Breast Cancer Detection in Mammography Images: A CNN-Based Approach with Feature Selection
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
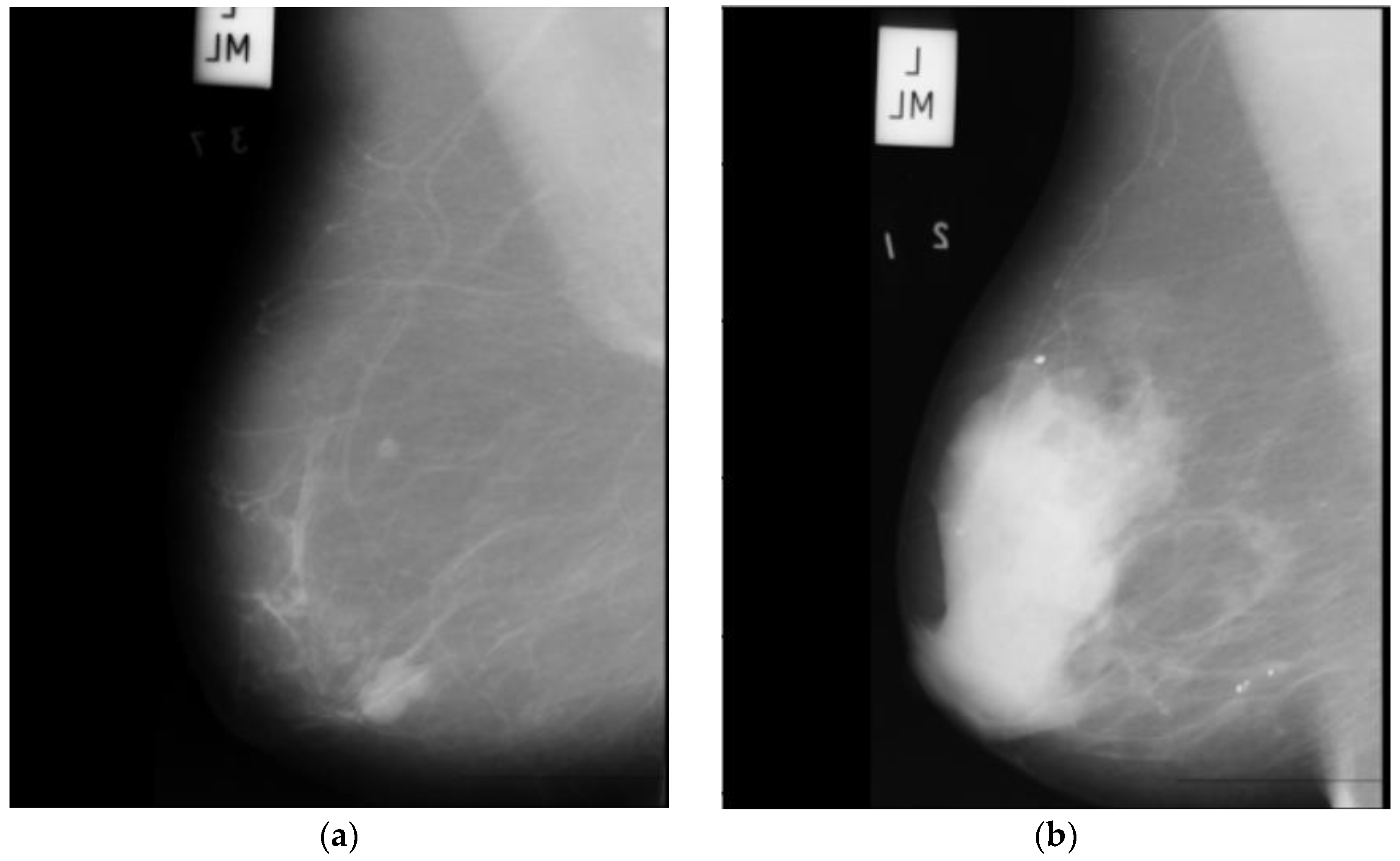
- The main dataset for this project is the radiological society of north america (RSNA) dataset from a recent Kaggle competition [22]. The dataset contains 54,713 images in dicom format from roughly 11,000 patients. For each patient, there are at least four images from different laterality and views. For each subject, two different views CC and MLO, and images from left and right laterality were provided. The images are of various sizes and formats, including jpeg and jpeg2000, and different types, such as monochrome-1 and monochrome-2. The dataset provides additional features some of which can be used for classification purposes: age, implant, BIRADS, and density. We base our work on this dataset, but since this dataset is new, it has not been used in any published research yet. Hence, for comparison purposes, we use two other well-known datasets MIAS and DDSM. This dataset is imbalanced as only 2 percent of the images are from cancer patients, which makes any classification method biased. To compensate for this, we use all positive cases and only 2320 images from negative cases. Figure 1 depicts two sample images from this dataset for cancer and normal cases.
- The mammographic image analysis society (MIAS) [23] dataset is a well-known and widely used dataset for the development and evaluation of CAD systems for BC detection. It consists of 322 mammographic images, with each image accompanied by a corresponding ground truth classification of benign or malignant tumors. The dataset is particularly valuable for researchers interested in developing machine learning algorithms for BC detection, as it includes examples of both normal and abnormal mammograms, as well as a range of breast densities and lesion types. Figure 2 depicts two sample images from this dataset for cancer and normal cases.
- The digital database for screening mammography (DDSM) [24] includes 55,890 images, of which 14% are positive, and the remaining 86% are negative. Images were tiled into 598 × 598 tiles, which were then resized to 299 × 299. A subset of this dataset which is for positive cases and is called CBIS-DDSM, has been annotated and the region of interest has been extracted by experts. In this research, we do not use the CBIS-DDSM and use the original DDSM dataset as we are classifying the images from normal subjects and cancer patients. Figure 3 depicts two sample images from this dataset for cancer and normal cases. Table 1 summarizes these three datasets.
