Engineering Nanopatterned Structures to Orchestrate Macrophage Phenotype by Cell Shape
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fabrication and Characterization of the Nanopatterned Surfaces
2.2. Effects of the Nanopatterned Structures on Fibronectin Distribution and Conformation
2.3. Cell Culture
2.4. Effects of the Nanopatterned Structures on Macrophage Cell Shape and Phenotype
2.4.1. Cell Morphology
2.4.2. Cytoskeleton and Focal Adhesion Observation
2.4.3. Flow Cytometry
2.4.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Assay
2.5. Western Blot
2.6. Statistical Analysis
3. Results and Discussion
3.1. Surface Characterization
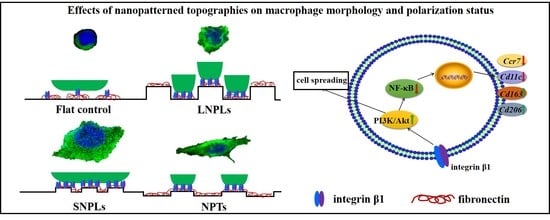
3.2. Effects of Nanopatterned Topographies on Macrophage Morphology
3.3. Correlation of Cell Shape and Macrophage Polarization Status
3.4. Biochemical Cues Underlying Cell-Nanopattern Interactions Govern Macrophage Polarization Status
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nel, A.E.; Madler, L.; Velegol, D.; Xia, T.; Hoek, E.M.V.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Wang, J.; Li, X.L.; Hu, N.; Voelcker, N.H.; Xie, X.; Elnathan, R. Emerging Roles of 1D Vertical Nanostructures in Orchestrating Immune Cell Functions. Adv. Mater. 2020, 32, 2001668. [Google Scholar] [CrossRef] [PubMed]
- Biggs, M.J.P.; Richards, R.G.; Dalby, M.J. Nanotopographical modification: A regulator of cellular function through focal adhesions. Nanomed.-Nanotechnol. 2010, 6, 619–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padmanabhan, J.; Kinser, E.R.; Stalter, M.A.; Duncan-Lewis, C.; Balestrini, J.L.; Sawyer, A.J.; Schroers, J.; Kyriakides, T.R. Engineering Cellular Response Using Nanopatterned Bulk Metallic Glass. ACS Nano 2014, 8, 4366–4375. [Google Scholar] [CrossRef] [PubMed]
- Vassey, M.J.; Figueredo, G.P.; Scurr, D.J.; Vasilevich, A.S.; Vermeulen, S.; Carlier, A.; Luckett, J.; Beijer, N.R.M.; Williams, P.; Winkler, D.A.; et al. Immune Modulation by Design: Using Topography to Control Human Monocyte Attachment and Macrophage Differentiation. Adv. Sci. 2020, 7, 1903392. [Google Scholar] [CrossRef]
- Gui, N.; Xu, W.; Myers, D.E.; Shukla, R.; Tang, H.P.; Qian, M. The effect of ordered and partially ordered surface topography on bone cell responses: A review. Biomater. Sci. 2018, 6, 250–264. [Google Scholar] [CrossRef]
- Yao, X.; Peng, R.; Ding, J.D. Cell-Material Interactions Revealed via Material Techniques of Surface Patterning. Adv. Mater. 2013, 25, 5257–5286. [Google Scholar] [CrossRef]
- Yim, E.K.F.; Pang, S.W.; Leong, K.W. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp. Cell Res. 2007, 313, 1820–1829. [Google Scholar] [CrossRef] [Green Version]
- Dalby, M.J.; Gadegaard, N.; Tare, R.; Andar, A.; Riehle, M.O.; Herzyk, P.; Wilkinson, C.D.W.; Oreffo, R.O.C. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 2007, 6, 997–1003. [Google Scholar] [CrossRef]
- Fu, J.P.; Wang, Y.K.; Yang, M.T.; Desai, R.A.; Yu, X.A.; Liu, Z.J.; Chen, C.S. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods 2010, 7, U733–U795. [Google Scholar] [CrossRef]
- Claes, L.; Recknagel, S.; Ignatius, A. Fracture healing under healthy and inflammatory conditions. Nat. Rev. Rheumatol. 2012, 8, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.N.; Ratner, B.D.; Goodman, S.B.; Amar, S.; Badylak, S.F. Macrophage polarization: An opportunity for improved outcomes in biomaterials and regenerative medicine. Biomaterials 2012, 33, 3792–3802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Li, K.; Liu, S.W.; Hu, T.; Razanau, I.; Wu, X.D.; Ao, H.Y.; Huang, L.P.; Xie, Y.T.; Zheng, X.B. Optimized Nanointerface Engineering of Micro/Nanostructured Titanium Implants to Enhance Cell-Nanotopography Interactions and Osseointegration. ACS Biomater. Sci. Eng. 2020, 6, 969–983. [Google Scholar] [CrossRef] [PubMed]
