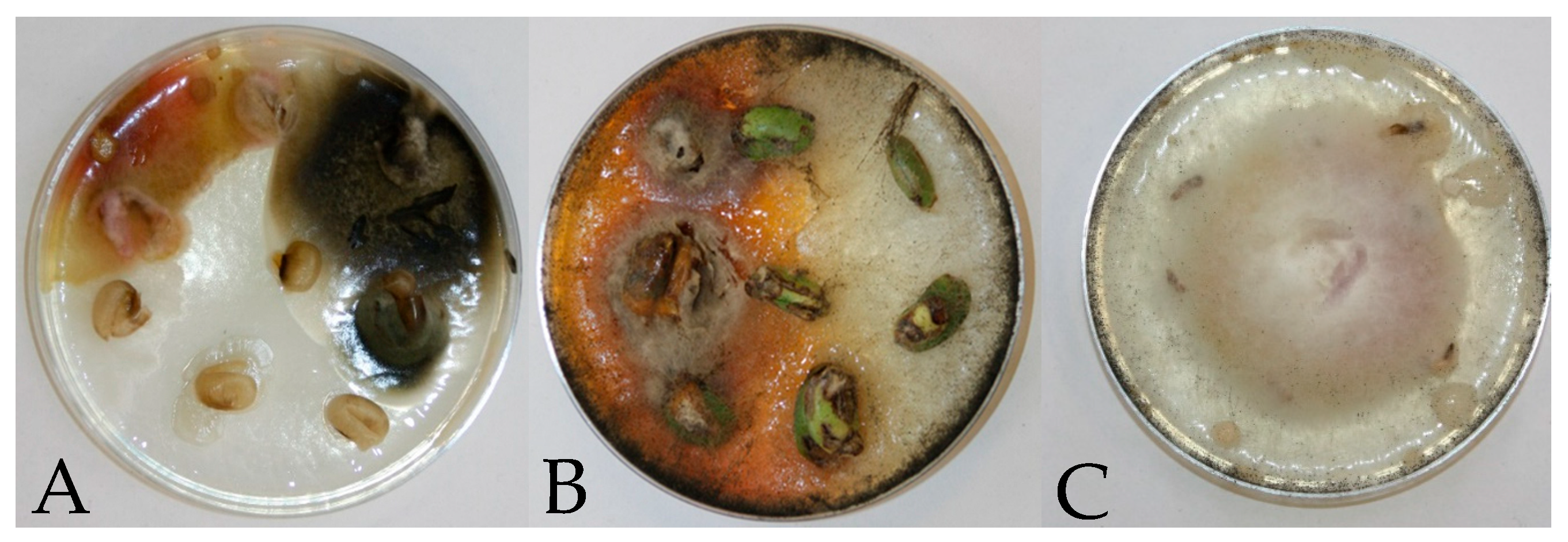
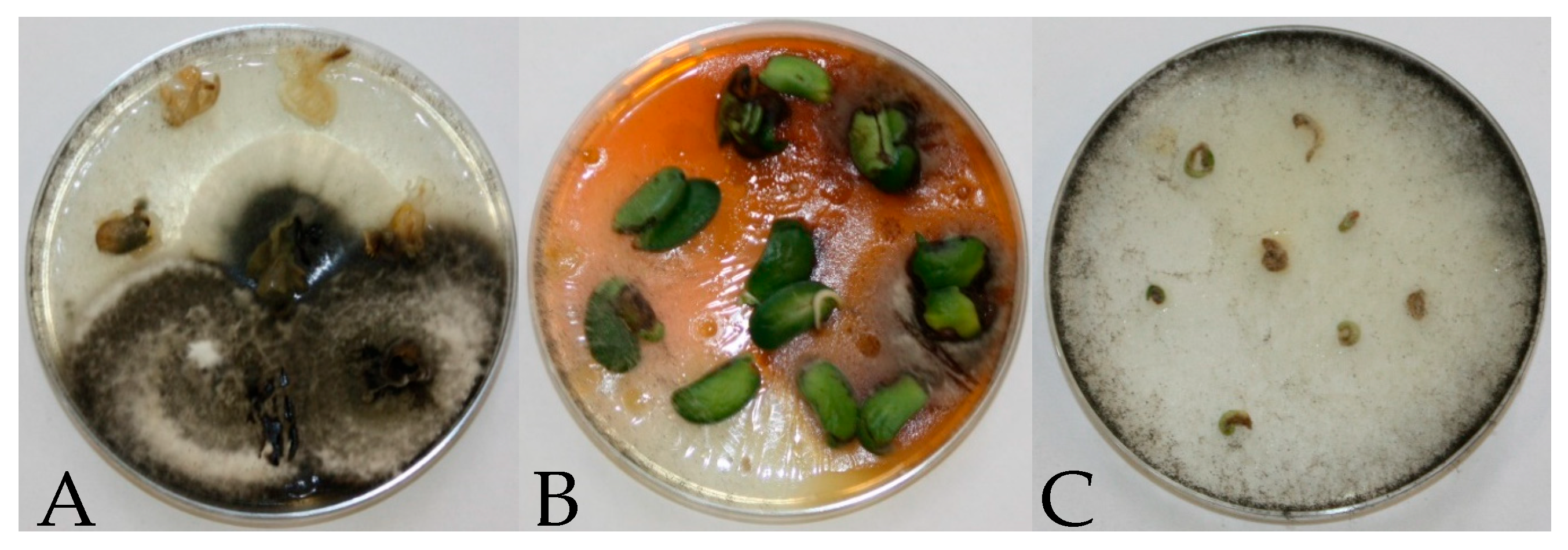
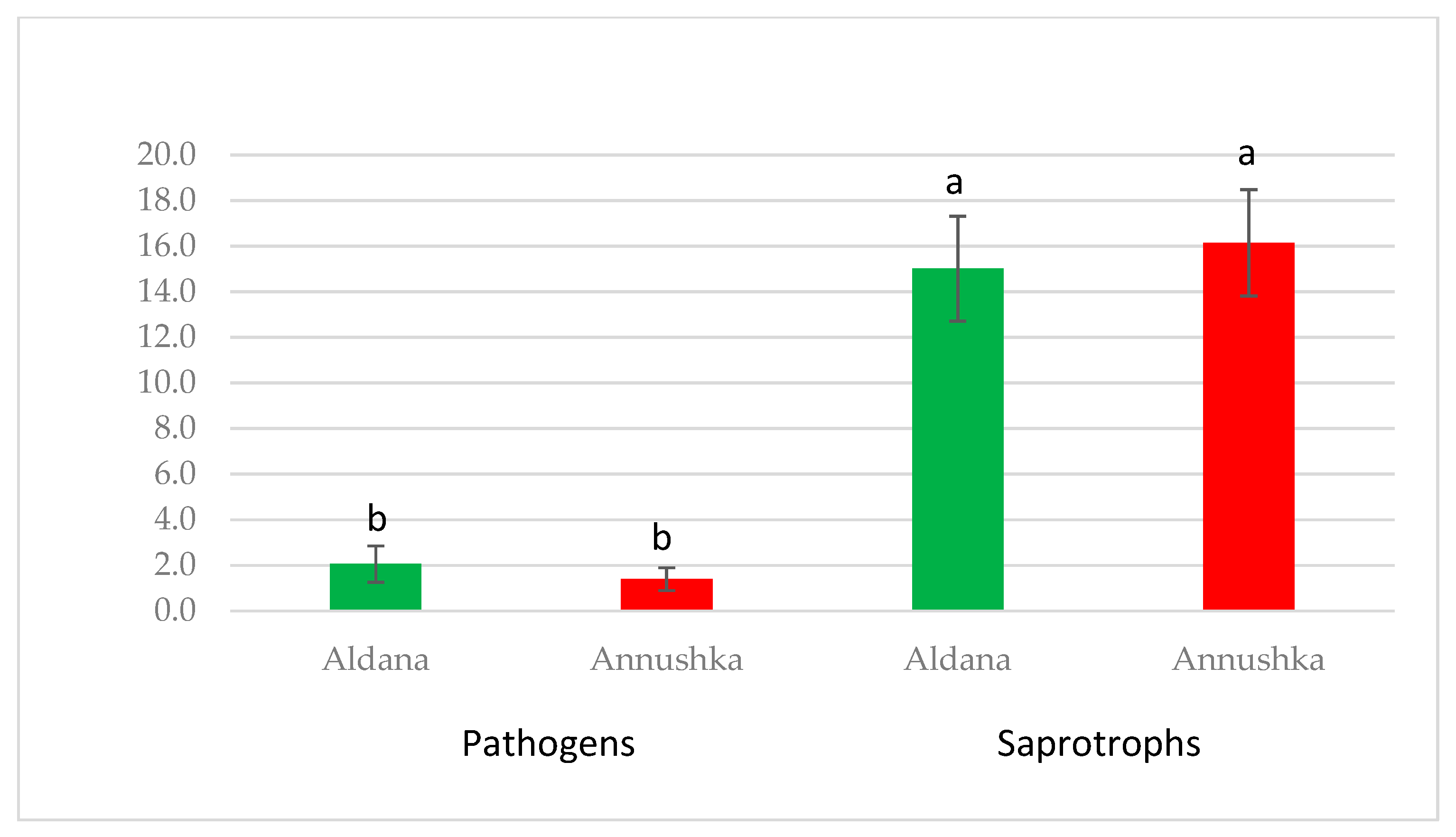
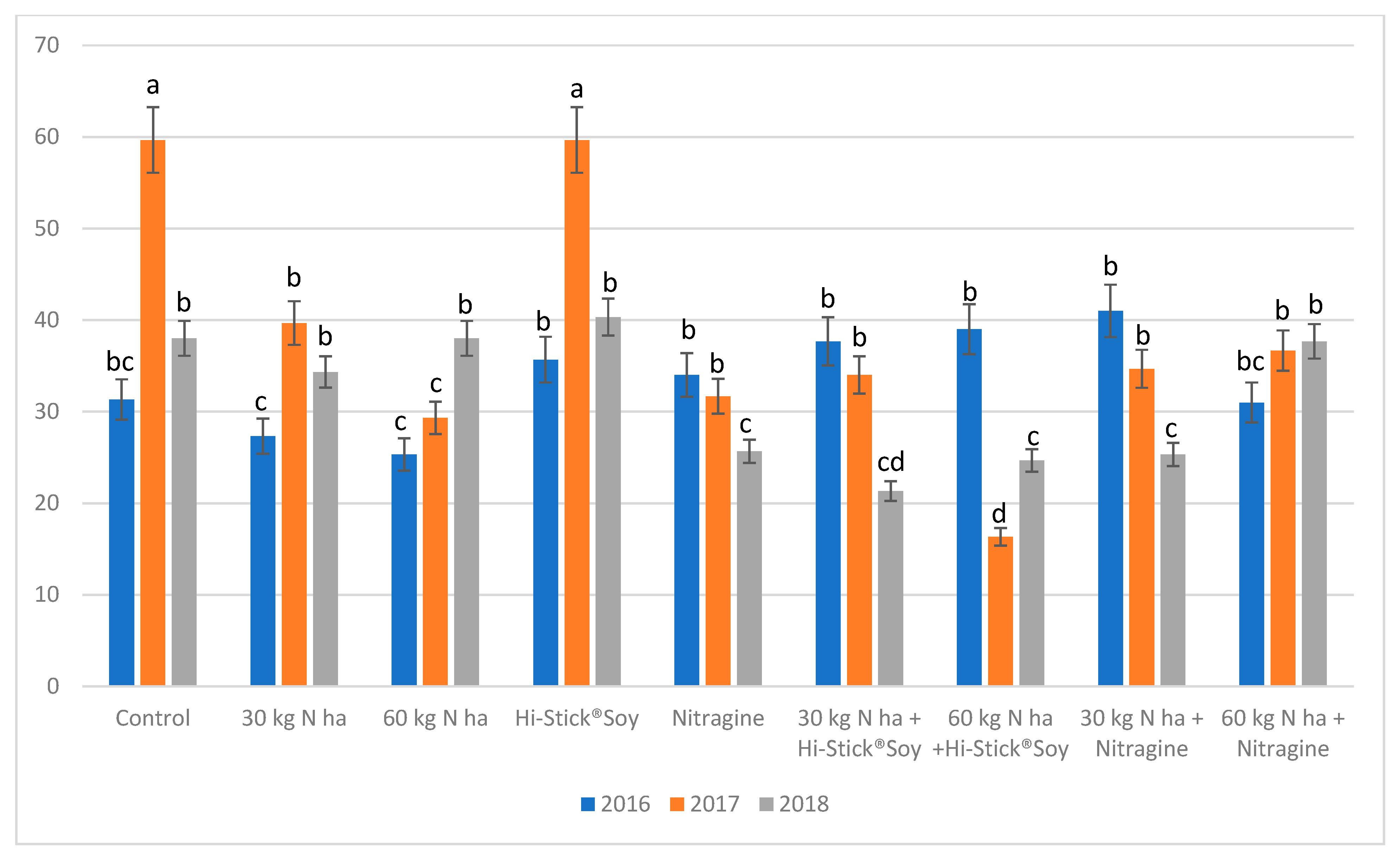
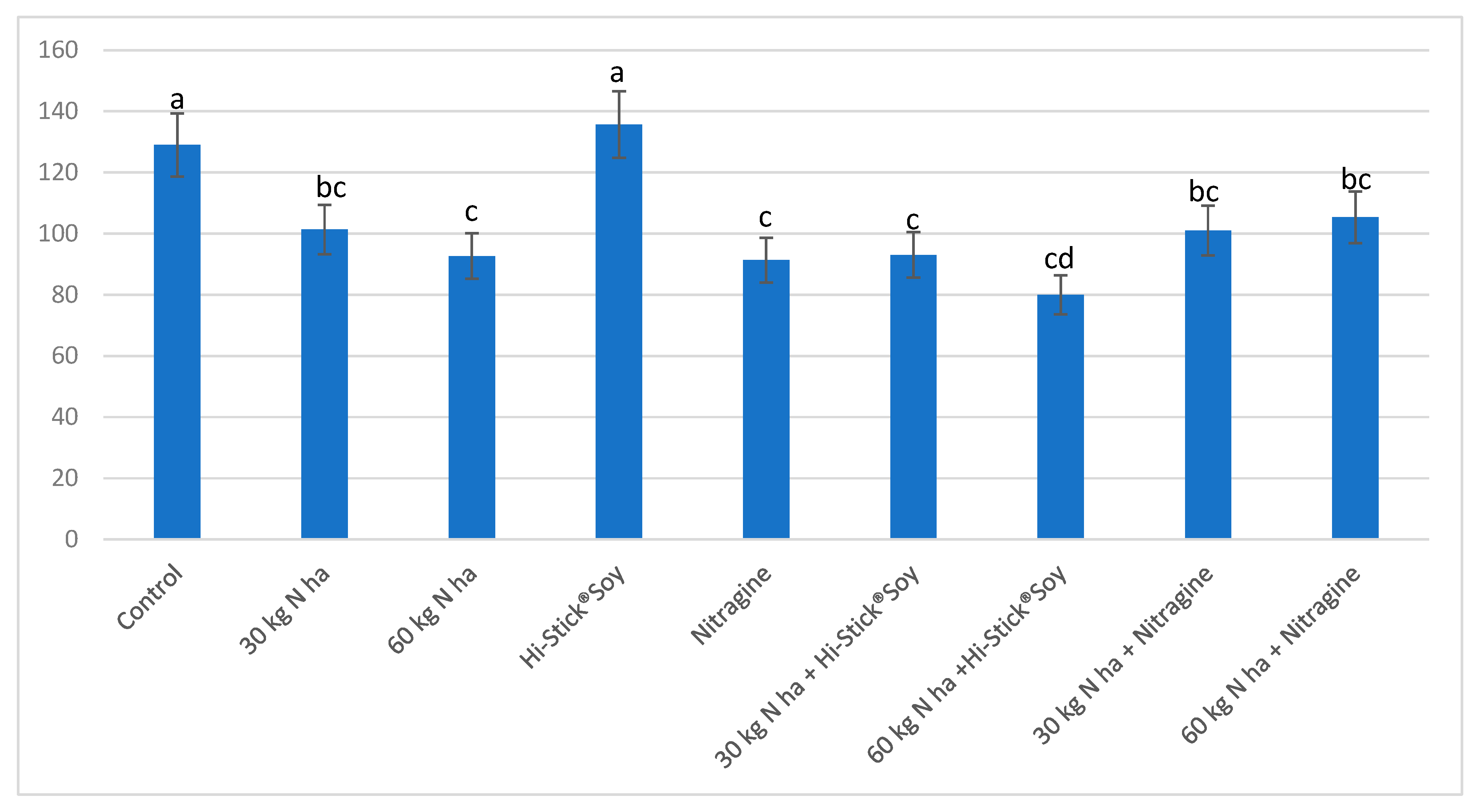
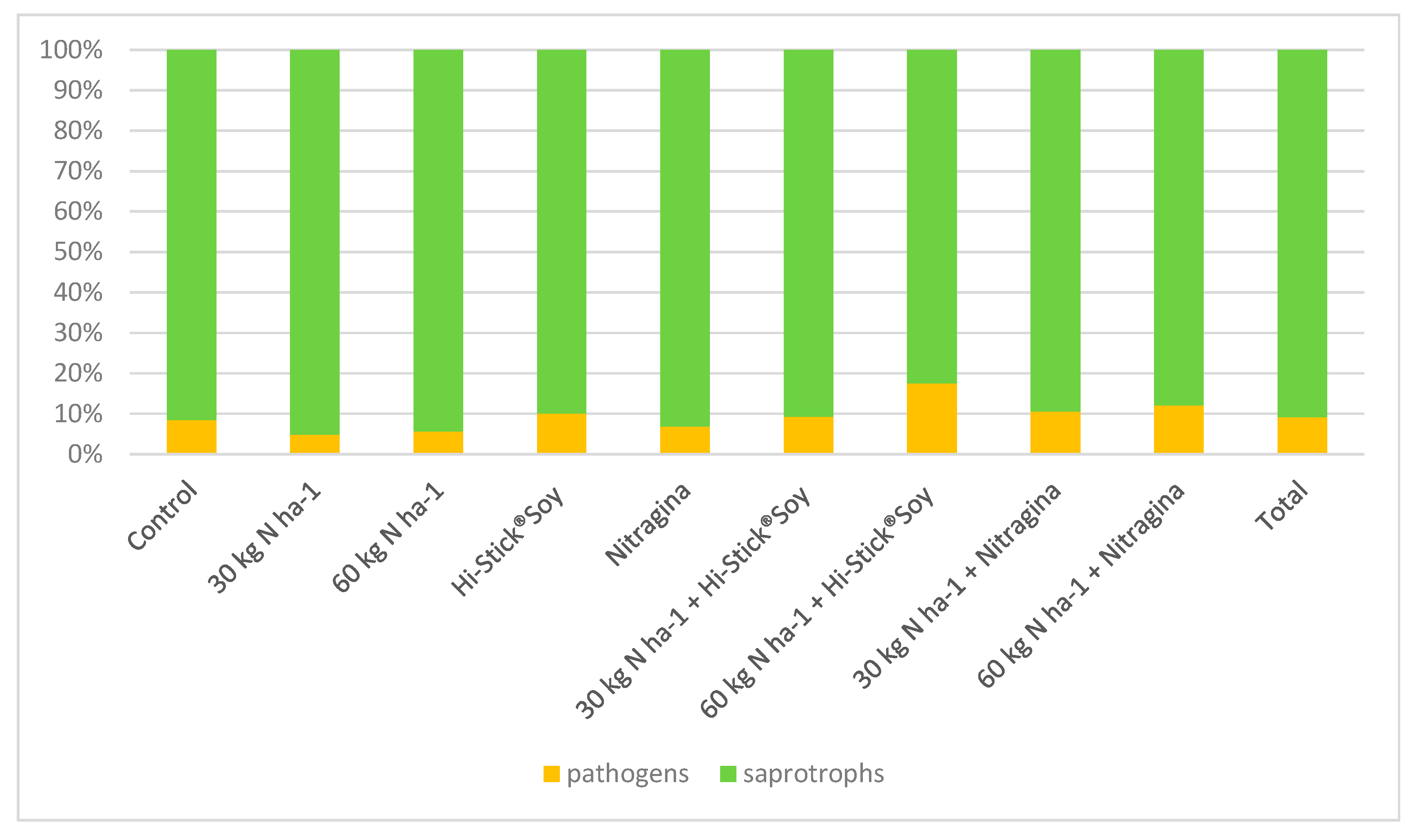

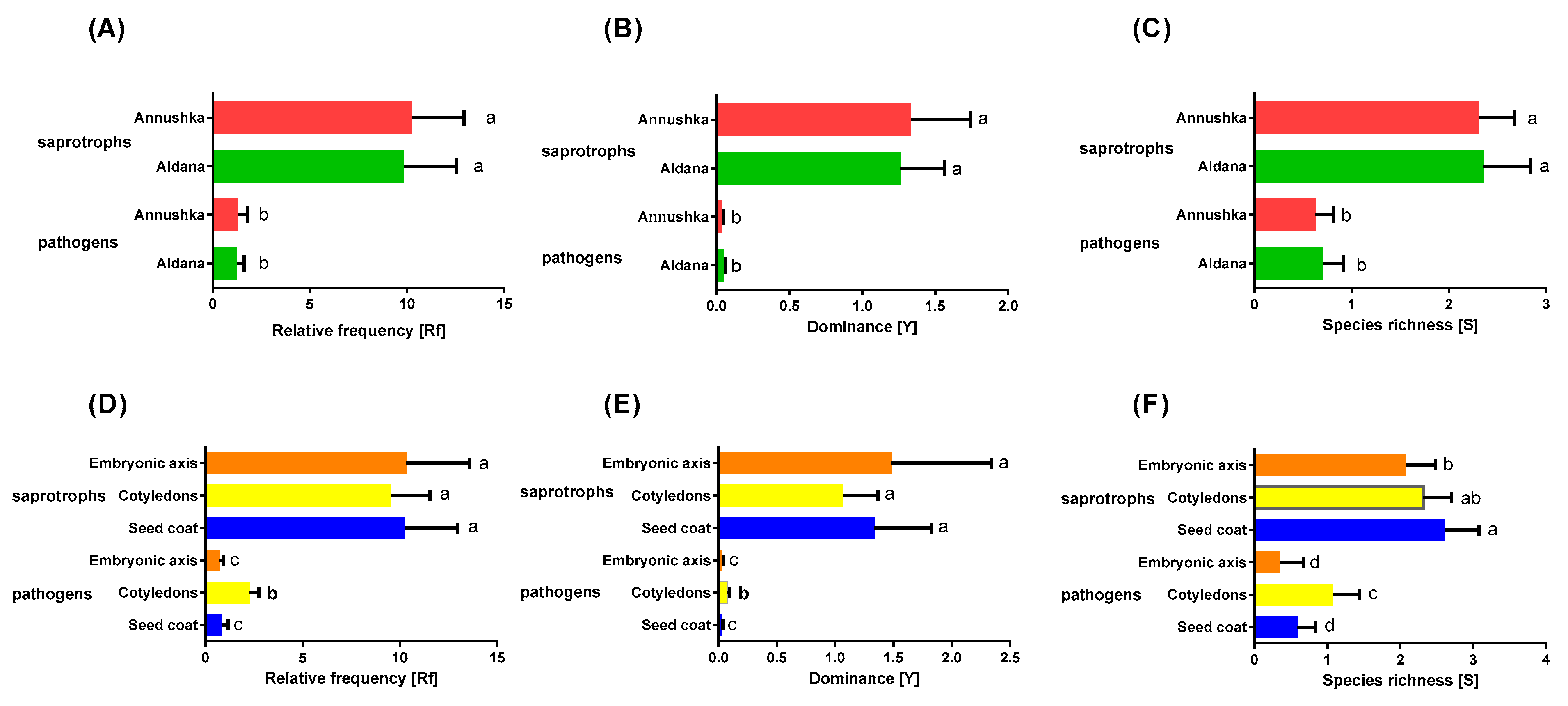
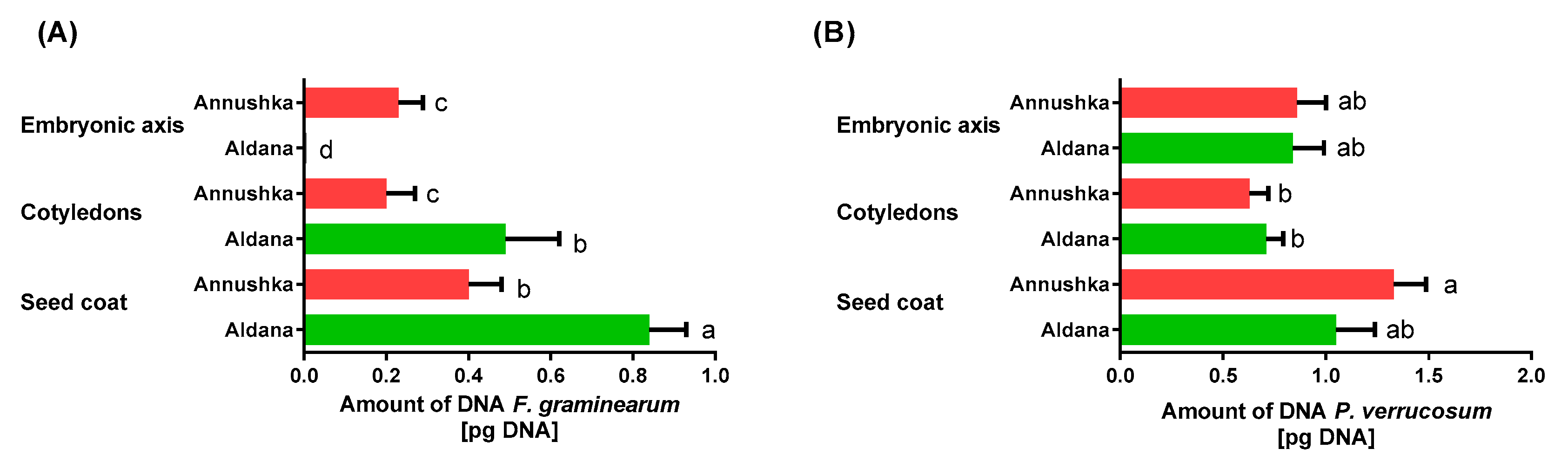
| Year/Month | 2016 | 2017 | 2018 | 1981–2015 * |
|---|---|---|---|---|
| Precipitation (mm) | ||||
| April | 33.1 | 52.1 | 28.6 | 30.0 |
| May | 70.8 | 34.0 | 41.0 | 59.0 |
| June | 66.3 | 109.9 | 64.7 | 72.0 |
| July | 138.6 | 106.1 | 140.7 | 85.0 |
| August | 71.9 | 54.8 | 31.2 | 66.0 |
| September | 17.1 | 211.6 | 29.1 | 55.0 |
| Temperature (°C) | ||||
| April | 8.8 | 6.7 | 11.9 | 7.8 |
| May | 14.8 | 13 | 16.5 | 13.3 |
| June | 13.0 | 16.7 | 17.9 | 15.9 |
| July | 18.5 | 17.2 | 19.9 | 18.3 |
| August | 17.5 | 18.8 | 20.5 | 17.9 |
| September | 14.8 | 13.3 | 15.3 | 13.1 |
| Anatomical Part/Year | 2016 | 2017 | 2018 | Total |
|---|---|---|---|---|
| Aldana | ||||
| Seed coat | 116 * | 173 | 162 | 451 |
| Cotyledons | 171 | 208 | 205 | 584 |
| Embryonic axis | 167 | 62 | 104 | 333 |
| Total isolates | 454 | 443 | 471 | 1368 |
| Annushka | ||||
| Seed coat | 133 | 204 | 169 | 506 |
| Cotyledons | 165 | 248 | 141 | 554 |
| Embryonic axis | 155 | 130 | 75 | 360 |
| Total isolates | 453 | 582 | 386 | 1420 |
| Total fungal isolates | 907 | 1025 | 856 | 2788 |
| Fungal Genus/Species | Treatment | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Seed coat | |||||||||
| Alternaria alternata (Fr.) Keissler | 1 * | 2 | 2 | 3 | 5 | 3 | |||
| Cladosporium cladosporioides (Fries.) de Vries | 2 | 2 | 3 | 3 | 2 | 4 | 2 | ||
| Colletotrichum spp. | 1 | ||||||||
| Fusarium spp. | 2 | ||||||||
| Penicillium spp. | 3 | 3 | 7 | 3 | 3 | 4 | 2 | ||
| Rhizopus nigricans Ehrenb. | 5 | 6 | 5 | 3 | 6 | 6 | 7 | 6 | 6 |
| Trichoderma spp. | 4 | ||||||||
| Total | 11 | 11 | 17 | 12 | 12 | 12 | 13 | 15 | 13 |
| % pathogens | 0.0 | 0.0 | 0.0 | 16.7 | 8.3 | 0.0 | 0.0 | 0.0 | 0.0 |
| % saprotrophs | 100.0 | 100.0 | 100.00 | 83.3 | 91.7 | 100.0 | 100.0 | 100.0 | 100.0 |
| Cotyledons | |||||||||
| Alternaria alternata (Fr.) Keissler | 7 | ||||||||
| Cladosporium cladosporioides (Fries.) de Vries | 3 | 3 | 2 | ||||||
| Fusarium culmorum (W.G. Smith) Sacc. | 8 | 4 | |||||||
| Fusarium spp. | 7 | ||||||||
| Penicillium spp. | 14 | 12 | 7 | 15 | 12 | 2 | 6 | 11 | |
| Rhizopus nigricans Ehrenb. | 7 | 8 | 5 | 7 | 8 | 5 | 6 | 5 | 6 |
| Verticillium spp. | 1 | ||||||||
| Total | 21 | 20 | 12 | 22 | 23 | 16 | 22 | 16 | 19 |
| % pathogens | 0.00 | 0.00 | 0.0 | 0.0 | 0.0 | 50.0 | 31.8 | 31.25 | 0.0 |
| % saprotrophs | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 50.0 | 68.2 | 68.75 | 100.0 |
| Embryonic axis | |||||||||
| Alternaria alternata (Fr.) Keissler | 1 | 7 | 1 | ||||||
| Cladosporium cladosporioides (Fries.) de Vries | 3 | 1 | 2 | 1 | 3 | 2 | 1 | ||
| Colletotrichum spp. | 3 | ||||||||
| Fusarium culmorum (W.G. Smith) Sacc. | 2 | ||||||||
| Fusarium spp. | 1 | 1 | |||||||
| Penicillium spp. | 8 | 4 | 9 | 4 | 24 | 8 | 18 | 11 | |
| Rhizopus nigricans Ehrenb. | 8 | 3 | 2 | 7 | 5 | 6 | 9 | 6 | 6 |
| Total | 19 | 7 | 5 | 19 | 13 | 33 | 26 | 25 | 20 |
| % pathogens | 0.0 | 0.0 | 20.0 | 5.3 | 23.1 | 0.0 | 0.0 | 0.0 | 10.0 |
| % saprotrophs | 100.0 | 100.0 | 80.0 | 94.7 | 76.9 | 100.0 | 100.0 | 100.0 | 90.0 |