2.2. Models
- AlexNet [25] is a deep CNN architecture that was introduced in 2012 and achieved a breakthrough in computer vision tasks such as image classification. It consists of eight layers, including five convolutional layers and three fully connected layers. The first convolutional layer uses a large receptive field to capture low-level features such as edges and textures, while subsequent layers use smaller receptive fields to capture increasingly complex and abstract features. AlexNet was the first deep network to successfully use the rectified linear unit (ReLU) activation functions, which have since become a standard activation function in deep learning. It also used dropout regularization to prevent overfitting during training. AlexNet’s success on the ImageNet dataset, which contains over one million images, demonstrated the potential of deep neural networks for image recognition tasks and paved the way for further advances in the field of computer vision.
- ResNet50 [26] is a deep CNN architecture that uses residual connections to enable learning from very deep architectures without suffering from the vanishing gradient problem. It consists of 50 layers, including convolutional layers, batch normalization layers, ReLU activation functions, and fully connected layers. ResNet50 also uses a skip connection that bypasses several layers in the network, allowing it to effectively learns both low-level and high-level features.
- EfficientNet [27] is a family of deep CNN architectures that were introduced in 2019 and have achieved state-of-the-art performance on a range of computer vision tasks. EfficientNet uses a compound scaling method to simultaneously optimize the depth, width, and resolution of the network, allowing it to achieve high accuracy while maintaining computational efficiency. EfficientNet consists of a backbone network that extracts features from input images and a head network that performs the final classification. The backbone network uses a combination of mobile inverted bottleneck convolutional layers and squeeze-and-excitation (SE) blocks to capture both spatial and channel-wise correlations in the input. The head network uses a combination of global average pooling and fully connected layers to perform the final classification.
- MobileNet [28] is a deep learning architecture suitable for efficient and accurate analysis of medical images, specifically in the context of BC diagnosis. With its emphasis on computational efficiency, MobileNet can effectively extract features from mammography images, enabling the detection of subtle patterns or abnormalities associated with breast cancer. By utilizing depthwise separable convolutions, MobileNet optimizes memory consumption and computational load, making it ideal for resource-constrained environments. The integration of the ReLU6 activation function further enhances efficiency and compatibility with medical imaging devices. Overall, MobileNet offers a valuable solution for BC analysis, providing accurate results while operating efficiently on limited computational resources.
- ConvNeXt [29] is an architecture that enhances the representational capacity of CNNs by leveraging parallel branches to capture diverse and complementary features, leading to improved performance on challenging visual recognition tasks. It has demonstrated excellent performance on various computer vision tasks, including image classification, object detection, and semantic segmentation. Its ability to capture complex relationships between features has made it a popular choice for tasks requiring a high-level understanding of visual data.
3. Proposed Method
- A.
- Preprocessing: In this research, the images obtained from various datasets exhibit variations in sizes and resolutions.
- 1.
- Normalization:The RSNA dataset consists of images in various formats, including 12 and 16 bits per pixel. Additionally, it has two different photometric interpretations known as MONOCHROME1 and MONOCHROME2. The former represents grayscale images with ascending pixel values from bright to dark, while the latter represents grayscale images with ascending pixel values from dark to bright. To ensure consistency within the RSNA dataset, we convert all MONOCHROME1 images to MONOCHROME2.In order to standardize the pixel values across the RSNA dataset, intensity normalization is performed. This involves scaling the pixel values to the range of 0 to 255, which is equivalent to 8 bits per pixel. By applying this normalization process, the pixel values across the dataset become more consistent and comparable.On the other hand, the DDSM and MIAS datasets already have pixel values within the range of 0 to 255, eliminating the need for additional normalization. Therefore, the pixel values in these datasets are deemed suitable, and no further adjustment is required.
- 2.
- Region of Interest Selection:To select the region of interest, we initially apply a global thresholding method to the image. Subsequently, we extract the contour of the largest object present in the image, which corresponds to the breast area. Utilizing this contour, we generated a mask that enables us to crop the image and isolate the specific region of interest for further analysis.
- 3.
- Image Alignment:In breast cancer datasets, there are two distinct laterality categories: left and right. To enhance consistency and improve accuracy in analysis, we align all laterality labels to the left side. This process involves horizontally flipping all left breast images to create a uniform orientation throughout the datasets. By standardizing the laterality representation, we ensure a consistent and reliable dataset for further research and analysis purposes.
- B.
