Metal–Peptide Complexes with Antimicrobial Potential for Cotton Fiber Protection
Abstract
:1. Introduction
2. Materials and Methods
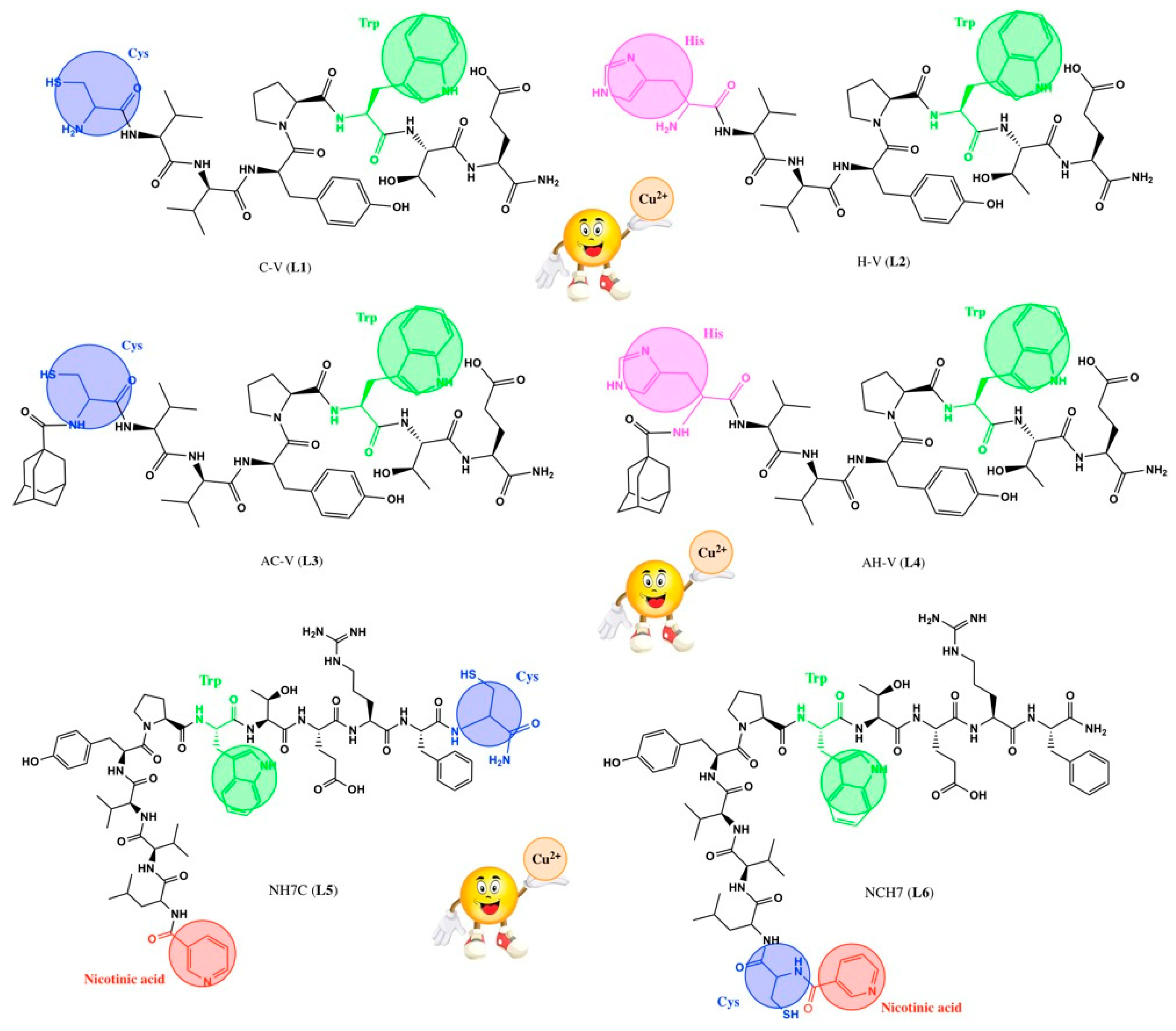
2.1. Synthesis of the Peptides
2.2. Physicochemical Characterization in Solution
2.2.1. Spectral Measurements
Apparatus
Solutions for UV-Vis and Fluorescence Analysis:
2.2.2. Electrochemical Measurements
2.3. Preparation of Complexes in Solid Phase
2.4. Coating and Fastness Test of Cotton Fibers with Alkaline Solutions of the Metallopeptide Complexes
2.5. Virology
2.5.1. Cytotoxicity Assay
2.5.2. Antiviral Activity Assay
2.5.3. Virucidal Assay
3. Results and Discussion
3.1. Solution State Characterization
3.1.1. Spectral Investigations: UV-Vis and Fluorimetry
3.1.2. Cyclic Voltammetry
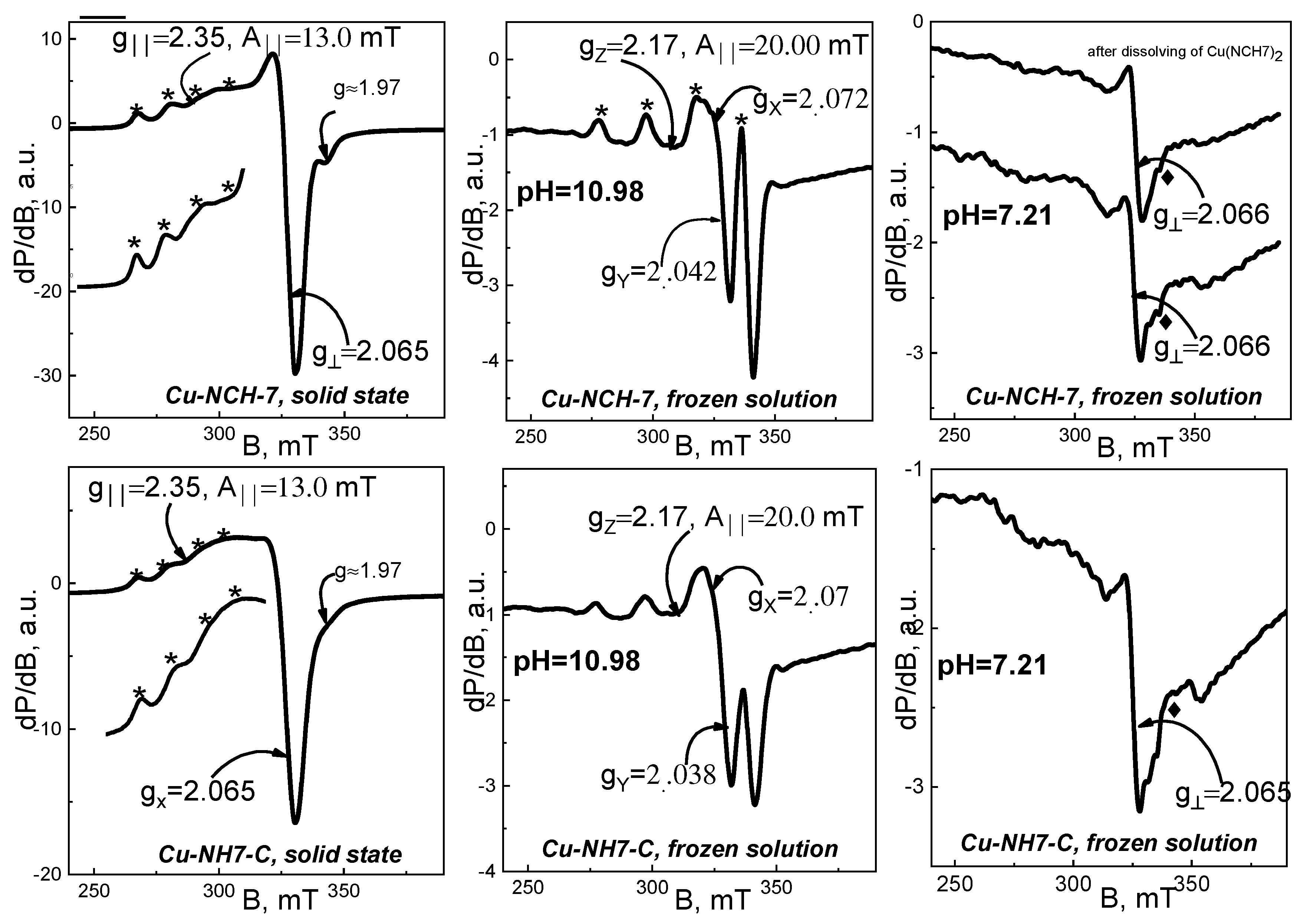
3.1.3. EPR Analysis and IR Spectroscopy
3.1.4. SEM Analysis
3.2. Virological Activity
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Smith, A.M. CHAPTER 1:Interaction of Metal Ions with Proteins as a Source of Inspiration for Biomimetic Materials. In Functional Metallosupramolecular Materials; Royal Society of Chemistry: London, UK, 2015; pp. 1–31. [Google Scholar] [CrossRef]
- Cowan, J.A. Inorganic Biochemistry: An Introduction, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 1997. [Google Scholar]