- McWhorter, F.Y.; Wang, T.T.; Nguyen, P.; Chung, T.; Liu, W.F. Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. USA 2013, 110, 17253–17258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaveri, T.D.; Dolgova, N.V.; Chu, B.H.; Lee, J.Y.; Wong, J.E.; Lele, T.P.; Ren, F.; Keselowsky, B.G. Contributions of surface topography and cytotoxicity to the macrophage response to zinc oxide nanorods. Biomaterials 2010, 31, 2999–3007. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Jones, J.A.; Xu, Y.G.; Low, H.Y.; Anderson, J.M.; Leong, K.W. Characterization of topographical effects on macrophage behavior in a foreign body response model. Biomaterials 2010, 31, 3479–3491. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Schwarz, U.S.; Riveline, D.; Goichberg, P.; Tzur, G.; Sabanay, I.; Mahalu, D.; Safran, S.; Bershadsky, A.; Addadi, L.; et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 2001, 3, 466–472. [Google Scholar] [CrossRef]
- Sjostrom, T.; Dalby, M.J.; Hart, A.; Tare, R.; Oreffo, R.O.C.; Su, B. Fabrication of pillar-like titania nanostructures on titanium and their interactions with human skeletal stem cells. Acta Biomater. 2009, 5, 1433–1441. [Google Scholar] [CrossRef]
- Lim, J.Y.; Hansen, J.C.; Siedlecki, C.A.; Hengstebeck, R.W.; Cheng, J.; Winograd, N.; Donahue, H.J. Osteoblast adhesion on poly(L-lactic acid)/polystyrene demixed thin film blends: Effect of nanotopography, surface chemistry, and wettability. Biomacromolecules 2005, 6, 3319–3327. [Google Scholar] [CrossRef]
- Lim, J.Y.; Dreiss, A.D.; Zhou, Z.Y.; Hansen, J.C.; Siedlecki, C.A.; Hengstebeck, R.W.; Cheng, J.; Winograd, N.; Donahue, H.J. The regulation of integrin-mediated osteoblast focal adhesion and focal adhesion kinase expression by nanoscale topography. Biomaterials 2007, 28, 1787–1797. [Google Scholar] [CrossRef] [PubMed]
- Krasteva, N.; Seifert, B.; Albrecht, W.; Weigel, T.; Schossig, M.; Altankov, G.; Groth, T. Influence of polymer membrane porosity on C3A hepatoblastoma cell adhesive interaction and function. Biomaterials 2004, 25, 2467–2476. [Google Scholar] [CrossRef] [PubMed]
- Biggs, M.J.P.; Richards, R.G.; Gadegaard, N.; McMurray, R.J.; Affrossman, S.; Wilkinson, C.D.W.; Oreffo, R.O.C.; Dalby, M.J. Interactions with nanoscale topography: Adhesion quantification and signal transduction in cells of osteogenic and multipotent lineage. J. Biomed. Mater. Res. A 2009, 91, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Hart, A.; Gadegaard, N.; Wilkinson, C.D.W.; Oreffo, R.O.C.; Dalby, M.J. Osteoprogenitor response to low-adhesion nanotopographies originally fabricated by electron beam lithography. J. Mater. Sci.-Mater. Med. 2007, 18, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Makaremi, S.; Luu, H.; Boyle, J.P.; Zhu, Y.J.; Cerson, C.; Bowdish, D.M.E.; Moran-Mirabal, J.M. The Topography of Silica Films Modulates Primary Macrophage Morphology and Function. Adv. Mater. Interfaces 2019, 6, 1900677. [Google Scholar] [CrossRef]
- Lee, H.S.; Stachelek, S.J.; Tomczyk, N.; Finley, M.J.; Composto, R.J.; Eckmann, D.M. Correlating macrophage morphology and cytokine production resulting from biomaterial contact. J. Biomed. Mater. Res. A 2013, 101, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Kianoush, F.; Nematollahi, M.; Waterfield, J.D.; Brunette, D.M. Regulation of RAW264.7 macrophage polarization on smooth and rough surface topographies by galectin-3. J. Biomed. Mater. Res. A 2017, 105, 2499–2509. [Google Scholar] [CrossRef]