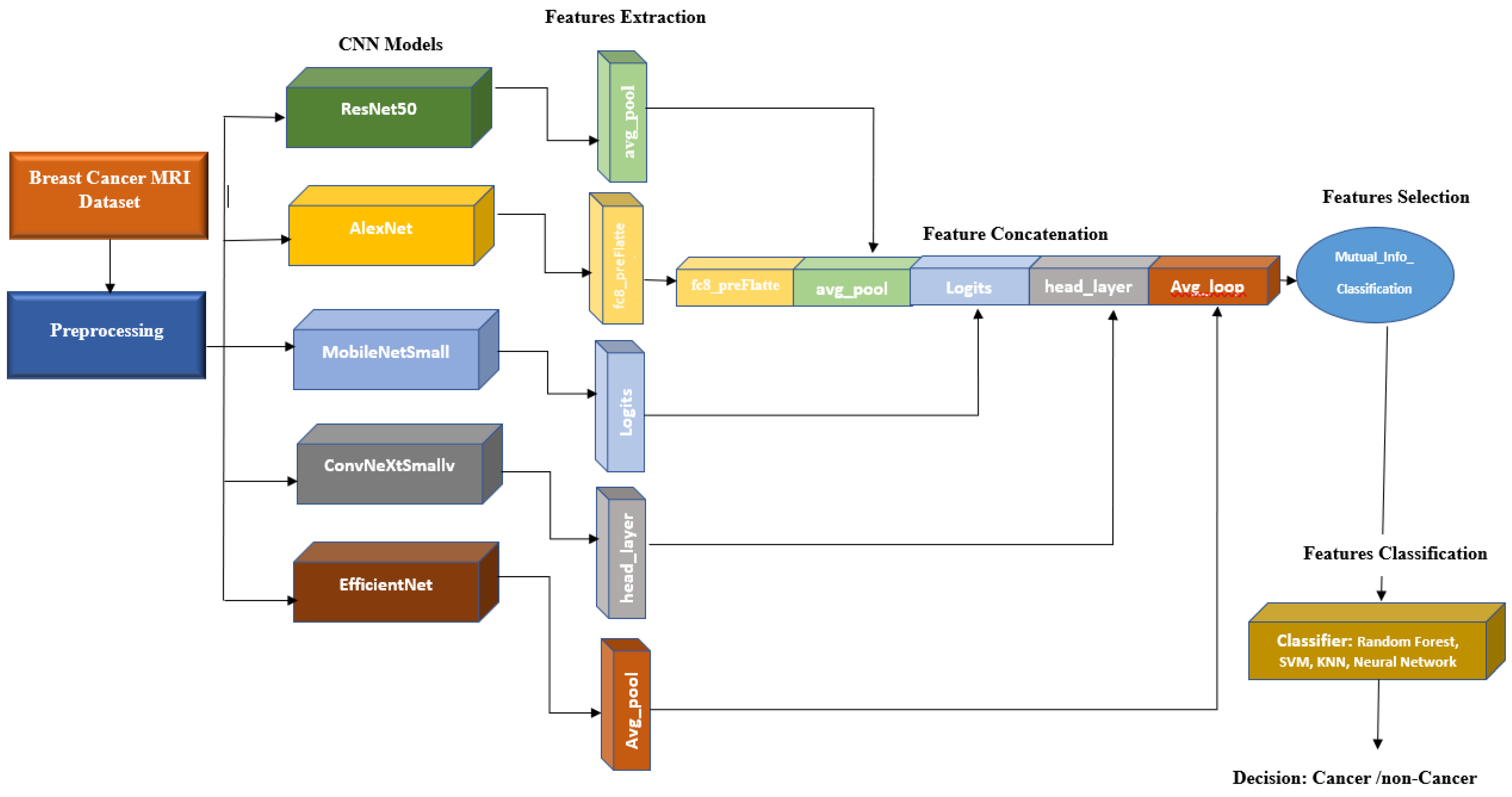
- Feature extraction: For feature extraction, we exploit the features computed by pre-trained CNN models described in Section 2.2. For each model, the features are extracted from the last layer before the last fully connected (FC) layer as the output of the final FC layer has been trained for 1000 classes of the ImageNet dataset, and hence, we skip this layer and extract the features from the last layer before the final FC layer. Table 2 depicts the layer before the final FC layer and the number of features extracted for each CNN model used in this paper.
- C.
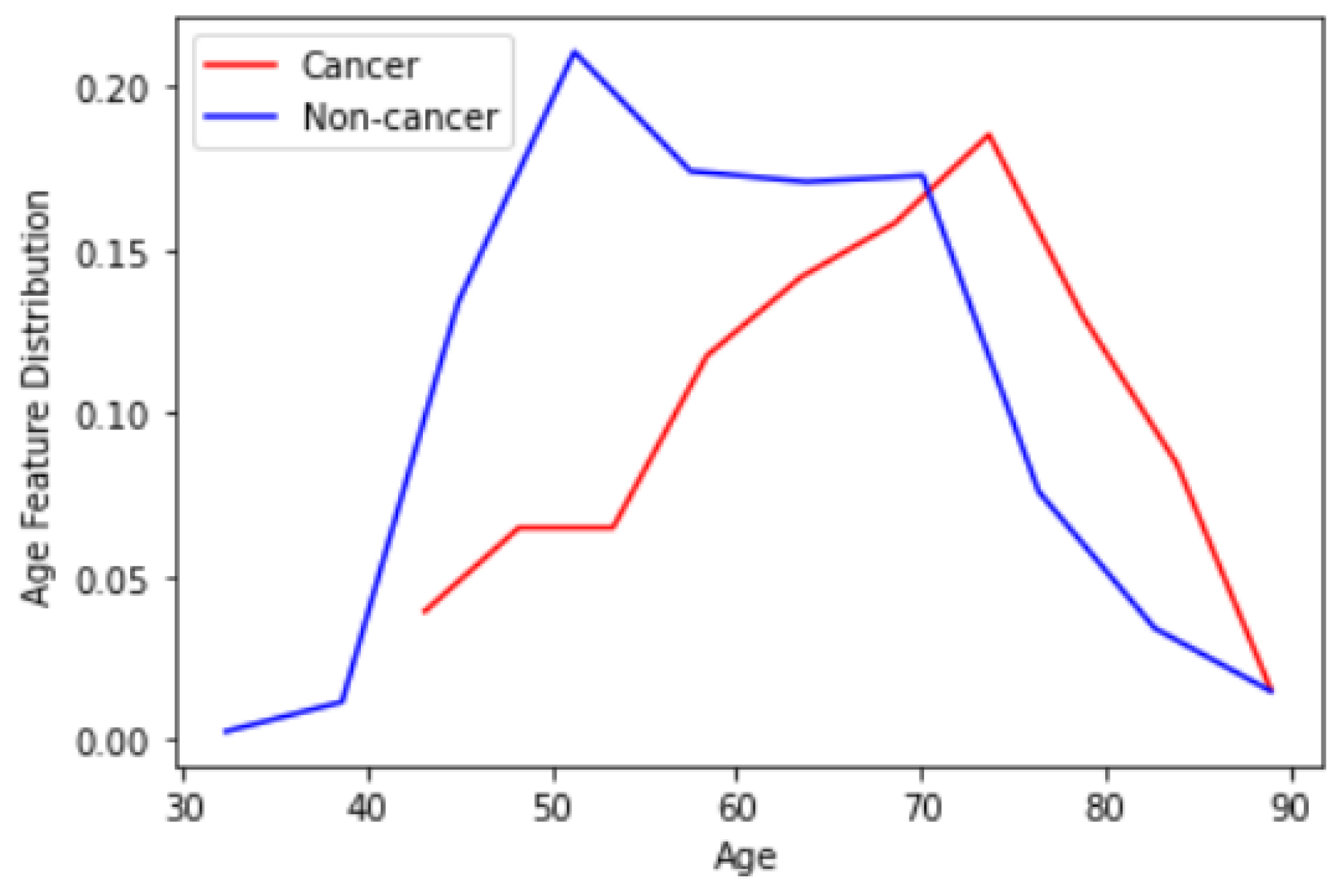
- Feature concatenation: The 1-dimensional (1D) features extracted in the previous step are concatenated to form a single 1D feature vector. Note that for each CNN model, we have extracted features from two different views CC and MLO. Hence, 10 1D vectors are concatenated here. This forms a vector with a size of 18,384 For the RSNA dataset that we use as the basis of our research, we have an additional useful feature for the patient age. Figure 5 depicts the distribution of the age feature provided by the RSNA dataset for both cancer and non-cancer subjects. As can be observed, age can also be considered a valuable feature. We can also simply normalize and add age to our feature vector to have 18,385 features in total.