- Pickart, L.; Margolina, A. Regenerative and Protective Actions of the GHK-Cu Peptide in the Light of the New Gene Data. Int. J. Mol. Sci. 2018, 19, 1987. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.; Sauce-Guevara, M.A.; Alarcon, E.I.; Mendez-Rojas, M.A.; Mendez-Rojas, M.A. Peptide Biomaterials for Tissue Regeneration. Front. Bioeng. Biotechnol. 2022, 10, 1116. [Google Scholar] [CrossRef]
- Zamoj, K.; Kamrowski, D.; Zdrowowicz, M.; Wyrzykowski, D.; Wiczk, W.; Chmurzynski, L.; Makowska, J. A Pentapeptide with Tyrosine Moiety as Fluorescent Chemosensor for Selective Nanomolar-Level Detection of Copper(II) Ions. Int. J. Mol. Sci. 2020, 21, 743. [Google Scholar] [CrossRef]
- Kozłowski, H.; Kowalik-Jankowska, T.; Jeżowska-Bojczuk, M. Chemical and biological aspects of Cu2+ interactions with peptides and aminoglycosides. Coord. Chem. Rev. 2005, 249, 2323–2334. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, X.; Dai, W.; Shao, Z. X-ray photoelectron spectroscopic and Raman analysis of silk fibroin-Cu(II) films. Biopolymers 2006, 82, 144–151. [Google Scholar] [CrossRef]
- Di Natale, C.; De Benedictis, I.; De Benedictis, A.; Marasco, D. Metal-Peptide Complexes as Promising Antibiotics to Fight Emerging Drug Resistance: New Perspectives in Tuberculosis. Antibiotics 2020, 9, 337. [Google Scholar] [CrossRef]
- Sasmal, P.K.; Streu, C.N.; Meggers, E. Metal Complex Catalysis in Living Biological Systems. Chem. Commun. 2013, 49, 1581–1587. [Google Scholar] [CrossRef]
- Alexander, J.L.; Thompson, Z.; Cowan, J.A. Antimicrobial Metallopeptides. ACS Chem. Biol. 2018, 13, 844–853. [Google Scholar] [CrossRef]