| Total number of fungal isolates obtained from seeds | 51 | 38 | 34 | 53 | 48 | 61 | 61 | 56 | 52 |
| Fungal Genus/Species | Treatment | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Seed coat | |||||||||
| Alternaria alternata (Fr.) Keissler | 5 * | 2 | |||||||
| Cladosporium cladosporioides (Fries.) de Vries | 3 | ||||||||
| Colletotrichum spp. | 2 | ||||||||
| Epicoccum nigrum Ehrenb. | 1 | ||||||||
| Penicillium spp. | 9 | 3 | 2 | 4 | 1 | 3 | 3 | 9 | |
| Rhizopus nigricans Ehrenb. | 6 | 10 | 14 | 6 | 12 | 10 | 11 | 8 | 9 |
| Total | 15 | 13 | 16 | 15 | 13 | 16 | 16 | 18 | 11 |
| % pathogens | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 18.1 |
| % saprotrophs | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 81.8 |
| Cotyledons | |||||||||
| Alternaria alternata (Fr.) Keissler | 2 | 4 | 3 | ||||||
| Colletotrichum spp. | 1 | ||||||||
| Fusarium avenaceum (Corda ex Fr.) Sacc. | 1 | ||||||||
| Fusarium culmorum (W.G. Smith) Sacc. | 1 | 1 | 3 | 2 | |||||
| Fusarium spp. | 2 | 2 | 2 | 2 | 3 | ||||
| Penicillium spp. | 4 | 3 | 7 | 3 | 7 | 8 | 2 | 4 | |
| Rhizopus nigricans Ehrenb. | 6 | 9 | 9 | 11 | 18 | 12 | 12 | 14 | 7 |
| Total | 15 | 16 | 16 | 19 | 22 | 21 | 26 | 16 | 14 |
| % pathogens | 20.0 | 0.0 | 0.0 | 10.5 | 18.2 | 9.5 | 23.1 | 0.0 | 21.4 |
| % saprotrophs | 80.0 | 100.0 | 100.0 | 89.5 | 81.8 | 90.5 | 76.9 | 100.0 | 78.5 |
| Embryonic axis | |||||||||
| Alternaria alternata (Fr.) Keissler | 3 | 1 | |||||||
| Cladosporium cladosporioides (Fries.) de Vries | 1 | 1 | 1 | ||||||
| Fusarium culmorum (W.G. Smith) Sacc. | 2 | ||||||||
| Penicillium spp. | 6 | 4 | 3 | 5 | 7 | 6 | 5 | 21 | |
| Rhizopus nigricans Ehrenb. | 6 | 11 | 7 | 15 | 12 | 9 | 8 | 9 | 12 |
| Total | 13 | 15 | 10 | 20 | 19 | 15 | 14 | 33 | 16 |
| % pathogens | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12.5 |
| % saprotrophs | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 87.5 |
| Total number of fungal isolates obtained from seeds | 43 | 44 | 42 | 54 | 54 | 52 | 56 | 67 | 41 |
| Fungal Genus/Species | Treatment | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Seed coat | |||||||||
| Alternaria alternata (Fr.) Keissler | 41 * | 7 | 17 | 6 | 5 | 2 | 7 | 8 | |
| Botrytis cinerea Pers ex Pers | 2 | ||||||||
| Cladosporium cladosporioides (Fries.) de Vries | 7 | 2 | 5 | 2 | |||||
| Diaporthe spp. | 1 | ||||||||
| Epicoccum nigrum Ehrenb. | 2 | ||||||||
| Fusarium avenaceum (Corda ex Fr.) Sacc. | 6 | 5 | |||||||
| Fusarium tricinctum (Corda) Sacc. | 4 | ||||||||
| Fusarium spp. | 3 | 3 | 2 | ||||||
| Penicillium spp. | 4 | 2 | 3 | ||||||
| Rhizopus nigricans Ehrenb. | 6 | 4 | 2 | 4 | 4 | 7 | |||
| Total | 53 | 17 | 24 | 20 | 4 | 5 | 12 | 21 | 17 |
| % pathogens | 22.6 | 0.0 | 0.0 | 40.0 | 50.0 | 0.0 | 33.3 | 0.0 | 0.0 |
| % saprotrophs | 77.4 | 100.0 | 100.0 | 60.0 | 50.0 | 100.0 | 66.7 | 100.0 | 100.0 |
| Cotyledons | |||||||||
| Alternaria alternata (Fr.) Keissler | 38 | 36 | 18 | 27 | 13 | 4 | 6 | 11 | 9 |
| Botrytis cinerea Pers. ex Pers. | 1 | ||||||||
| Cladosporium cladosporioides (Fries.) de Vries | 4 | 1 | |||||||
| Diaporthe spp. | 1 | ||||||||
| Fusarium avenaceum (Corda ex Fr.) Sacc. | 2 | 2 | 2 | ||||||
| Fusarium equiseti (Corda) Sacc. | 2 | 4 | 2 | ||||||
| Fusarium spp. | 2 | ||||||||
| Penicillium spp. | 3 | 1 | 2 | 2 | 2 | ||||