- D.
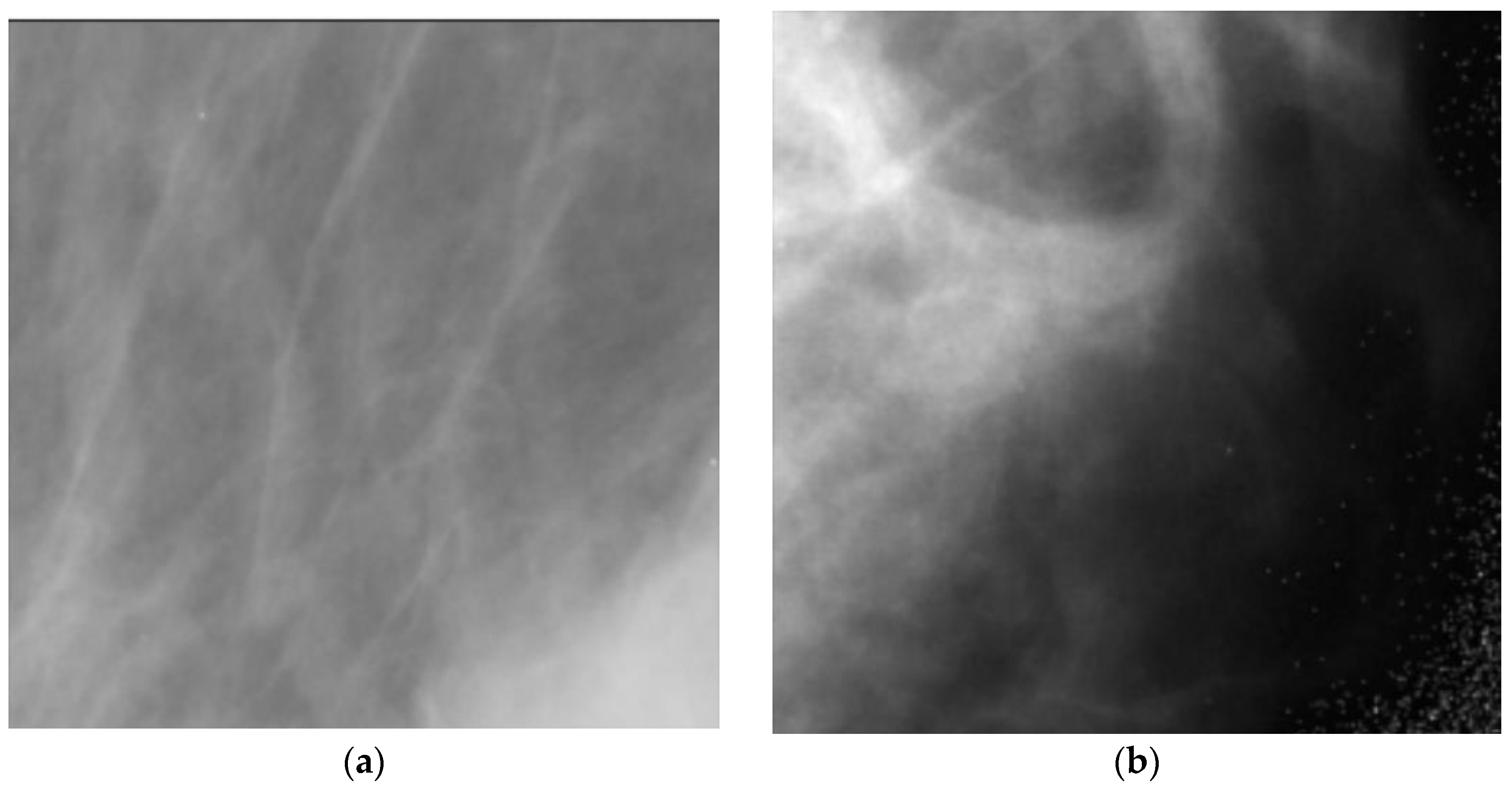
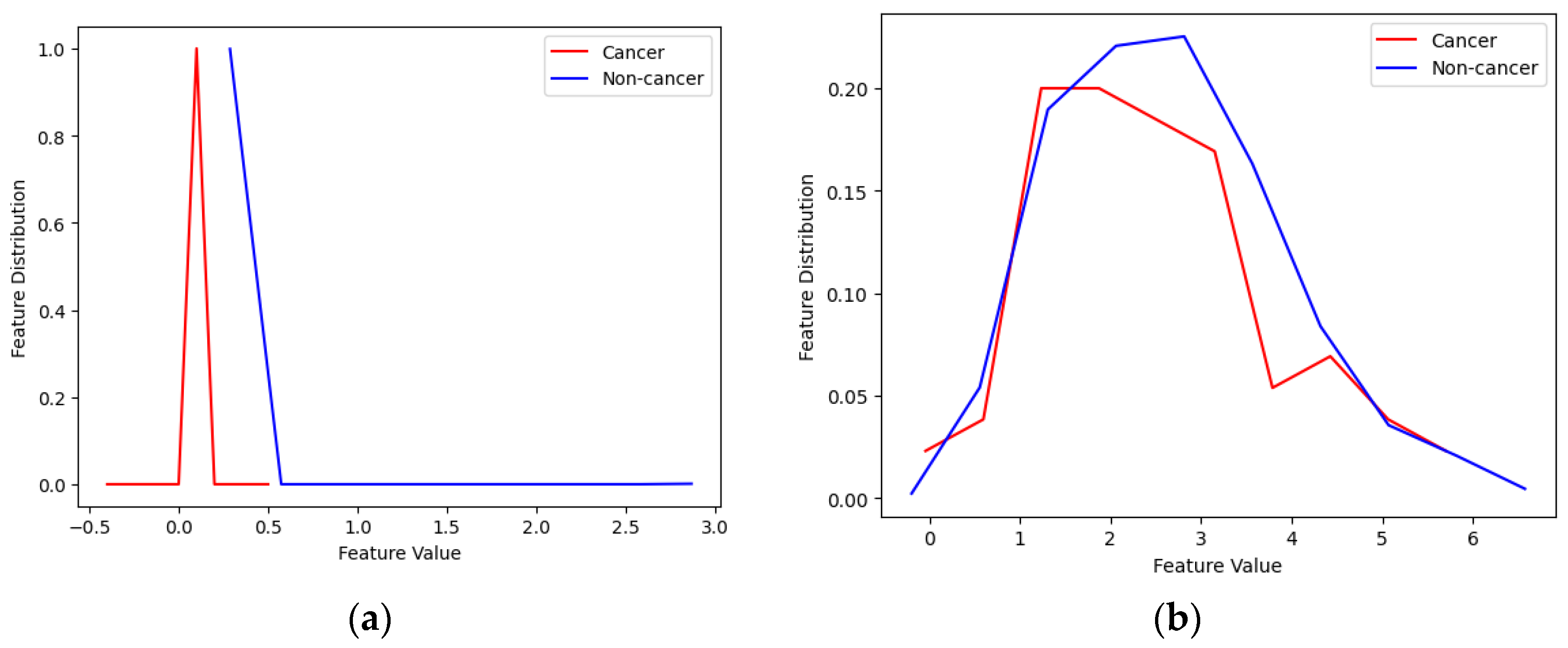
- Feature selection: The majority of the features are redundant and do not carry any useful information and only increase the complexity of the system. Figure 6 illustrates 2 samples of good and weak features. As one can see from the figure, in the case of weak features, the distribution of the feature for normal and cancerous subjects are similar showing that there is no useful information in this feature and the calculated mutual information between them is zero. For the case of good features, normal and cancerous subjects have obviously different distributions showing that these features carry useful information, although small, that can improve the performance of classifiers used in the next step. To compute mutual information we use the method in [30]. We empirically found a 0.02 threshold gives us the best results. Note that we have also adopted feature selection based on mutual information empirically and after using various feature selection methods. The number of features for each dataset before and after feature selection is presented in Table 3.
- E.
- Feature classification: After selecting the best features, we need to classify them. For this purpose, we tried multiple machine learning algorithms such as k-NN, random forest (RF), SVM, and NN. In our study, we utilize an RF algorithm with specific parameters to enhance breast cancer detection. We construct an ensemble of 100 trees, setting the minimum number of samples required to split a node as 2. Additionally, we limit the maximum number of features considered for each tree to 5 and the maximum tree depth to 4. These parameter settings are chosen to optimize the performance of our model and improve the accuracy of breast cancer detection in our X-ray image datasets.In our SVM classifier implementation, we utilize a linear kernel and set the regularization parameter “C” to a value of 1. The linear kernel allows us to learn a linear decision boundary, while the “C” parameter balances the trade-off between training accuracy and the complexity of the decision boundary.In the k-NN classifier, we set k = 5, and for the NN classifier, we used two fully connected (FC) layers with a hidden layer including 96 neurons and a single-neuron classification layer. For the classification layer, we use a sigmoid activation function that classifies non-cancer cases from cancerous ones.
4. Results and Discussion
4.1. Evaluation Metrics [31]
- True positives (TP): Instances where the predicted class and actual class are both positive. This indicates that the classifier accurately classified the instance with a positive label.