- Makowska, J.; Żamojć, K.; Wyrzykowski, D.; Uber, D.; Wierzbicka, M.; Wiczk, W.; Chmurzyński, L. Binding of Cu(II) ions to peptides studied by fluorescence spectroscopy and isothermal titration calorimetry. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 153, 451–456. [Google Scholar] [CrossRef]
- Maret, W. Metalloproteomics, metalloproteomes, and the annotation of metalloproteins. Metallomics 2010, 2, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Mori, Y.; Mori, S.; Irie, T.; Nagai, H.; Goto, T.; Tatsu, Y.; Imaishi, H.; Morigaki, K. Microarray of human P450 with an integrated oxygen sensing film for high-throughput detection of metabolic activities. Anal. Chem. 2012, 84, 5292–5297. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yeung, N.; Sieracki, N.; Marshall, N.M. Design of functional metalloproteins. Nature 2009, 460, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Rongfeng, Z.; Wang, Q.; Wu, J.; Wu, J.; Schmuck, C.; Tian, H. Peptide self-assembly triggered by metal ions. Chem. Soc. Rev. 2015, 44, 5200–5219. [Google Scholar] [CrossRef]
- Todorov, P.; Peneva, P.; Tchekalarova, J.; Georgieva, S. Potential anticonvulsant activity of novel VV-hemorphin-7 analogues containing unnatural amino acids: Synthesis and characterization. Amino Acids 2020, 52, 567–585. [Google Scholar] [CrossRef] [PubMed]
