Atomic Force Microscopy Study of the Effect of an Electric Field, Applied to a Pyramidal Structure, on Enzyme Biomolecules
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Enzyme
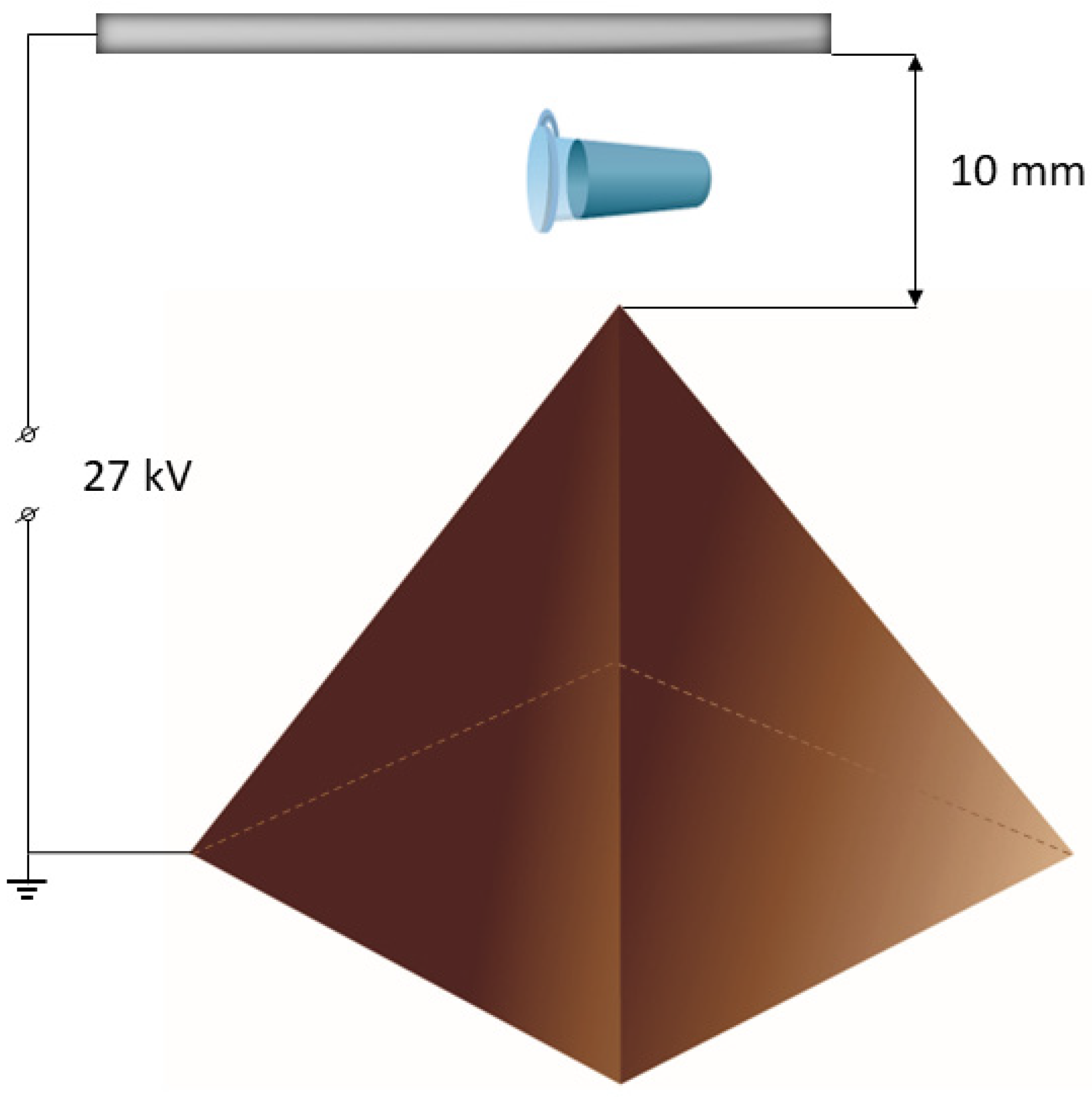
2.2. Experimental Setup
2.3. AFM Sample Preparation
2.4. AFM Measurements
2.5. Spectrophotometry Measurements
3. Results
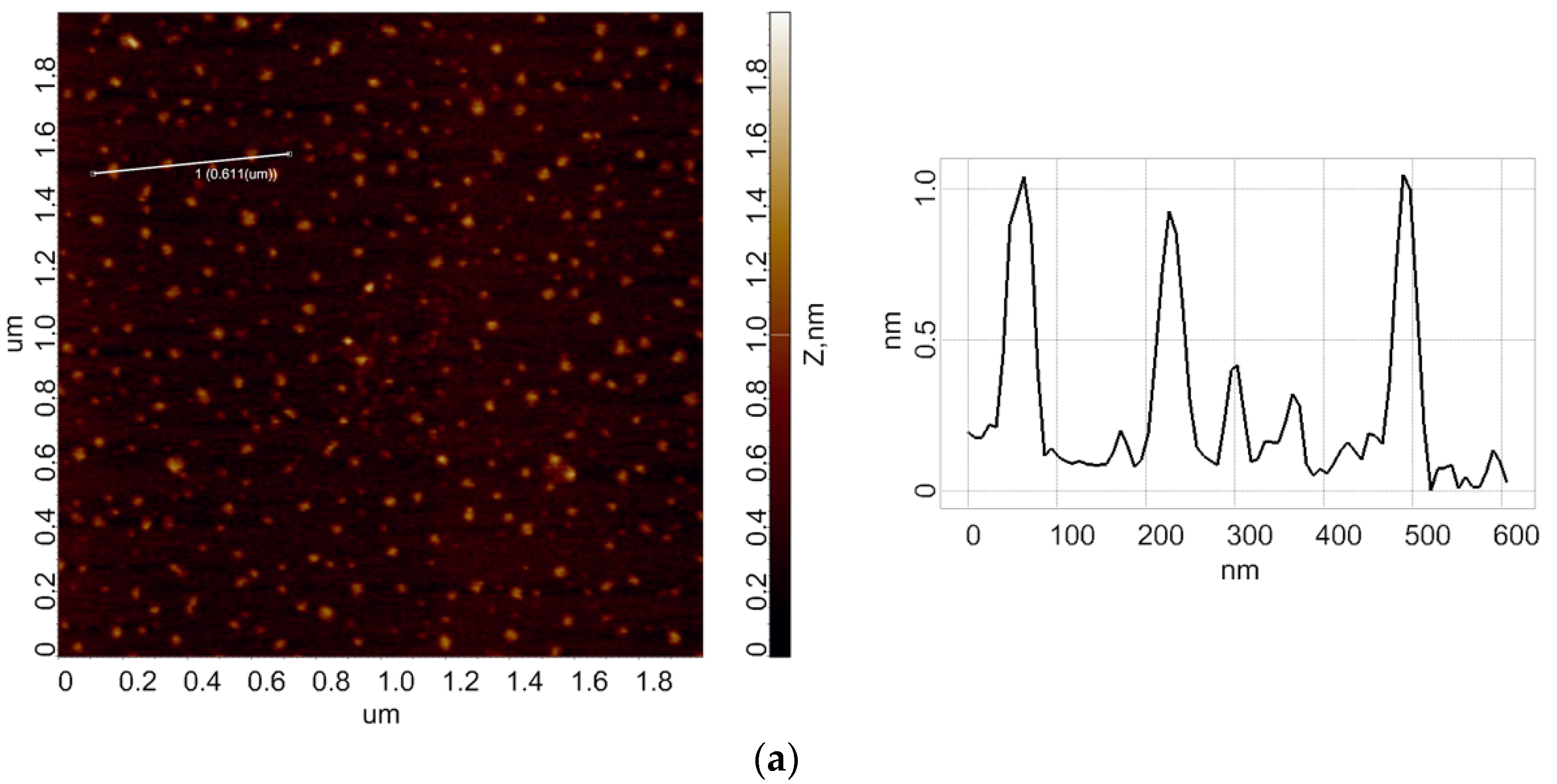
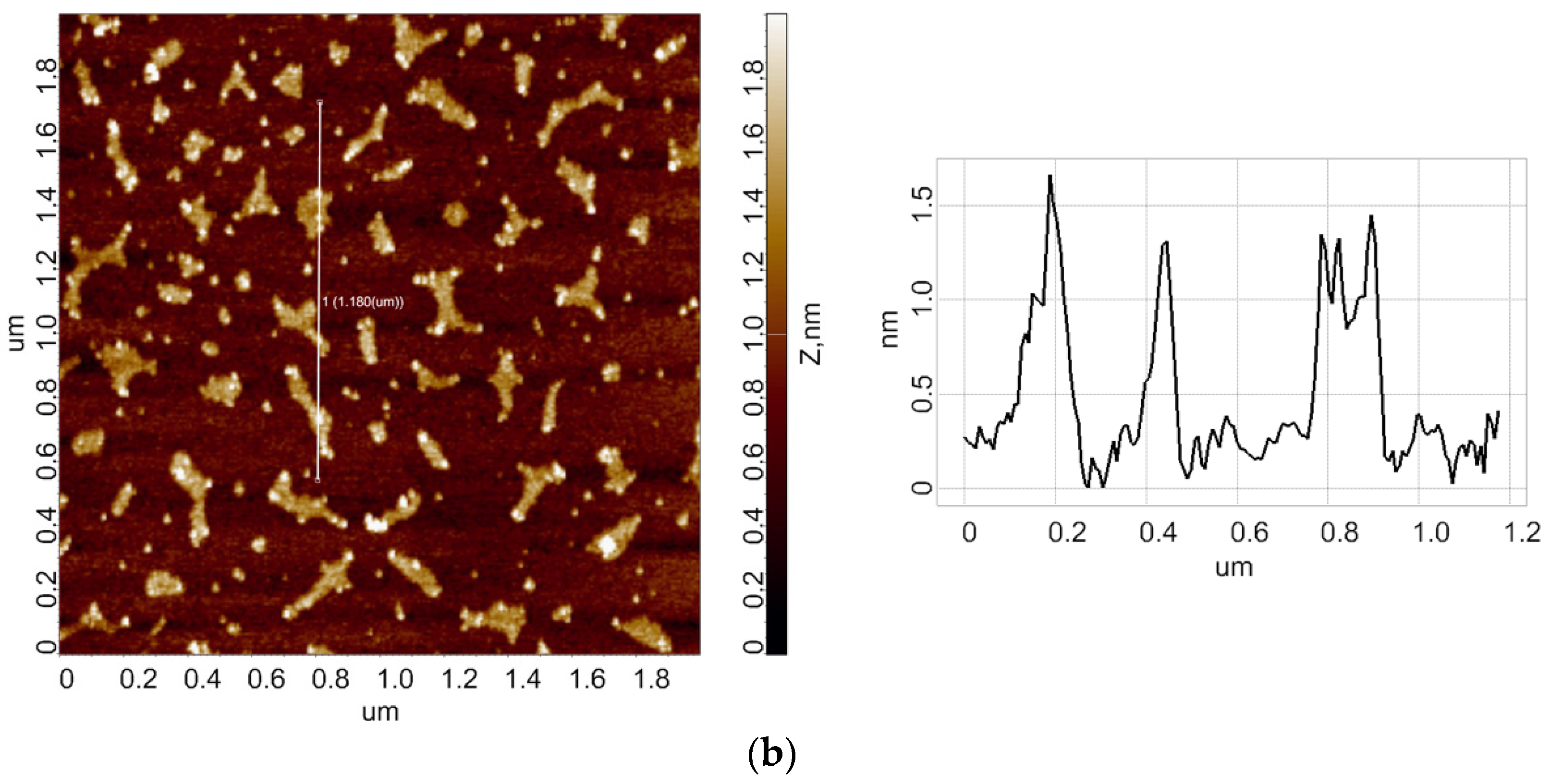
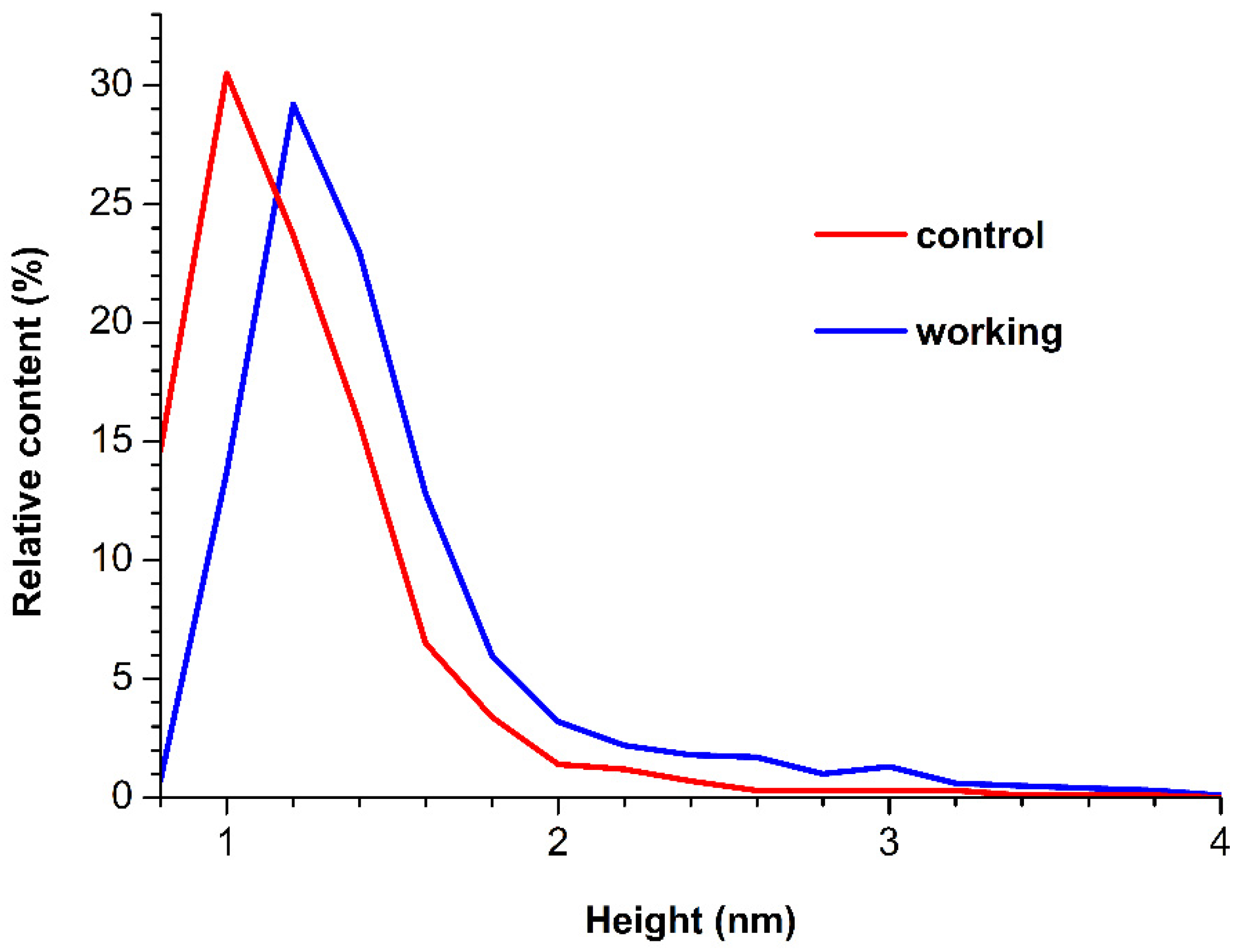
3.1. Atomic Force Microscopy
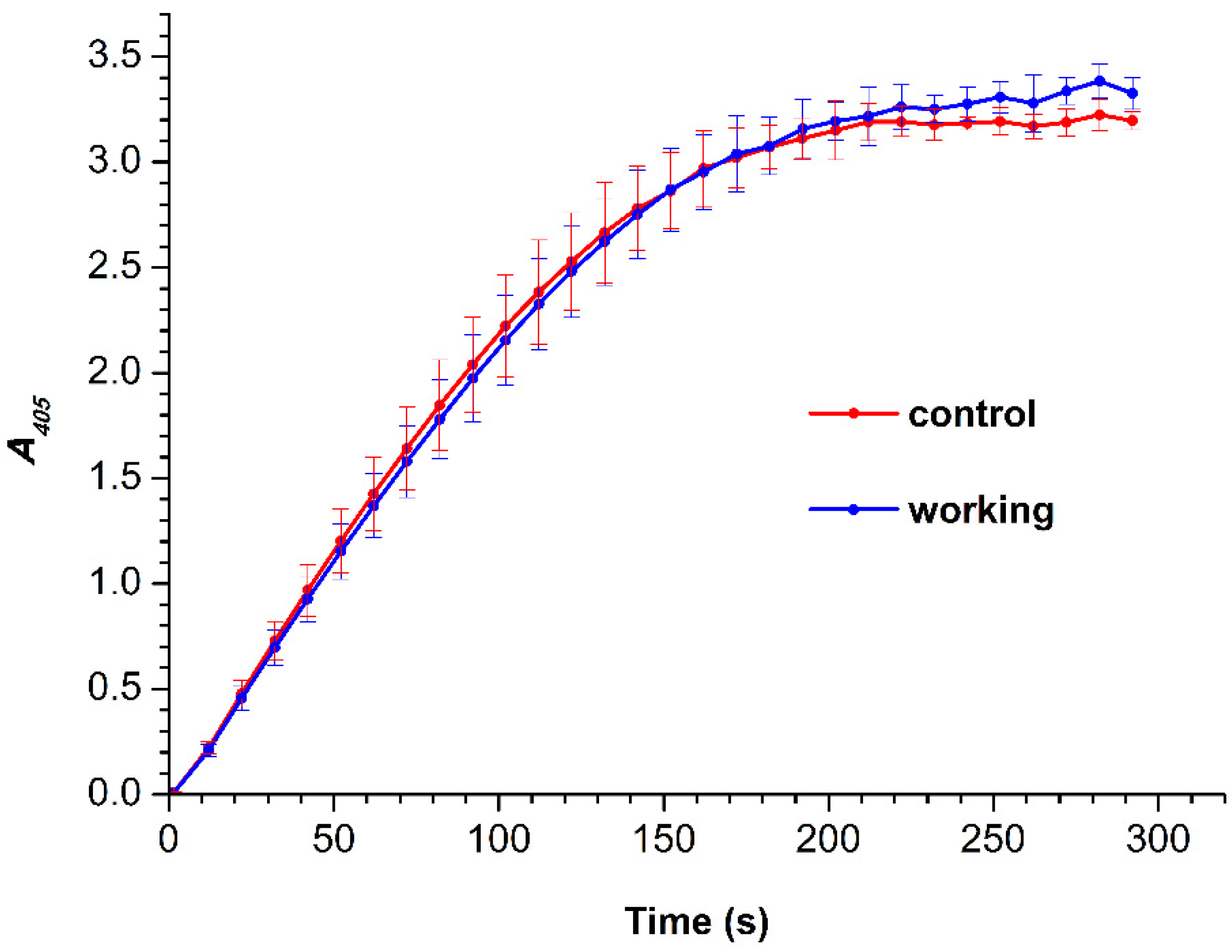
3.2. Spectrophotometry
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Latorre, M.E.; Bonelli, P.R.; Rojas, A.M.; Gerschenson, L.N. Microwave Inactivation of Red Beet (Beta vulgaris L. Var. Conditiva) Peroxidase and Polyphenoloxidase and the Effect of Radiation on Vegetable Tissue Quality. J. Food Eng. 2012, 109, 676–684. [Google Scholar] [CrossRef]
- Lopes, L.C.; Barreto, M.T.; Gonçalves, K.M.; Alvarez, H.M.; Heredia, M.F.; de Souza, R.O.M.; Cordeiro, Y.; Dariva, C.; Fricks, A.T. Stability and Structural Changes of Horseradish Peroxidase: Microwave versus Conventional Heating Treatment. Enzym. Microb. Technol. 2015, 69, 10–18. [Google Scholar] [CrossRef]