- McWhorter, F.Y.; Davis, C.T.; Liu, W.F. Physical and mechanical regulation of macrophage phenotype and function. Cell Mol. Life Sci. 2015, 72, 1303–1316. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Awuah, D.; Rostam, H.M.; Emes, R.D.; Kandola, N.K.; Onion, D.; Htwe, S.S.; Rajchagool, B.; Cha, B.H.; Kim, D.; et al. Unbiased Analysis of the Impact of Micropatterned Biomaterials on Macrophage Behavior Provides Insights beyond Predefined Polarization States. ACS Biomater. Sci. Eng. 2017, 3, 969–978. [Google Scholar] [CrossRef]
- Malheiro, V.; Lehner, F.; Dinca, V.; Hoffmann, P.; Maniura-Weber, K. Convex and concave micro-structured silicone controls the shape, but not the polarization state of human macrophages. Biomater. Sci. 2016, 4, 1562–1573. [Google Scholar] [CrossRef] [Green Version]
- Shao, D.D.; Li, K.; You, M.Y.; Liu, S.W.; Hu, T.; Huang, L.P.; Xie, Y.T.; Zheng, X.B. Macrophage polarization by plasma sprayed ceria coatings on titanium-based implants: Cerium valence state matters. Appl. Surf. Sci. 2020, 504, 144070. [Google Scholar] [CrossRef]
- Lv, L.; Li, K.; Xie, Y.T.; Cao, Y.Z.; Zheng, X.B. Enhanced osteogenic activity of anatase TiO2 film: Surface hydroxyl groups induce conformational changes in fibronectin. Mater. Sci. Eng. C-Mater. 2017, 78, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Vanterpool, F.A.; Cantini, M.; Seib, F.P.; Salmeron-Sanchez, M. A material-based platform to modulate fibronectin activity and focal adhesion assembly. BioRes. Open Access 2014, 3, 286–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Hu, D.D.; Xie, Y.T.; Huang, L.P.; Zheng, X.B. Sr-doped nanowire modification of Ca-Si-based coatings for improved osteogenic activities and reduced inflammatory reactions. Nanotechnology 2018, 29, 084001. [Google Scholar] [CrossRef]
- Abshire, M.Y.; Thomas, K.S.; Owen, K.A.; Bouton, A.H. Macrophage motility requires distinct alpha5beta1/FAK and alpha4beta1/paxillin signaling events. J. Leukoc. Biol. 2011, 89, 251–257. [Google Scholar] [CrossRef] [Green Version]
- Massia, S.P.; Hubbell, J.A. An RGD spacing of 440 nm is sufficient for integrin alpha V beta 3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation. J. Cell Biol. 1991, 114, 1089–1100. [Google Scholar] [CrossRef]
- Selhuber-Unkel, C.; Lopez-Garcia, M.; Kessler, H.; Spatz, J.P. Cooperativity in Adhesion Cluster Formation during Initial Cell Adhesion. Biophys. J. 2008, 95, 5424–5431. [Google Scholar] [CrossRef] [Green Version]
- Schvartzman, M.; Palma, M.; Sable, J.; Abramson, J.; Hu, X.A.; Sheetz, M.P.; Wind, S.J. Nanolithographic Control of the Spatial Organization of Cellular Adhesion Receptors at the Single-Molecule Level. Nano Lett. 2011, 11, 1306–1312. [Google Scholar] [CrossRef] [Green Version]
- Gautrot, J.E.; Malmstrom, J.; Sundh, M.; Margadant, C.; Sonnenberg, A.; Sutherland, D.S. The Nanoscale Geometrical Maturation of Focal Adhesions Controls Stem Cell Differentiation and Mechanotransduction. Nano Lett. 2014, 14, 3945–3952. [Google Scholar] [CrossRef]
- Lv, L.; Xie, Y.T.; Li, K.; Hu, T.; Lu, X.; Cao, Y.Z.; Zheng, X.B. Unveiling the Mechanism of Surface Hydrophilicity-Modulated Macrophage Polarization. Adv. Healthc. Mater. 2018, 7, 1800675. [Google Scholar] [CrossRef]