| Rhizopus nigricans Ehrenb. | 3 | 3 | 3 | 3 | 1 | ||||
| Total | 43 | 38 | 18 | 41 | 21 | 4 | 8 | 21 | 14 |
| % pathogens | 4.7 | 5.3 | 0.0 | 9.8 | 19.1 | 0.0 | 0.0 | 19.1 | 14.3 |
| % saprotrophs | 95.3 | 94.7 | 100.0 | 90.2 | 80.9 | 100.0 | 100.0 | 80.9 | 85.7 |
| Embryonic axis | |||||||||
| Alternaria alternata (Fr.) Keissler | 7 | 4 | 5 | ||||||
| Botrytis cinerea Pers. ex Pers. | 4 | ||||||||
| Cladosporium cladosporioides (Fries.) de Vries | 2 | ||||||||
| Diaporthe spp. | 1 | ||||||||
| Fusarium avenaceum (Corda ex Fr.) Sacc. | 2 | ||||||||
| Penicillium spp. | 2 | 4 | 1 | 2 | 2 | 4 | 9 | ||
| Rhizopus nigricans Ehrenb. | 2 | 1 | 1 | 7 | |||||
| Trichoderma spp. | 2 | ||||||||
| Total | 12 | 2 | 6 | 11 | 3 | 4 | 3 | 5 | 16 |
| % pathogens | 41.7 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| % saprotrophs | 58.3 | 0.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| Total number of fungal isolates obtained from seeds | 108 | 57 | 48 | 72 | 28 | 13 | 23 | 47 | 47 |
| Fungal Genus/Species | Treatment | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Seed coat | |||||||||
| Alternaria alternata (Fr.) Keissler | 9 * | 4 | 9 | 43 | 2 | 19 | 2 | 4 | |
| Aureobasidium spp. | 4 | ||||||||
| Btrytis cinerea Pers. ex Pers. | 1 | ||||||||
| Cladosporium cladosporioides (Fries.) de Vries | 3 | ||||||||
| Diaporthe spp. | 1 | ||||||||
| Fusarium avenaceum (Corda ex Fr.) Sacc. | 2 | 4 | 2 | ||||||
| Penicillium spp. | 2 | 2 | 2 | 5 | 21 | 7 | 2 | 9 | |
| Rhizopus nigricans Ehrenb. | 1 | 6 | 14 | 2 | 2 | 2 | 3 | ||
| Trichoderma spp. | 15 | ||||||||
| Total | 14 | 12 | 13 | 51 | 24 | 44 | 11 | 8 | 27 |
| % pathogens | 0.0 | 0.0 | 15.4 | 11.8 | 0.0 | 4.5 | 0.0 | 0.0 | 0.0 |
| % saprotrophs | 100.0 | 100.0 | 84.6 | 88.2 | 100.0 | 95.5 | 100.0 | 100.0 | 100.0 |
| Cotyledons | |||||||||
| Alternaria alternata (Fr.) Keissler | 31 | 21 | 10 | 29 | 18 | 22 | 4 | 21 | 3 |
| Fusarium avenaceum (Corda ex Fr.) Sacc. | 2 | 6 | 4 | 2 | 2 | ||||
| Fusarium equiseti (Corda) Sacc. | 2 | ||||||||
| Fusarium poae (Peck) Wollenw. | 2 | ||||||||
| Fusarium tricinctum (Corda) Sacc. | 2 | 2 | |||||||
| Fusarium spp. | 2 | ||||||||
| Penicillium spp. | 8 | 2 | 2 | 4 | 5 | 12 | |||
| Phoma glomerata (Corda) Wollenw. | 1 | ||||||||
| Rhizopus nigricans Ehrenb. | 6 | 4 | 2 | 5 | 4 | 2 | |||
| Stemphylium botryosum Wallr. | 6 | ||||||||
| Total | 48 | 31 | 18 | 33 | 25 | 32 | 8 | 34 | 19 |
| % pathogens | 4.2 | 12.9 | 33.3 | 12.1 | 8.0 | 12.5 | 0.0 | 0.0 | 21.1 |
| % saprotrophs | 95.8 | 87.1 | 66.7 | 87.9 | 92.0 | 87.5 | 100.0 | 100.0 | 78.9 |
| Embryonic axis | |||||||||
| Alternaria alternata (Fr.) Keissler | 5 | 20 | 12 | 8 | 2 | ||||
| Botrytis cinerea Pers. ex Pers. | 1 | ||||||||
| Fusarium avenaceum (Corda ex Fr.) Sacc. | 2 | ||||||||
| Fusarium spp. | 2 | 2 | |||||||
| Penicillium spp. | 2 | 8 | 9 | 2 | 3 | 5 | 15 | 12 | |
| Rhizopus nigricans Ehrenb. | 4 | ||||||||
| Trichoderma spp. | 11 | 5 | |||||||
| Total | 9 | 19 | 9 | 23 | 18 | 13 | 7 | 15 | 17 |
| % pathogens | 44.4 | 0.0 | 0.0 | 13.0 | 0.0 | 15.4 | 0.0 | 0.0 | 0.0 |
| % saprotrophs | 55.6 | 100.0 | 100.0 | 87.0 | 100.0 | 84.6 | 100.0 | 100.0 | 100.0 |
| Total number of fungal isolates obtained from seeds | 71 | 62 | 40 | 107 | 67 | 89 | 26 | 57 | 63 |
| Fungal Genus/Species | Treatment | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Seed coat | |||||||||