- Yao, Y.; Zhang, B.; Pang, H.; Wang, Y.; Fu, H.; Chen, X.; Wang, Y. The Effect of Radio Frequency Heating on the Inactivation and Structure of Horseradish Peroxidase. Food Chem. 2023, 398, 133875. [Google Scholar] [CrossRef]
- Fortune, J.A.; Wu, B.-I.; Klibanov, A.M. Radio Frequency Radiation Causes No Nonthermal Damage in Enzymes and Living Cells. Biotechnol. Prog. 2010, 26, 1772–1776. [Google Scholar] [CrossRef]
- Caliga, R.; Maniu, C.L.; Mihasan, M. ELF-EMF Exposure Decreases the Peroxidase Catalytic Efficiency In Vitro. Open Life Sci. 2016, 11, 71–77. [Google Scholar] [CrossRef]
- Wasak, A.; Drozd, R.; Jankowiak, D.; Rakoczy, R. The Influence of Rotating Magnetic Field on Bio-Catalytic Dye Degradation Using the Horseradish Peroxidase. Biochem. Eng. J. 2019, 147, 81–88. [Google Scholar] [CrossRef]
- Emamdadi, N.; Gholizadeh, M.; Housaindokht, M.R. Investigation of Static Magnetic Field Effect on Horseradish Peroxidase Enzyme Activity and Stability in Enzymatic Oxidation Process. Int. J. Biol. Macromol. 2020, 170, 189–195. [Google Scholar] [CrossRef]
- Sun, J.; Sun, F.; Xu, B.; Gu, N. The Quasi-One-Dimensional Assembly of Horseradish Peroxidase Molecules in Presence of the Alternating Magnetic Field. Colloids Surf. A Physicochem. Eng. Asp. 2010, 360, 94–98. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Ivanova, I.A.; Valueva, A.A.; Tatur, V.Y.; Smelov, M.V.; Ivanova, N.D.; Ziborov, V.S. AFM Imaging of Protein Aggregation in Studying the Impact of Knotted Electromagnetic Field on A Peroxidase. Sci. Rep. 2020, 10, 9022. [Google Scholar] [CrossRef]