- Blishchenko, E.Y.; Sazonova, O.V.; Kalinina, O.A.; Yatskin, O.N.; Philippova, M.M.; Surovoy, A.Y.; Karelin, A.A.; Ivanov, V.T. Family of hemorphins: Co-relations between amino acid sequences and effects in cell cultures. Peptides 2002, 23, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Jinsmaa, Y.; Yoshikawa, M. Release of Hemorphin-5 from Human Hemoglobin by Pancreatic Elastase. Biosci. Biotechnol. Biochem. 2002, 66, 1130–1132. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Alzeyoudi, R.; Almutawa, S.A.; Alnajjar, A.N.; Vijayan, R. Molecular basis of the therapeutic properties of hemorphins. Pharmacol. Res. 2020, 158, 104855. [Google Scholar] [CrossRef]
- Ali, A.; Baby, B.; Soman, S.S.; Vijayan, R. Molecular insights into the interaction of hemorphin and its targets. Sci. Rep. 2020, 9, 14747. [Google Scholar] [CrossRef]
- Nyberg, F.; Sanderson, K.; Glämsta, E.L. The hemorphins: A new class of opioid peptides derived from the blood protein hemoglobin. Biopolymers 1997, 43, 147–156. [Google Scholar] [CrossRef]
- Glämsta, E.L.; Marklund, A.; Hellman, U.; Wernstedt, C.; Terenius, L.; Nyberg, F. Isolation and characterization of a hemo-globinderived opioid peptide from the human pituitary gland. Regul. Pept. 1991, 34, 169–179. [Google Scholar] [CrossRef]
- Todorov, P.; Georgieva, S.; Staneva, D.; Peneva, P.; Grozdanov, P.; Nikolova, I.; Vasileva-Tonkova, E.; Grabchev, I. Study of Novel Peptides for Antimicrobial Protection in Solution and on Cotton Fabric. Molecules 2022, 27, 4770. [Google Scholar] [CrossRef] [PubMed]
- Sóvágó, I.; Várnagy, K.; Lihi, N.; Grenács, Á. Coordinating properties of peptides containing histidyl residues. Coord. Chem. Rev. 2016, 327–328, 43–54. [Google Scholar] [CrossRef]