| Alternaria alternata (Fr.) Keissler | 3 * | 5 | 5 | 5 | 9 | ||||
| Cladosporium cladosporioides (Fries.) de Vries | 18 | ||||||||
| Fusarium culmorum(W.G. Smith) Sacc. | 2 | 2 | 2 | ||||||
| Fusarium oxysporum Schltdl. | 2 | 2 | 4 | ||||||
| Fusarium solani (Mart.) Sacc. | 2 | 2 | |||||||
| Penicillium spp. | 9 | 22 | 23 | 10 | 15 | 4 | 3 | ||
| Periconia spp. | 2 | ||||||||
| Rhizopus nigricans Ehrenb. | 4 | 1 | 3 | 3 | |||||
| Trichoderma spp. | |||||||||
| Total | 18 | 30 | 33 | 12 | 22 | 18 | 15 | 11 | 3 |
| % pathogens | 11.1 | 0.0 | 6.1 | 16.7 | 9.1 | 0.0 | 53.3 | 18.2 | 0.0 |
| % saprotrophs | 88.9 | 100.0 | 95.9 | 83.3 | 90.9 | 100.0 | 46.7 | 81.8 | 100.0 |
| Cotyledons | |||||||||
| Alternaria alternata (Fr.) Keissler | 24 | 4 | 4 | 17 | 3 | 5 | 5 | ||
| Cladosporium cladosporioides (Fries.) de Vries | 2 | 4 | 2 | 2 | |||||
| Fusarium avenaceum (Corda ex Fr.) Sacc. | 2 | ||||||||
| Fusarium culmorum (W.G. Smith) Sacc. | 2 | 2 | 2 | ||||||
| Fusarium oxysporum Schltdl. | 3 | 3 | 4 | 4 | 2 | 7 | |||
| Fusarium solani (Mart.) Sacc. | 4 | 1 | 4 | 1 | |||||
| Penicillium spp. | 18 | 24 | 14 | 4 | 8 | 11 | 4 | 5 | |
| Rhizopus nigricans Ehrenb. | 2 | 2 | |||||||
| Total | 35 | 22 | 33 | 18 | 24 | 18 | 22 | 13 | 20 |
| % pathogens | 25.7 | 0.0 | 15.1 | 0.0 | 12.5 | 33.3 | 36.4 | 15.4 | 40.0 |
| % saprotrophs | 74.3 | 100.0 | 84.9 | 100.0 | 87.5 | 66.7 | 63.6 | 84.6 | 60.0 |
| Embryonic axis | |||||||||
| Alternaria alternata (Fr.) Keissler | 4 | 5 | 3 | 2 | |||||
| Cladosporium cladosporioides (Fries.) de Vries | 4 | 4 | 2 | ||||||
| Fusarium culmorum (W.G. Smith) Sacc. | 2 | ||||||||
| Fusarium oxysporum Schltdl. | 3 | 2 | 2 | ||||||
| Fusarium solani (Mart.) Sacc. | 2 | ||||||||
| Penicillium spp. | 16 | 15 | 17 | 7 | 6 | 4 | |||
| Rhizopus nigricans Ehrenb. | 1 | 1 | 2 | ||||||
| Total | 4 | 17 | 21 | 25 | 7 | 4 | 16 | 6 | 4 |
| % pathogens | 0.0 | 0.0 | 0.0 | 20.00 | 0.0 | 0.0 | 25.0 | 0.0 | 50.0 |
| % saprotrophs | 100.0 | 100.0 | 100.0 | 80.0 | 100.0 | 100.0 | 75.0 | 100.0 | 50.0 |
| Total number of fungal isolates obtained from seeds | 57 | 69 | 87 | 55 | 53 | 40 | 53 | 30 | 27 |
| Fungal Genus/Species | Treatment | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Seed coat | |||||||||
| Alternaria alternata (Fr.) Keissler | 18 * | 4 | 10 | 5 | 3 | 2 | |||
| Cladosporium cladosporioides (Fries.) de Vries | 2 | 2 | 3 | ||||||
| Fusarium oxysporum Schltdl. | 2 | 2 | 2 | 5 | 2 | ||||
| Fusarium solani (Mart.) Sacc. | 2 | 3 | |||||||
| Penicillium spp. | 4 | 6 | 4 | 27 | 8 | 2 | 1 | 8 | 31 |
| Rhizopus nigricans Ehrenb. | 3 | 1 | 1 | 4 | 1 | 1 | |||
| Total | 24 | 12 | 9 | 37 | 11 | 10 | 10 | 17 | 39 |
| % pathogens | 8.3 | 16.7 | 0.0 | 0.0 | 0.0 | 20.0 | 20.0 | 47.1 | 5.1 |
| % saprotrophs | 91.7 | 83.3 | 100.0 | 100.0 | 100.0 | 80.0 | 80.0 | 52.9 | 94.9 |
| Cotyledons | |||||||||
| Alternaria alternata (Fr.) Keissler | 5 | 2 | 2 | 5 | |||||
| Aspergillus spp. | 1 | ||||||||
| Cladosporium cladosporioides (Fries.) de Vries | 1 | 3 | |||||||
| Fusarium oxysporum Schltdl. | 2 | 4 | 2 | 2 | 3 | ||||
| Fusarium solani (Mart.) Sacc. | 2 | 2 | 5 | ||||||
| Penicillium spp. | 14 | 16 | 2 | 11 | 9 | 4 | 22 | 11 | |
| Rhizopus nigricans Ehrenb. | 2 | 2 | 2 | 2 | 3 | ||||
| Total | 25 | 21 | 4 | 14 | 12 | 8 | 9 | 26 | 22 |