- Ivanov, Y.; Pleshakova, T.; Shumov, I.; Kozlov, A.; Romanova, T.; Valueva, A.; Tatur, V.; Stepanov, I.; Ziborov, V. Investigation of the Influence of Liquid Motion in a Flow-Based System on an Enzyme Aggregation State with an Atomic Force Microscopy Sensor: The Effect of Water Flow. Appl. Sci. 2020, 10, 4560. [Google Scholar] [CrossRef]
- Ziborov, V.S.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Ivanova, I.A.; Valueva, A.A.; Tatur, V.Y.; Negodailov, A.N.; Lukyanitsa, A.; Ivanov, Y.D. Investigation of the Influence of Liquid Motion in a Flow-Based System on an Enzyme Aggregation State with an Atomic Force Microscopy Sensor: The Effect of Glycerol Flow. Appl. Sci. 2020, 10, 4825. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Valueva, A.A.; Ivanova, I.A.; Ershova, M.O.; Larionov, D.I.; Repnikov, V.V.; Ivanova, N.D.; et al. AFM and FTIR Investigation of the Effect of Water Flow on Horseradish Peroxidase. Molecules 2021, 26, 306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Cheng, L.; Wang, L.; Guan, Z. Inactivation Effects of PEF on Horseradish Peroxidase (HRP) and Pectinesterase (PE). IEEE Trans. Plasma Sci. 2006, 34, 2630–2636. [Google Scholar] [CrossRef]