- False positives (FP): Instances where the predicted class is positive but the actual class is negative. This means that the classifier incorrectly classified the instance with a positive label. In the context of breast abnormality classification, an FP response corresponds to a type I error according to statisticians. For example, it could refer to a calcification image being classified as a mass lesion or a benign mass lesion being classified as a malignant mammogram in the diagnosis.
- True negatives (TN): Instances where the predicted class and actual class are both negative. This indicates that the classifier correctly classified the instance with a negative label.
- False negatives (FN): Instances where the predicted class is negative but the actual class is positive. This means that the classifier incorrectly classified the instance with a negative label. In the context of breast abnormality classification, an FN response is considered a type II error. For instance, it could refer to a mass mammogram being classified as calcification or a malignant mass lesion being classified as a benign mammogram in the diagnosis. Type II errors are particularly significant in their consequences.
- Accuracy: This metric represents the overall number of correctly classified instances. In the case of the abnormality classifier, accuracy signifies the correct classification of image patches containing either mass or calcification. Similarly, accuracy shows the correct classification of image patches as either malignant or benign in the pathology classifier.
- Sensitivity or Recall: This metric represents the proportion of positive image patches that are correctly classified. In the abnormality type classifier, sensitivity indicates the fraction of image patches that are truly mass lesions and are correctly classified. Similarly, the abnormality pathology classifier shows the fraction of truly malignant image patches that are correctly classified. Given the significance of type II errors, this metric is valuable for evaluating performance.
- Precision: This metric reflects the proportion of positive predictions that are correctly categorized. It is calculated using the following formula:
- F1 Score: This measure combines the impact of recall and precision using the harmonic mean, giving equal penalties to extreme values. It is commonly calculated using the formula:
4.2. Performance Evaluation of the Proposed Model for Different Classifiers
4.3. Comparison of the Proposed System with State-of-the-Art Methods
4.4. Cross-Dataset Validation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Lei, S.; Zheng, R.; Zhang, S.; Wang, S.; Chen, R.; Sun, K.; Zeng, H.; Zhou, J.; Wei, W. Global patterns of breast cancer incidence and mortality: A population-based cancer registry data analysis from 2000 to 2020. Cancer Commun. 2021, 41, 1183–1194. [Google Scholar] [CrossRef]
- Marks, J.S.; Lee, N.C.; Lawson, H.W.; Henson, R.; Bobo, J.K.; Kaeser, M.K. Implementing recommendations for the early detection of breast and cervical cancer among low-income women. Morb. Mortal. Wkly. Rep. Recomm. Rep. 2000, 49, 35–55. [Google Scholar]
- Du-Crow, E. Computer-Aided Detection in Mammography; The University of Manchester: Manchester, UK, 2022. [Google Scholar]
- Evans, A.; Trimboli, R.M.; Athanasiou, A.; Balleyguier, C.; Baltzer, P.A.; Bick, U.; Herrero, J.C.; Clauser, P.; Colin, C.; Cornford, E.; et al. Breast ultrasound: Recommendations for information to women and referring physicians by the European Society of Breast Imaging. Insights Imaging 2018, 9, 449–461. [Google Scholar] [CrossRef]
- Schueller, G.; Schueller-Weidekamm, C.; Helbich, T.H. Accuracy of ultrasound-guided, large-core needle breast biopsy. Eur. Radiol. 2008, 18, 1761–1773. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Liang, C.; Wang, H. Multiview robust graph-based clustering for cancer subtype identification. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 20, 544–556. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, G.; Peng, J.; Deng, R.; Fu, X. Towards Adaptive Consensus Graph: Multi-view Clustering via Graph Collaboration. IEEE Trans. Multimed. 2022, 1–13. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Zhang, Z.; Fu, X.; Zhuo, L.; Xu, M.; Wang, M. Kernelized multiview subspace analysis by self-weighted learning. IEEE Trans. Multimed. 2020, 23, 3828–3840. [Google Scholar] [CrossRef]
- Wang, H.; Yao, M.; Jiang, G.; Mi, Z.; Fu, X. Graph-Collaborated Auto-Encoder Hashing for Multi-view Binary Clustering. arXiv 2023, arXiv:2301.02484. [Google Scholar]
- Bai, J.; Posner, R.; Wang, T.; Yang, C.; Nabavi, S. Applying deep learning in digital breast tomosynthesis for automatic breast cancer detection: A review. Med. Image Anal. 2021, 71, 102049. [Google Scholar] [CrossRef]