| % pathogens | 16.0 | 19.0 | 0.0 | 0.0 | 0.0 | 0.0 | 22.2 | 15.4 | 36.4 |
| % saprotrophs | 84.0 | 81.0 | 100.0 | 100.0 | 100.0 | 100.0 | 77.8 | 84.6 | 63.6 |
| Embryonic axis | |||||||||
| Alternaria alternata (Fr.) Keissler | 4 | 3 | |||||||
| Cladosporium cladosporioides (Fries.) de Vries | 3 | ||||||||
| Fusarium oxysporum Schltdl. | 1 | 2 | |||||||
| Fusarium solani (Mart.) Sacc. | 2 | ||||||||
| Penicillium spp. | 9 | 15 | 1 | 6 | 1 | 1 | 19 | ||
| Rhizopus nigricans Ehrenb. | 4 | 2 | 1 | 2 | |||||
| Total | 8 | 1 | 14 | 15 | 1 | 6 | 2 | 3 | 25 |
| % pathogens | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 50.0 | 0.0 | 16.0 |
| % saprotrophs | 100.0 | 0.0 | 100.0 | 100.0 | 100.0 | 100.0 | 50.0 | 100.0 | 84.0 |
| Total number of fungal colonies isolated from seeds | 57 | 34 | 27 | 66 | 24 | 24 | 21 | 46 | 86 |
| Parameter | Cultivar (C) | Anatomical part (APSS) | Treatment (T) | Year (Y) | C × APSS | C × T | C × Y | APSS × T | APSS × Y | T × Y | C × APSS × T | C × APSS × Y | C × T × Y | APSS × T × Y | C × APSS × T × Y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of fungal isolates | ns | ** | * | ** | ** | ** | * | * | ** | * | ** | ** | * | ** | ** | |
| Relative frequency [Rf] | pathogens | ns | * | ns | * | * | ns | * | * | * | * | * | * | * | * | * |
| saprotrophs | ns | * | ns | * | * | ns | * | * | * | * | * | * | * | * | * | |
| Dominance [Y] | pathogens | ns | * | ns | * | * | ns | * | * | * | * | * | * | * | * | * |
| saprotrophs | ns | * | ns | * | * | ns | * | * | * | * | * | * | * | * | * | |
| Species richness [S] | pathogens | ns | * | ns | * | * | ns | * | * | * | * | * | * | * | * | * |
| saprotrophs | ns | ** | ns | ** | ** | ns | ** | ** | ** | ** | ** | ** | ** | ** | * | |
| F. graminearum DNA | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | * | * | |
| P. verrucosum DNA | * | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | |
| Fungal Genera | Seed Coat | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | Total | % | 2016 | 2017 | 2018 | Total | % | Total | % | |
| Aldana | Annushka | |||||||||||
| Alternaria | 16 * | 93 | 27 | 136 | 30.16 | 7 | 92 | 42 | 141 | 27.86 | 277 | 28.95 |
| Aureobasidium | 0 | 4 | 4 | 0.79 | 4 | 0.42 | ||||||
| Botrytis | 2 | 2 | 0.44 | 1 | 1 | 0.20 | 3 | 0.31 | ||||
| Cladosporium | 18 | 16 | 18 | 52 | 11.53 | 3 | 3 | 7 | 13 | 2.57 | 65 | 6.79 |
| Colletotrichum | 1 | 1 | 0.22 | 2 | 2 | 0.40 | 3 | 0.31 | ||||
| Diaporthe | 1 | 1 | 0.22 | 1 | 1 | 0.20 | 2 | 0.21 | ||||
| Epicoccum | 2 | 2 | 0.44 | 1 | 1 | 0.20 | 3 | 0.31 | ||||
| Fusarium | 2 | 23 | 18 | 43 | 9.53 | 8 | 18 | 26 | 5.14 | 69 | 7.21 | |
| Penicillium | 25 | 9 | 86 | 120 | 26.61 | 34 | 50 | 91 | 175 | 34.58 | 295 | 30.82 |
| Periconia | 2 | 2 | 0.44 | 0 | 2 | 0.21 | ||||||
| Rhizopus | 50 | 27 | 11 | 88 | 19.52 | 86 | 30 | 11 | 127 | 25.10 | 215 | 22.48 |
| Trichoderma | 4 | 4 | 0.89 | 15 | 15 | 2.96 | 19 | 1.98 | ||||
| Total | 116 | 173 | 162 | 451 | 100 | 133 | 204 | 169 | 506 | 100 | 957 | 100 |
| Fungal Genera | Cotyledons | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | Total | % | 2016 | 2017 | 2018 | Total | % | Total | % | |
| Aldana | Annushka | |||||||||||
| Alternaria | 7 * | 162 | 62 | 231 | 39.55 | 9 | 159 | 14 | 182 | 32.85 | 413 | 36.29 |
| Aspergillus | 0 | 1 | 1 | 0.18 | 1 | 0.09 | ||||||
| Cladosporium | 8 | 5 | 10 | 23 | 3.94 | 4 | 4 | 0.72 | 27 | 2.37 | ||