- Hamidzadeh, K.; Christensen, S.M.; Dalby, E.; Chandrasekaran, P.; Mosser, D.M. Macrophages and the Recovery from Acute and Chronic Inflammation. Annu. Rev. Physiol. 2017, 79, 567–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dan, H.C.; Cooper, M.J.; Cogswell, P.C.; Duncan, J.A.; Ting, J.P.Y.; Baldwin, A.S. Akt-dependent regulation of NF-κB is controlled by mTOR and Raptor in association with IKK. Gene Dev. 2008, 22, 1490–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Lee, J.Y.; Hwang, D.H. The phosphatidylinositol 3-kinase/Akt pathway negatively regulates Nod2-mediated NF-kappa B pathway. Biochem. Pharmacol. 2008, 75, 1515–1525. [Google Scholar] [CrossRef]
- Park, Y.C.; Lee, C.H.; Kang, H.S.; Chung, H.T.; Kim, H.D. Wortmannin, a specific inhibitor of phosphatidylinositol-3-kinase, enhances LPS-induced NO production from murine peritoneal macrophages. Biochem. Biophys. Res. Commun. 1997, 240, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Guha, M.; Mackman, N. The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. J. Biol. Chem. 2002, 277, 32124–32132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimenez, C.; Portela, R.A.; Mellado, M.; Rodriguez-Frade, J.M.; Collard, J.; Serrano, A.; Martinez, A.C.; Avila, J.; Carrera, A.C. Role of the PI3K regulatory subunit in the control of actin organization and cell migration. J. Cell Biol. 2000, 151, 249–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mettouchi, A.; Klein, S.; Guo, W.; Lopez-Lago, M.; Lemichez, E.; Westwick, J.K.; Giancotti, F.G. Integrin-specific activation of Rac controls progression through the G(1) phase of the cell cycle. Mol. Cell 2001, 8, 115–127. [Google Scholar] [CrossRef]






| Target Gene | Direction | 5′-3′ Primer Sequence |
|---|---|---|
| Itga5 | F | 5′-CTTCTCCGTGGAGTTTTACCG-3′ |
| R | 5′-GCTGTCAAATTGAATGGTGGTG-3′ | |
| Itgb1 | F | 5′-CGTGGTTGCCGGAATTGTTC-3′ |
| R | 5′-ACCAGCTTTACGTCCATAGTTTG-3′ | |
| Cd11c | F | 5’ -CTGGATAGCCTTTCTTCTGCTG- 3’ |
| R | 5’ -GCACACTGTGTCCGAACTCA- 3’ | |
| Ccr7 | F | 5′-TGTACGAGTCGGTGTGCTTC-3′ |
| R | 5′-GGTAGGTATCCGTCATGGTCTTG-3′ | |
| Cd163 | F | 5′-ATGGGTGGACACAGAATGGTT-3′ |
| R | 5′-CAGGAGCGTTAGTGACAGCAG-3′ | |
| Cd206 | F | 5′-CTCTGTTCAGCTATTGGACGC-3′ |
| R | 5′-CGGAATTTCTGGGATTCAGCTTC-3′ | |
| Il6 | F | 5′-ACTCACCTCTTCAGAACGAATTG-3′ |
| R | 5′-CCATCTTTGGAAGGTTCAGGTTG-3′ | |
| Tnfa | F | 5′-CCTCTCTCTAATCAGCCCTCTG-3′ |
| R | 5′-GAGGACCTGGGAGTAGATGAG-3′ | |
| Il1b | F | 5′-TGGAGAGTGTGGATCCCAAG-3′ |
| R | 5′-GGTGCTGATGTACCAGTTGG-3′ | |
| Il10 | F | 5′-GACTTTAAGGGTTACCTGGGTTG-3′ |
| R | 5′-TCACATGCGCCTTGATGTCTG-3′ | |
| Il1ra | F | 5′-CATTGAGCCTCATGCTCTGTT-3′ |
| R | 5′-CGCTGTCTGAGCGGATGAA-3′ | |
| GAPDH | F | 5′-TGACCACAGTCCATGCCATC-3′ |
| R | 5′-GACGGACACATTGGGGGTAG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Lv, L.; Shao, D.; Xie, Y.; Cao, Y.; Zheng, X. Engineering Nanopatterned Structures to Orchestrate Macrophage Phenotype by Cell Shape. J. Funct. Biomater. 2022, 13, 31. https://doi.org/10.3390/jfb13010031
Li K, Lv L, Shao D, Xie Y, Cao Y, Zheng X. Engineering Nanopatterned Structures to Orchestrate Macrophage Phenotype by Cell Shape. Journal of Functional Biomaterials. 2022; 13(1):31. https://doi.org/10.3390/jfb13010031
Chicago/Turabian StyleLi, Kai, Lin Lv, Dandan Shao, Youtao Xie, Yunzhen Cao, and Xuebin Zheng. 2022. "Engineering Nanopatterned Structures to Orchestrate Macrophage Phenotype by Cell Shape" Journal of Functional Biomaterials 13, no. 1: 31. https://doi.org/10.3390/jfb13010031
APA StyleLi, K., Lv, L., Shao, D., Xie, Y., Cao, Y., & Zheng, X. (2022). Engineering Nanopatterned Structures to Orchestrate Macrophage Phenotype by Cell Shape. Journal of Functional Biomaterials, 13(1), 31. https://doi.org/10.3390/jfb13010031