- Yang, Z.H.; Xu, Q. Study of High Voltage Electric Field on the Mechanism of Horseradish Peroxidase Activity. Adv. Mater. Res. 2011, 236–238, 2383–2386. [Google Scholar] [CrossRef]
- Deamer, D.; Akeson, M.; Branton, D. Three Decades of Nanopore Sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, J.; Hinkers, H.; Sundermeier, C.; Seifert, W.; Morell, O.M.; Knoll, M. Miniaturized Real-Time Monitoring System for L-Lactate and Glucose Using Microfabricated Multi-Enzyme Sensors. Biosens. Bioelectron. 2000, 15, 515–522. [Google Scholar] [CrossRef]
- Dufrêne, Y.; Ando, T.; Garcia, R.; Alsteens, Y.F.D.D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Muller, D.J. Imaging Modes of Atomic Force Microscopy for Application in Molecular and Cell Biology. Nat. Nanotechnol. 2017, 12, 295–307. [Google Scholar] [CrossRef]
- Kodera, N.; Noshiro, D.; Dora, S.K.; Mori, T.; Habchi, J.; Blocquel, D.; Gruet, A.; Dosnon, M.; Salladini, E.; Bignon, C.; et al. Structural and Dynamics Analysis of Intrinsically Disordered Proteins by High-Speed Atomic Force Microscopy. Nat. Nanotechnol. 2020, 16, 181–189. [Google Scholar] [CrossRef]
- Kumaki, J.; Nishikawa, Y.; Hashimoto, T. Visualization of Single-Chain Conformations of a Synthetic Polymer with Atomic Force Microscopy. J. Am. Chem. Soc. 1996, 118, 3321–3322. [Google Scholar] [CrossRef]