- Li, Z.; Liu, F.; Yang, W.; Peng, S.; Zhou, J. A survey of convolutional neural networks: Analysis, applications, and prospects. IEEE Trans. Neural Netw. Learn. Syst. 2022, 33, 6999–7019. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga-Gomez, J.; Al Masry, Z.; Benaggoune, K.; Meraghni, S.; Zerhouni, N. A CNN-based methodology for breast cancer diagnosis using thermal images. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2021, 9, 131–145. [Google Scholar] [CrossRef]
- Eroğlu, Y.; Yildirim, M.; Çinar, A. Convolutional Neural Networks based classification of breast ultrasonography images by hybrid method with respect to benign, malignant, and normal using mRMR. Comput. Biol. Med. 2021, 133, 104407. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Yang, F.; Liu, L.; Li, X. Automatic segmentation of breast lesions for interaction in ultrasonic computer-aided diagnosis. Inf. Sci. 2015, 314, 293–310. [Google Scholar] [CrossRef]
- Huang, Q.; Huang, Y.; Luo, Y.; Yuan, F.; Li, X. Segmentation of breast ultrasound image with semantic classification of superpixels. Med. Image Anal. 2020, 61, 101657. [Google Scholar] [CrossRef]
- Zhou, J.; Luo, L.; Dou, Q.; Chen, H.; Chen, C.; Li, G.; Jiang, Z.; Heng, P. Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J. Magn. Reson. Imaging 2019, 50, 1144–1151. [Google Scholar] [CrossRef]
- Yurttakal, A.H.; Erbay, H.; Ikizceli, T.; Karaçavuş, S. Detection of breast cancer via deep convolution neural networks using MRI images. Multimed. Tools Appl. 2019, 79, 15555–15573. [Google Scholar] [CrossRef]
- Rahman, A.S.; Belhaouari, S.B.; Bouzerdoum, A.; Baali, H.; Alam, T.; Eldaraa, A.M. Breast mass tumor classification using deep learning. In Proceedings of the 2020 IEEE International Conference on Informatics, IoT, and Enabling Technologies (ICIoT), Doha, Qatar, 2 February 2020; pp. 271–276. [Google Scholar]
- Sun, L.; Wang, J.; Hu, Z.; Xu, Y.; Cui, Z. Multi-view convolutional neural networks for mammographic image classification. IEEE Access 2019, 7, 126273–126282. [Google Scholar] [CrossRef]
- Heravi, E.J.; Aghdam, H.H.; Puig, D. Classification of Foods Using Spatial Pyramid Convolutional Neural Network. InCCIA 2016, 288, 163–168. [Google Scholar] [CrossRef]
- Carr, C.; Kitamura, F.; Partridge, G.; Kalpathy-Cramer, J.; Mongan, J.; Andriole, K.; Lavender Vazirabad, M.; Riopel, M.; Ball, R.; Dane, S.; et al. RSNA Screening Mammography Breast Cancer Detection, Kaggle 2022. Available online: https://kaggle.com/competitions/rsna-breast-cancer-detection (accessed on 1 December 2022).
- Suckling, J.; Parker, J.; Dance, D.; Astley, S.; Hutt, I.; Boggis, C.; Ricketts, I.; Stamatakis, E.; Cerneaz, N.; Kok, S.; et al. Mammographic Image Analysis Society (Mias) Database v1. 21. Available online: https://www.repository.cam.ac.uk/handle/1810/250394 (accessed on 2 February 2023).
- Heath, M.; Bowyer, K.; Kopans, D.; Kegelmeyer, P.; Moore, R.; Chang, K.; Munishkumaran, S. Current status of the digital database for screening mammography. Digit. Mammogr. Nijmegen 1998, 1998, 457–460. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Tan, M.; Le, Q. Efficientnetv2: Smaller models and faster training. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual Event, 1 July 2021; pp. 10096–10106. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2022, New Orleans, LA, USA, 19–20 June 2022; pp. 11976–11986. [Google Scholar]
- Ross, B.C. Mutual Information between Discrete and Continuous Data Sets. PLoS ONE 2014, 9, e87357. [Google Scholar] [CrossRef] [PubMed]
- Azour, F.; Boukerche, A. An efficient transfer and ensemble learning based computer aided breast abnormality diagnosis system. IEEE Access 2022, 11, 21199–21209. [Google Scholar] [CrossRef]
- Rampun, A.; Scotney, B.W.; Morrow, P.J.; Wang, H.; Winder, J. Breast density classification using local quinary patterns with various neighbourhood topologies. J. Imaging 2018, 4, 14. [Google Scholar] [CrossRef]
- Vijayarajeswari, R.; Parthasarathy, P.; Vivekanandan, S.; Basha, A.A. Classification of mammogram for early detection of breast cancer using SVM classifier and Hough transform. Measurement 2019, 146, 800–805. [Google Scholar] [CrossRef]
- Arafa, A.A.A.; El-Sokary, N.; Asad, A.; Hefny, H. Computer-aided detection system for breast cancer based on GMM and SVM. Arab. J. Nucl. Sci. Appl. 2019, 52, 142–150. [Google Scholar] [CrossRef]
- Diaz, R.A.; Swandewi, N.N.; Novianti, K.D. Malignancy determination breast cancer based on mammogram image with k-nearest neighbor. In Proceedings of the 2019 1st International Conference on Cybernetics and Intelligent System (ICORIS), Denpasar, Indonesia, 22 August 2019; Volume 1, pp. 233–237. [Google Scholar]
- Agrawal, S.; Rangnekar, R.; Gala, D.; Paul, S.; Kalbande, D. Detection of breast cancer from mammograms using a hybrid approach of deep learning and linear classification. In Proceedings of the 2018 International Conference on Smart City and Emerging Technology (ICSCET), Mumbai, India, 5 January 2018; pp. 1–6. [Google Scholar]