| Penicillium | 79 | 10 | 88 | 177 | 30.31 | 38 | 33 | 89 | 160 | 28.88 | 337 | 29.61 |
| Rhizopus | 57 | 13 | 4 | 74 | 12.67 | 98 | 23 | 11 | 132 | 23.84 | 206 | 18.10 |
| Stemphylium | 0 | 6 | 6 | 1.08 | 6 | 0.53 | ||||||
| Botrytis | 1 | 1 | 0.17 | 0 | 1 | 0.09 | ||||||
| Colletotrichum | 0 | 1 | 1 | 0.18 | 1 | 0.09 | ||||||
| Diaporthe | 1 | 1 | 0.17 | 0 | 1 | 0.09 | ||||||
| Fusarium | 19 | 16 | 41 | 76 | 13.02 | 19 | 26 | 22 | 67 | 12.09 | 143 | 12.56 |
| Phoma | 0 | 1 | 1 | 0.18 | 1 | 0.09 | ||||||
| Verticillium | 1 | 1 | 0.17 | 0 | 1 | 0.09 | ||||||
| Total | 171 | 208 | 205 | 584 | 100 | 165 | 248 | 141 | 554 | 100 | 1138 | 100 |
| Fungal Genera | Embryonic axis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | Total | % | 2016 | 2017 | 2018 | Total | % | Total | % | |
| Aldana | Annushka | |||||||||||
| Alternaria | 9 * | 16 | 14 | 39 | 11.71 | 4 | 47 | 7 | 58 | 16.12 | 97 | 14.00 |
| Cladosporium | 13 | 2 | 10 | 25 | 7.51 | 3 | 3 | 6 | 1.67 | 31 | 4.47 | |
| Penicillium | 86 | 24 | 65 | 175 | 52.55 | 57 | 56 | 52 | 165 | 45.83 | 340 | 49.07 |
| Rhizopus | 52 | 11 | 4 | 67 | 20.12 | 89 | 4 | 9 | 102 | 28.34 | 169 | 24.39 |
| Trichoderma | 2 | 2 | 0.60 | 16 | 16 | 4.44 | 18 | 2.60 | ||||
| Botrytis | 4 | 4 | 1.20 | 1 | 1 | 0.27 | 5 | 0.72 | ||||
| Colletotrichum | 3 | 3 | 0.90 | 0 | 3 | 0.43 | ||||||
| Diaporthe | 1 | 1 | 0.30 | 0 | 1 | 0.14 | ||||||
| Fusarium | 4 | 2 | 11 | 17 | 5.11 | 2 | 6 | 4 | 12 | 3.33 | 29 | 4.18 |
| Total | 167 | 62 | 104 | 333 | 100 | 155 | 130 | 75 | 360 | 100 | 693 | 100 |
| Ten-Day Periods | Pathogens | Saprotrophs | |||||
|---|---|---|---|---|---|---|---|
| Relative Frequency [Rf] | Dominance [Y] | Species Richness [S] | Relative Frequency [Rf] | Dominance [Y] | Species Richness [S] | ||
| 1–10 April | Mean daily temperature | ns | ns | ns | ns | ns | 0.21 * |
| Total precipitation | ns | ns | ns | ns | ns | ns | |
| 11–20 April | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | ns | ns | ns | ns | ns | ns | |
| 21–30 April | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | 0.24 * | 0.23 * | 0.18 * | ns | ns | −0.22 * | |
| 1–10 May | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | ns | ns | ns | ns | ns | ns | |
| 11–20 May | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | ns | ns | ns | ns | ns | ns | |
| 21–31 May | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | ns | ns | −0.19 * | ns | ns | 0.23 * | |
| 1–10 June | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | ns | ns | 0.16 * | ns | ns | −0.17 * | |
| 11–20 June | Mean daily temperature | ns | ns | ns | ns | ns | −0.24 * |
| Total precipitation | 0.29 * | 0.26 * | 0.19 * | ns | ns | ns | |
| 21–30 June | Mean daily temperature | 0.31 * | 0.20 * | 0.29 * | ns | ns | 0.24 |
| Total precipitation | 0.19 * | 0.21 * | 0.16 * | ns | ns | −0.17 * | |
| 1–10 July | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | −0.24 * | −0.23 * | −0.18 * | 0.31 * | 0.28 * | 0.27 * | |
| 11–20 July | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | ns | ns | ns | ns | ns | ns | |
| 21–31 July | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | −0.28 * | −0.26 * | −0.19 * | 0.25 * | 0.21 * | 0.24 * | |
| 1–10 August | Mean daily temperature | 0.31 * | 0.26 * | 0.23 * | ns | ns | −0.24 * |