- Yip, T.T.; Nakagawa, Y.; Porath, J. Evaluation of the interaction of peptides with Cu(II), Ni(II), and Zn(II) by high-performance immobilized metal ion affinity chromatography. Anal. Biochem. 1989, 183, 159–171. [Google Scholar] [CrossRef]
- Shang, S.M. 13-Process control in dyeing of textiles. In Woodhead Publishing Series in Textiles, Process Control in Textile Manufacturing; Majumdar, A., Das, A., Alagirusamy, R., Kothari, V., Eds.; Woodhead Publishing: Sawston, Cambridge, 2013; pp. 300–338. ISBN 9780857090270. [Google Scholar] [CrossRef]
- Mohini, K.; Tejashree, L.; Vijay, N. Data set on analysis of dyeing property of natural dye from Thespesia populnea bark on different. Data Brief 2018, 16, 401–410. [Google Scholar] [CrossRef]
- Chowdhury, S.M.; Talukder, S.A.; Khan, A.M.; Afrin, N.; Ali, M.A.; Islam, R.; Parves, R.; Al Mamun, A.; Sufian, M.A.; Hossain, M.N.; et al. Antiviral Peptides as Promising Therapeutics against SARS-CoV-2. J. Phys. Chem. B 2020, 124, 9785–9792. [Google Scholar] [CrossRef] [PubMed]
- Kowalik-Jankowska, T.; Rajewska, A.; Jankowska, E.; Grzonka, Z. Copper(II) binding by fragments of α-synuclein containing M1-D2- and –H50 –residues; a combined potentiometric and spectroscopic study. Dalton Trans. 2006, 42, 5068–5076. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 443–530, ISBN-10: 0-387-31278-1, ISBN-13: 978-0387-31278-1. [Google Scholar]