- Ierardi, V.; Ferrera, F.; Millo, E.; Damonte, G.; Filaci, G.; Valbusa, U. Bioactive Surfaces for Antibody-Antigen Complex Detection by Atomic Force Microscopy. J. Phys. Conf. Ser. 2013, 439, 012001. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Bukharina, N.S.; Frantsuzov, P.A.; Pleshakova, T.O.; Kanashenko, S.L.; Medvedeva, N.V.; Argentova, V.V.; Zgoda, V.G.; Munro, A.W.; Archakov, A.I. AFM Study of Cytochrome CYP102A1 Oligomeric State. Soft Matter 2012, 8, 4602–4608. [Google Scholar] [CrossRef]
- Pleshakova, T.O.; Bukharina, N.S.; Archakov, A.; Ivanov, Y.D. Atomic Force Microscopy for Protein Detection and Their Physicochemical Characterization. Int. J. Mol. Sci. 2018, 19, 1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramanaviciene, A.; Snitka, V.; Mieliauskiene, R.; Kazlauskas, R.; Ramanavicius, A. AFM Study of Complement System Assembly Initiated by Antigen-Antibody Complex. Open Chem. 2006, 4, 194–206. [Google Scholar] [CrossRef]
- Crampton, N.; Bonass, W.A.; Kirkham, J.; Thomson, N.H. Formation of Aminosilane-Functionalized Mica for Atomic Force Microscopy Imaging of DNA. Langmuir 2005, 21, 7884–7891. [Google Scholar] [CrossRef]