| Total precipitation | −0.25 * | −0.23 * | −0.27 * | 0.27 | 0.23 | 0.29 | |
| 11–20 August | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | ns | ns | ns | ns | ns | ns | |
| 21–31 August | Mean daily temperature | ns | ns | ns | ns | ns | ns |
| Total precipitation | ns | ns | ns | ns | ns | ns | |
| Treatment | Penicillium spp.—Culture-Based Method | P. verrucosum—qPCR (rRNA) Assay | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | 2016 | 2017 | 2018 | |||||||||||||
| O * | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | |
| Control | + | + | + | _ | _ | _ | + | _ | _ | + | _ | + | _ | _ | + | + | _ | _ |
| 30 kg N ha−1 | + | + | + | + | _ | + | + | + | + | + | _ | + | + | _ | + | + | + | + |
| 60 kg N ha−1 | + | + | _ | _ | _ | + | + | + | + | + | _ | + | + | + | + | + | + | + |
| HiStick®Soy | _ | + | + | _ | + | + | + | + | + | + | + | + | _ | + | _ | + | + | + |
| Nitragina | + | + | + | _ | + | + | + | + | + | + | _ | + | _ | _ | + | + | + | + |
| 30 kg N ha−1 + HiStick®Soy | _ | _ | + | _ | _ | + | _ | + | - | + | _ | + | _ | _ | + | _ | _ | _ |
| 60 kg N ha−1 + HiStick®Soy | + | + | + | + | + | + | + | + | + | + | _ | + | + | + | + | + | + | + |
| 30 kg N ha−1 + Nitragina | + | + | + | + | + | + | _ | + | + | + | _ | + | + | + | + | + | + | _ |
| 60 kg N ha−1 + Nitragina | + | + | + | _ | + | + | + | + | _ | + | _ | + | + | + | + | + | _ | + |
| Treatment | Penicillium spp.—Culture-Based Method | P. verrucosum—qPCR (rRNA) Assay | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | 2016 | 2017 | 2018 | |||||||||||||
| O * | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | |
| Control | + | + | + | _ | + | + | + | + | _ | + | _ | + | + | _ | _ | + | _ | + |
| 30 kg N ha−1 | + | + | + | + | + | + | + | + | _ | + | + | + | + | + | + | + | _ | _ |
| 60 kg N ha−1 | + | + | + | + | _ | + | + | + | + | + | _ | + | + | + | + | + | + | + |
| HiStick®Soy | + | + | + | + | _ | _ | + | + | + | + | _ | + | + | _ | + | + | + | + |
| Nitragina | + | + | + | + | _ | + | + | + | + | + | + | + | + | _ | + | + | + | + |
| 30 kg N ha−1 + HiStick®Soy | + | + | + | + | + | + | + | + | + | + | _ | + | + | + | + | + | + | + |
| 60 kg N ha−1 + HiStick®Soy | + | + | + | + | + | + | + | _ | + | + | + | + | + | + | + | + | + | + |
| 30 kg N ha−1 + Nitragina | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 60 kg N ha−1 + Nitragina | _ | + | _ | + | + | + | + | + | + | + | + | + | + | + | _ | + | + | _ |
| Treatment | Fusarium spp.—Culture-Based Method | Fusarium spp.—PCR Assay | F. graminearum—qPCR (Tri5 gene) Assay | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | 2016 | 2017 | 2018 | 2016 | 2017 | 2018 | |||||||||||||||||||
| O * | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | |
| Control | _ | _ | _ | + | _ | _ | + | + | _ | _ | _ | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ |
| 30 kg N ha−1 | _ | _ | _ | _ | + | + | _ | - | _ | _ | + | _ | _ | + | _ | _ | _ | _ | _ | _ | _ | + | _ | _ | _ | _ | _ |
| 60 kg N ha−1 | _ | _ | + | _ | _ | _ | + | + | _ | _ | _ | + | _ | + | _ | _ | + | _ | _ | + | _ | _ | _ | _ | _ | + | _ |
| HiStick®Soy | + | _ | + | + | _ | _ | + | - | + | _ | _ | + | + | _ | _ | _ | _ | _ | _ | _ | _ | + | _ | _ | _ | _ | _ |
| Nitragina | _ | _ | _ | + | + | _ | + | + | _ | _ | + | _ | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ |
| 30 kg N ha−1 + HiStick®Soy | _ | + | _ | _ | _ | _ | _ | + | _ | _ | _ | + | _ | _ | + | _ | _ | _ | _ | _ | _ | _ | + | _ | _ | _ | _ |
| 60 kg N ha−1 + HiStick®Soy | _ | + | _ | + | _ | _ | + | + | + | _ | + | _ | + | _ | _ | _ | + | _ | _ | _ | _ | _ | + | _ | _ | _ | _ |
| 30 kg N ha−1 + Nitragina | _ | + | _ | _ | + | _ | + | + | _ | _ | _ | + | _ | + | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | + | _ | _ |
| 60 kg N ha−1 + Nitragina | _ | _ | + | _ | + | _ | _ | + | + | _ | _ | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | + | _ | _ | _ | _ | _ |
| Treatment | Fusarium spp.—Culture-Based Method | Fusarium spp.—PCR Assay | F. graminearum—qPCR (Tri5 gene) Assay | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | 2016 | 2017 | 2018 | 2016 | 2017 | 2018 | |||||||||||||||||||
| O * | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | O | L | Z | |
| Control | _ | + | _ | _ | + | + | + | + | _ | _ | + | + | _ | + | _ | _ | _ | _ | _ | _ | + | _ | _ | + | _ | _ | _ |
| 30 kg N ha−1 | _ | _ | _ | _ | + | _ | + | + | + | _ | _ | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | _ | + | _ | _ | _ | _ |
| 60 kg N ha−1 | _ | _ | _ | + | _ | _ | _ | _ | _ | _ | _ | + | _ | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ |
| HiStick®Soy | _ | + | _ | + | _ | + | _ | _ | _ | _ | + | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | + | _ | _ | _ | _ | _ |
| Nitragina | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | + | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | + | _ | _ | _ | _ | _ |
| 30 kg N ha−1 + HiStick®Soy | _ | + | _ | + | + | + | + | _ | _ | _ | + | _ | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | + | _ | _ | _ | _ |
| 60 kg N ha−1 + HiStick®Soy | _ | _ | _ | _ | _ | _ | + | + | _ | _ | + | _ | _ | + | _ | _ | _ | _ | + | _ | + | _ | _ | _ | _ | _ | _ |
| 30 kg N ha−1 + Nitragina | _ | + | _ | _ | _ | _ | + | + | _ | + | + | + | _ | + | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ |
| 60 kg N ha−1 + Nitragina | _ | + | + | _ | + | _ | + | + | + | _ | + | _ | _ | + | _ | _ | -_ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olszewski, J.; Dzienis, G.; Okorski, A.; Goś, W.; Pszczółkowska, A. Fungal Colonization of the Anatomical Parts of Soybean Seeds Supplied with Different Nitrogen Rates and Inoculated with Bradyrhizobium japonicum. Agriculture 2025, 15, 857. https://doi.org/10.3390/agriculture15080857
Olszewski J, Dzienis G, Okorski A, Goś W, Pszczółkowska A. Fungal Colonization of the Anatomical Parts of Soybean Seeds Supplied with Different Nitrogen Rates and Inoculated with Bradyrhizobium japonicum. Agriculture. 2025; 15(8):857. https://doi.org/10.3390/agriculture15080857
Chicago/Turabian StyleOlszewski, Jacek, Grzegorz Dzienis, Adam Okorski, Weronika Goś, and Agnieszka Pszczółkowska. 2025. "Fungal Colonization of the Anatomical Parts of Soybean Seeds Supplied with Different Nitrogen Rates and Inoculated with Bradyrhizobium japonicum" Agriculture 15, no. 8: 857. https://doi.org/10.3390/agriculture15080857
APA StyleOlszewski, J., Dzienis, G., Okorski, A., Goś, W., & Pszczółkowska, A. (2025). Fungal Colonization of the Anatomical Parts of Soybean Seeds Supplied with Different Nitrogen Rates and Inoculated with Bradyrhizobium japonicum. Agriculture, 15(8), 857. https://doi.org/10.3390/agriculture15080857