- Williams, R.T.; Bridges, J.W. Fluorescence of solutions: A review. J. Clin. Path. 1964, 17, 371. [Google Scholar] [CrossRef] [PubMed]
- Enriquez, J.; Yu, M.; Bouley, B.; Xie, D.; Que, E.L. Copper(II) complexes for cysteine detection using 19F magnetic resonance. Dalton Trans. 2018, 47, 15024. [Google Scholar] [CrossRef]
- Tabak, M.; Sartor, G.; Cavatorta, P. On the interactions of metal ions with tryptophan and glyciltryptophan: A fluorescence study. J. Lumin. 1989, 43, 355–361. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, J.; Zhou, G. Determination of Cysteine using the Fluorescence from a L-Tyrosine-Copper(II) Complex. Anal. Lett. 2017, 50, 1168–1181. [Google Scholar] [CrossRef]
- Huang, Z.; Du, J.; Zhang, J.; Yu, X.Q.; Pu, L. A simple and efficient fluorescent sensor for histidine. Chem. Commun. 2012, 48, 3412–3414. [Google Scholar] [CrossRef]
- Zong, X.H.; Zhou, P.; Shao, Z.; Chen, S.M.; Chen, X.; Hu, B.W.; Deng, F.; Yao, W.H. Effect of pH and copper(II) on the conformation transitions of silk fibroin based on EPR, NMR, and Raman spectroscopy. Biochemistry 2004, 43, 11932–11941. [Google Scholar] [CrossRef] [PubMed]
- Alston, R.W.; Lasagna, M.; Grimsley, G.R.; Scholtz, J.M.; Reinhart, G.D.; Pace, C.N. Peptide sequence and conformation strongly influence tryptophan fluorescence. Biophys. J. 2008, 94, 2280–2287. [Google Scholar] [CrossRef] [PubMed]
- Silva, K.I.; Michael, B.C.; Geib, S.J.; Saxena, S. ESEEM analysis of multi-histidine Cu(II)-coordination in model complexes, peptides, and amyloid-β. J. Phys. Chem. B 2014, 118, 8935–8944. [Google Scholar] [CrossRef] [PubMed]