- Limanskaya, L.A.; Limanskii, A.P. Compaction of Single Supercoiled DNA Molecules Adsorbed onto Amino Mica. Russ. J. Bioorg. Chem. 2006, 32, 444–459. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Ivanova, I.A.; Valueva, A.A.; Ershova, M.O.; Tatur, V.Y.; Stepanov, I.N.; Repnikov, V.V.; et al. AFM Study of Changes in Properties of Horseradish Peroxidase after Incubation of Its Solution near a Pyramidal Structure. Sci. Rep. 2021, 11, 9907. [Google Scholar] [CrossRef]
- Davies, P.; Rennke, H.; Cotran, R. Influence of Molecular Charge upon the Endocytosis and Intracellular Fate of Peroxidase Activity in Cultured Arterial Endothelium. J. Cell Sci. 1981, 49, 69–86. [Google Scholar] [CrossRef]
- Welinder, K.G. Amino Acid Sequence Studies of Horseradish Peroxidase. Amino and Carboxyl Termini, Cyanogen Bromide and Tryptic Fragments, the Complete Sequence, and Some Structural Characteristics of Horseradish Peroxidase C. Eur. J. Biochem. 1979, 96, 483–502. [Google Scholar] [CrossRef]
- Tams, J.; Welinder, K.G. Mild Chemical Deglycosylation of Horseradish Peroxidase Yields a Fully Active, Homogeneous Enzyme. Anal. Biochem. 1995, 228, 48–55. [Google Scholar] [CrossRef]
- Ignatenko, O.V.; Sjölander, A.; Hushpulian, D.M.; Kazakov, S.V.; Ouporov, I.V.; Chubar, T.A.; Poloznikov, A.A.; Ruzgas, T.; Tishkov, V.I.; Gorton, L.; et al. Electrochemistry of Chemically Trapped Dimeric and Monomeric Recombinant Horseradish Peroxidase. Adv. Biosens. Bioelectron. 2013, 2, 25–34. [Google Scholar]