- Li, B.; Ge, Y.; Zhao, Y.; Guan, E.; Yan, W. Benign and malignant mammographic image classification based on convolutional neural networks. In Proceedings of the 2018 10th International Conference on Machine Learning and Computing, New York, NY, USA, 26 February 2018; pp. 247–251. [Google Scholar]
- Platania, R.; Shams, S.; Yang, S.; Zhang, J.; Lee, K.; Park, S.J. Automated breast cancer diagnosis using deep learning and region of interest detection (bc-droid). In Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Boston, MA, USA, 20 August 2017; pp. 536–543. [Google Scholar]
- Swiderski, B.; Kurek, J.; Osowski, S.; Kruk, M.; Barhoumi, W. Deep learning and non-negative matrix factorization in recognition of mammograms. In Proceedings of the Eighth International Conference on Graphic and Image Processing (ICGIP 2016), Tokyo, Japan, 8 February 2017; Volume 10225, pp. 53–59. [Google Scholar]

| Dataset | Number of Images | Image Types | Image Size |
|---|---|---|---|
| RSNA | 54,713 | Variable | Variable |
| MIAS | 322 | PGM | 1024 × 1024 |
| DDSM | 55,890 | JPEG | 598 × 598 |
| CNN Models | Layer Name 1 | Number of Features |
|---|---|---|
| ResNet50 | avg_pool | 2048 |
| AlexNet | fc8_preflatten | 4096 |
| MobileNetSmall | Logits | 1000 |
| ConvNeXtSmall | head_layer | 768 |
| EfficientNet | avg_pool | 1280 |
| Dataset | Before Feature Selection | After Feature Selection |
|---|---|---|
| RSNA | 18,385 1 | 452 |
| MIAS | 9192 | 212 |
| DDSM | 9192 | 206 |
| CNN Models | Acc | Sn | Pr | AUC | F-Score |
|---|---|---|---|---|---|
| AlexNet | 81% | 84% | 87% | 0.82 | 0.86 |
| Resnet50 | 84% | 90% | 86% | 0.89 | 0.88 |
| MobileNetSmall | 77% | 85% | 81% | 0.81 | 0.83 |
| ConvNexSmall | 79% | 87% | 83% | 0.83 | 0.85 |
| EfficientNet | 86% | 92% | 88% | 0.92 | 0.90 |
| Concat. Model | 92% | 96% | 92% | 0.96 | 0.94 |
| CNN Models | Acc | Sn | Pr | AUC | F-Score |
|---|---|---|---|---|---|
| AlexNet | 73% | 70% | 72% | 0.70 | 0.71 |
| Resnet50 | 72% | 75% | 71% | 0.73 | 0.73 |
| MobileNetSmall | 64% | 71% | 67% | 0.68 | 0.69 |
| ConvNexSmall | 66% | 74% | 70% | 0.71 | 0.72 |
| EfficientNet | 71% | 78% | 74% | 0.76 | 0.76 |
| Concat. Model | 78% | 81% | 79% | 0.82 | 0.80 |
| CNN Models | Acc | Sn | Pr | AUC | F-Score |
|---|---|---|---|---|---|
| AlexNet | 71% | 67% | 69% | 0.68 | 0.68 |
| Resnet50 | 69% | 70% | 67% | 0.71 | 0.68 |
| MobileNetSmall | 60% | 67% | 63% | 0.64 | 0.65 |
| ConvNexSmall | 62% | 69% | 65% | 0.67 | 0.67 |
| EfficientNet | 73% | 74% | 70% | 0.75 | 0.72 |
| Concat. Model | 78% | 79% | 77% | 0.80 | 0.78 |
| CNN Models | Acc | Sn | Pr | AUC | F-Score |
|---|---|---|---|---|---|
| AlexNet | 62% | 61% | 63% | 0.62 | 0.62 |
| Resnet50 | 64% | 66% | 63% | 0.65 | 0.64 |
| MobileNetSmall | 60% | 63% | 59% | 0.60 | 0.61 |
| ConvNexSmall | 62% | 65% | 61% | 0.63 | 0.63 |
| EfficientNet | 68% | 70% | 66% | 0.68 | 0.68 |
| Concat. Model | 73% | 75% | 72% | 0.74 | 0.73 |
| Method | Dataset | Number of Images | ACC | Sn | Pr |
|---|---|---|---|---|---|
| SVM & Hough [32] | MIAS & InBreast | 322&206 | 86.13% | 80.67% | 92.81% |
| LQP & SVM [33] | MIAS | 95 | 94% | NA | NA |
| GMM & SVM [34] | Mini-MIAS dataset | 90 | 92.5% | NA | NA |
| KNN [35] | Mini-MIAS | 120 | 92% | NA | NA |
| Voting Classifier [36] | MIAS | 322 | 85% | NA | NA |
| CNN-4d [37] | Mini-MIAS | 547 | 89.05% | 90.63% | 83.67% |
| CNN [38] | DDSM | 10,480 | 93.5% | NA | NA |
| CNNs [39] | DDSM | 11,218 | 85.82% | 82.28% | 86.59% |
| Our Method + NN | RSNA | 54,713 | 92% | 96% | 92% |
| Our Method + NN | MIAS | 322 | 94.5% | 96.32% | 91.80% |
| Our Method + NN | DDSM | 55,890 | 96% | 94.70% | 97% |
| Train Dataset | Test Dataset | ACC | Sn | Pr |
|---|---|---|---|---|
| RSNA | MIAS | 79.13% | 82.67% | 80.81% |
| RSNA | DDSM | 74% | 77.50% | 76% |
| MIAS | RSNA | 76.5% | 78.80% | 78% |
| MIAS | DDSM | 80.70% | 82% | 82.80% |
| DDSM | RSNA | 72% | 75.50% | 76% |
| DDSM | MIAS | 79% | 80% | 79.87% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jafari, Z.; Karami, E. Breast Cancer Detection in Mammography Images: A CNN-Based Approach with Feature Selection. Information 2023, 14, 410. https://doi.org/10.3390/info14070410
Jafari Z, Karami E. Breast Cancer Detection in Mammography Images: A CNN-Based Approach with Feature Selection. Information. 2023; 14(7):410. https://doi.org/10.3390/info14070410
Chicago/Turabian StyleJafari, Zahra, and Ebrahim Karami. 2023. "Breast Cancer Detection in Mammography Images: A CNN-Based Approach with Feature Selection" Information 14, no. 7: 410. https://doi.org/10.3390/info14070410
APA StyleJafari, Z., & Karami, E. (2023). Breast Cancer Detection in Mammography Images: A CNN-Based Approach with Feature Selection. Information, 14(7), 410. https://doi.org/10.3390/info14070410