- Kiselyova, O.I.; Yaminsky, I.; Ivanov, Y.D.; Kanaeva, I.P.; Kuznetsov, V.Y.; Archakov, A.I. AFM Study of Membrane Proteins, Cytochrome P450 2B4, and NADPH–Cytochrome P450 Reductase and Their Complex Formation. Arch. Biochem. Biophys. 1999, 371, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pleshakova, T.O.; Kaysheva, A.L.; Shumov, I.D.; Ziborov, V.S.; Bayzyanova, J.M.; Konev, V.A.; Uchaikin, V.F.; Archakov, A.I.; Ivanov, Y.D. Detection of Hepatitis C Virus Core Protein in Serum Using Aptamer-Functionalized AFM Chips. Micromachines 2019, 10, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, S.A.; Bray, R.C.; Smith, A.T. PH-Dependent Properties of a Mutant Horseradish Peroxidase Isoenzyme C in Which Arg38 Has Been Replaced with Lysine. Eur. J. Biochem. 1994, 224, 1029–1037. [Google Scholar] [CrossRef] [PubMed]
- Enzymatic Assay of Peroxidase (EC 1.11.1.7) 2,20-Azino-Bis(3-Ethylbenzthiazoline-6-Sulfonic Acid) as a Substrate Sigma Prod. No. P-6782. Available online: https://www.sigmaaldrich.com/RU/en/technical-documents/protocol/protein-biology/enzymeactivity-assays/enzymatic-assay-of-peroxidase-abts-as-substrate (accessed on 18 February 2022).
- Pershin, S.M. Conversion of Ortho-Para H2O Isomers in Water and a Jump in Erythrocyte Fluidity through a Microcapillary at a Temperature of 36.6 ± 0.3 °C. Phys. Wave Phenom. 2009, 17, 241–250. [Google Scholar] [CrossRef]
- Morón, M. Protein Hydration Shell Formation: Dynamics of Water in Biological Systems Exhibiting Nanoscopic Cavities. J. Mol. Liq. 2021, 337, 116584. [Google Scholar] [CrossRef]
- Ziborov, V.S.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Valueva, A.A.; Ivanova, I.A.; Ershova, M.O.; Larionov, D.I.; Evdokimov, A.N.; Tatur, V.Y.; et al. The Impact of Fast-Rise-Time Electromagnetic Field and Pressure on the Aggregation of Peroxidase upon Its Adsorption onto Mica. Appl. Sci. 2021, 11, 11677. [Google Scholar] [CrossRef]
- Pershin, S.M. Physical Basis of the Anomalous Properties of Water-Quantum Differences between the Ortho and Para Spin Isomers of H2O. Available online: http://www.biophys.ru/lib/sci/water/250-h2o-00029 (accessed on 1 October 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanov, Y.D.; Tatur, V.Y.; Shumov, I.D.; Kozlov, A.F.; Valueva, A.A.; Ivanova, I.A.; Ershova, M.O.; Ivanova, N.D.; Stepanov, I.N.; Lukyanitsa, A.A.; et al. Atomic Force Microscopy Study of the Effect of an Electric Field, Applied to a Pyramidal Structure, on Enzyme Biomolecules. J. Funct. Biomater. 2022, 13, 234. https://doi.org/10.3390/jfb13040234
Ivanov YD, Tatur VY, Shumov ID, Kozlov AF, Valueva AA, Ivanova IA, Ershova MO, Ivanova ND, Stepanov IN, Lukyanitsa AA, et al. Atomic Force Microscopy Study of the Effect of an Electric Field, Applied to a Pyramidal Structure, on Enzyme Biomolecules. Journal of Functional Biomaterials. 2022; 13(4):234. https://doi.org/10.3390/jfb13040234
Chicago/Turabian StyleIvanov, Yuri D., Vadim Y. Tatur, Ivan D. Shumov, Andrey F. Kozlov, Anastasia A. Valueva, Irina A. Ivanova, Maria O. Ershova, Nina D. Ivanova, Igor N. Stepanov, Andrei A. Lukyanitsa, and et al. 2022. "Atomic Force Microscopy Study of the Effect of an Electric Field, Applied to a Pyramidal Structure, on Enzyme Biomolecules" Journal of Functional Biomaterials 13, no. 4: 234. https://doi.org/10.3390/jfb13040234
APA StyleIvanov, Y. D., Tatur, V. Y., Shumov, I. D., Kozlov, A. F., Valueva, A. A., Ivanova, I. A., Ershova, M. O., Ivanova, N. D., Stepanov, I. N., Lukyanitsa, A. A., & Ziborov, V. S. (2022). Atomic Force Microscopy Study of the Effect of an Electric Field, Applied to a Pyramidal Structure, on Enzyme Biomolecules. Journal of Functional Biomaterials, 13(4), 234. https://doi.org/10.3390/jfb13040234