- Synhaivska, O.; Mermoud, Y.; Baghernejad, M.; Alshanski, I.; Hurevich, M.; Yitzchaik, S.; Wipf, M. Detection of Cu2+ Ions with GGH Peptide Realized with Si-Nanoribbon ISFET. Sensors 2019, 19, 4022. [Google Scholar] [CrossRef] [PubMed]
- Vuceta, J.; Morgan, J. Hydrolysis of Cu (II). Limnol. Oceanogr. 1977, 22, 742–746. [Google Scholar] [CrossRef]
- Kowalik-Jankowska, T.; Ruta-Dolejsz, M.; Wiśniewska, K.; Łankiewicz, L. Cu(II) interaction with N-terminal fragments of human and mouse β-amyloid peptide. J. Inorg. Biochem. 2001, 86, 535–545. [Google Scholar] [CrossRef]
- Kowalik-Jankowska, T.; Ruta-Dolejsz, M.; Wiśniewska, K.; Łankiewicz, L. Coordination of copper(II) ions by the 11–20 and 11–28 fragments of human and mouse β-amyloid peptide. J. Inorg. Biochem. 2002, 92, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Székely, E.; Csire, G.; Balogh, B.D.; Erdei, J.Z.; Király, J.M.; Kocsi, J.; Pinkóczy, J.; Várnagy, K. The Role of Side Chains in the Fine-Tuning of the Metal-Binding Ability of Multihistidine Peptides. Molecules 2022, 27, 3435. [Google Scholar] [CrossRef]
- Devi, S.; Singh, B.; Paul, A.K.; Tyagi, S. Highly sensitive and selective detection of trinitrotoluene using cysteine-capped gold nanoparticles. Anal. Methods 2016, 8, 4398–4405. [Google Scholar] [CrossRef]
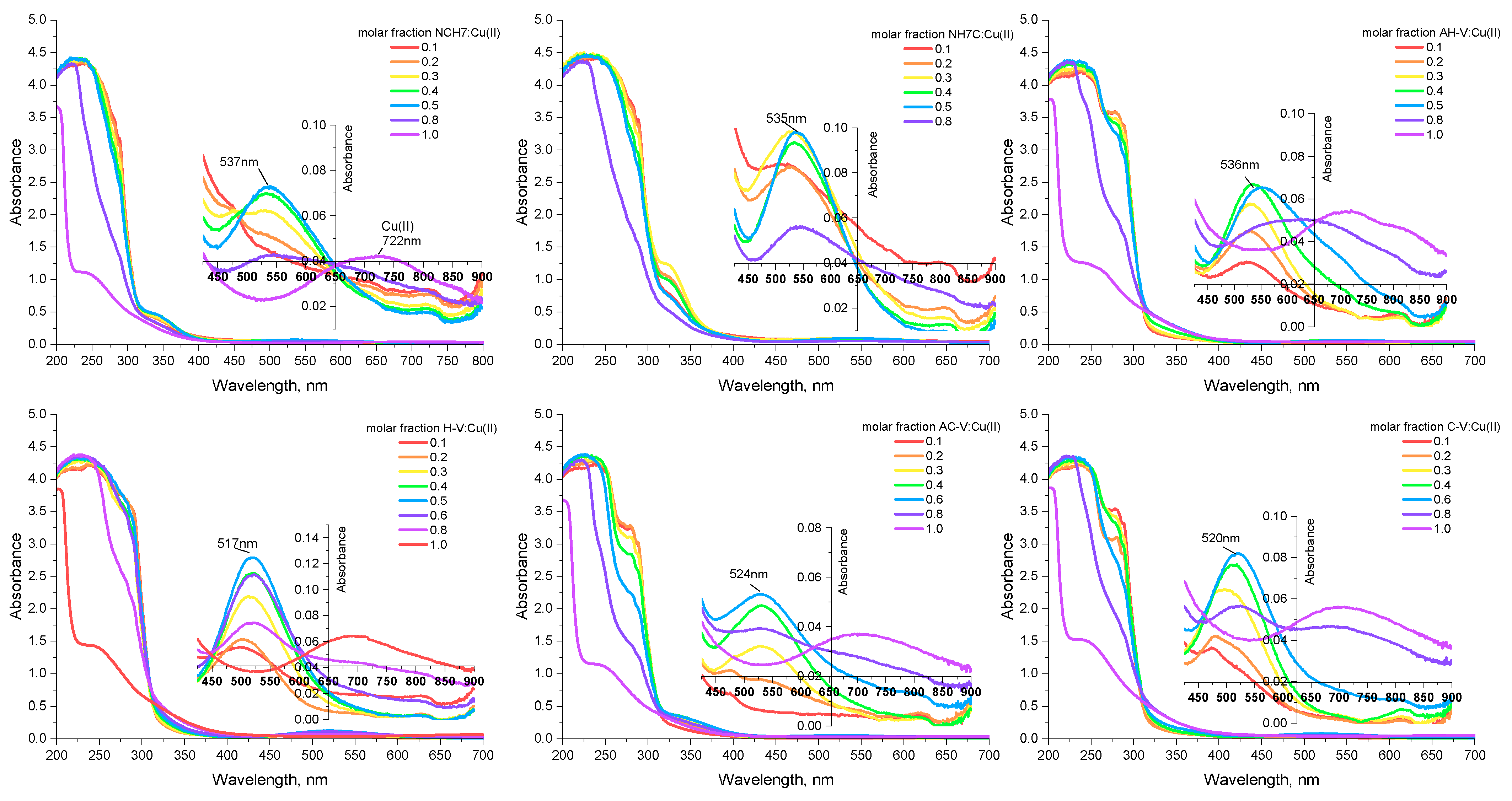
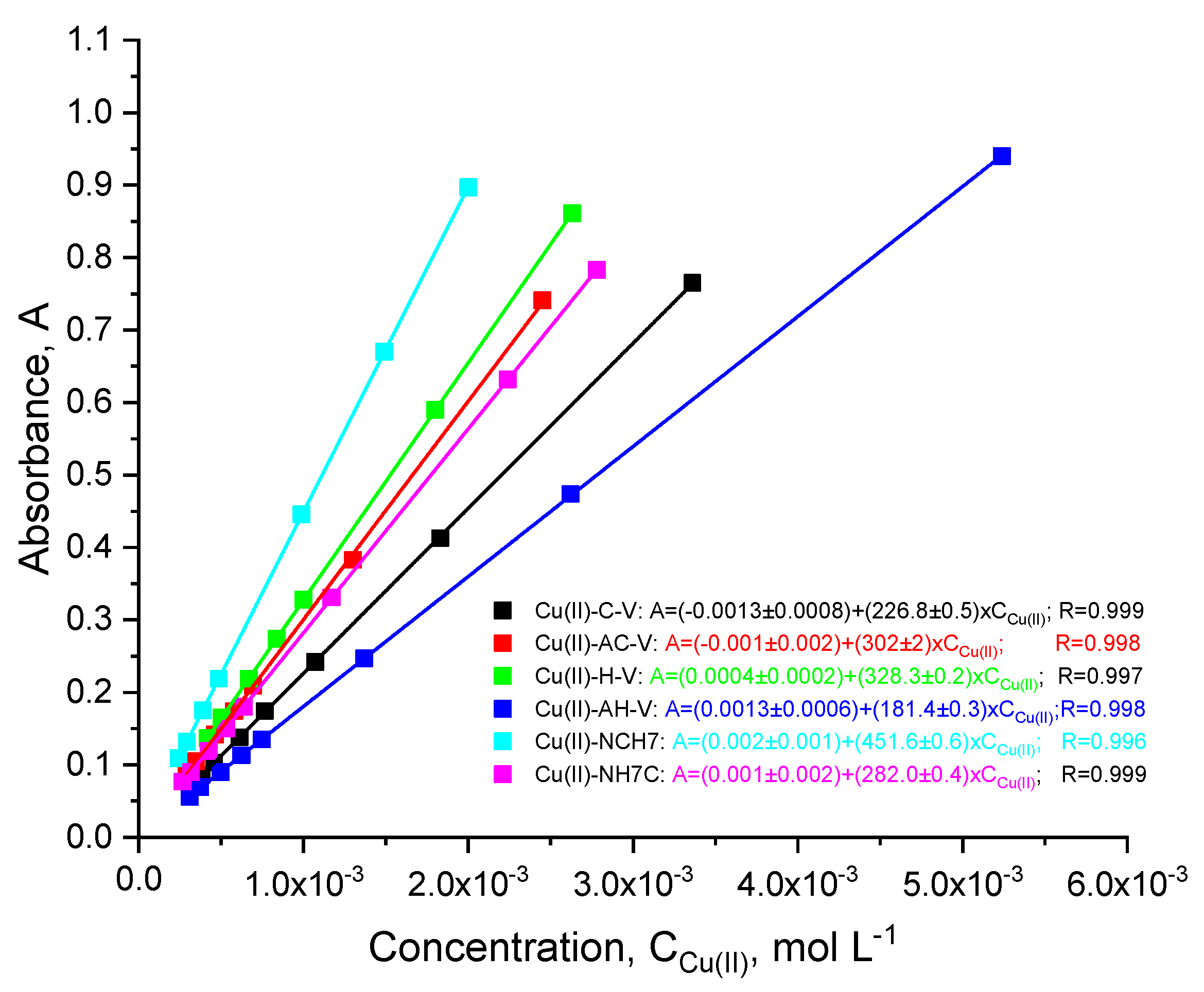








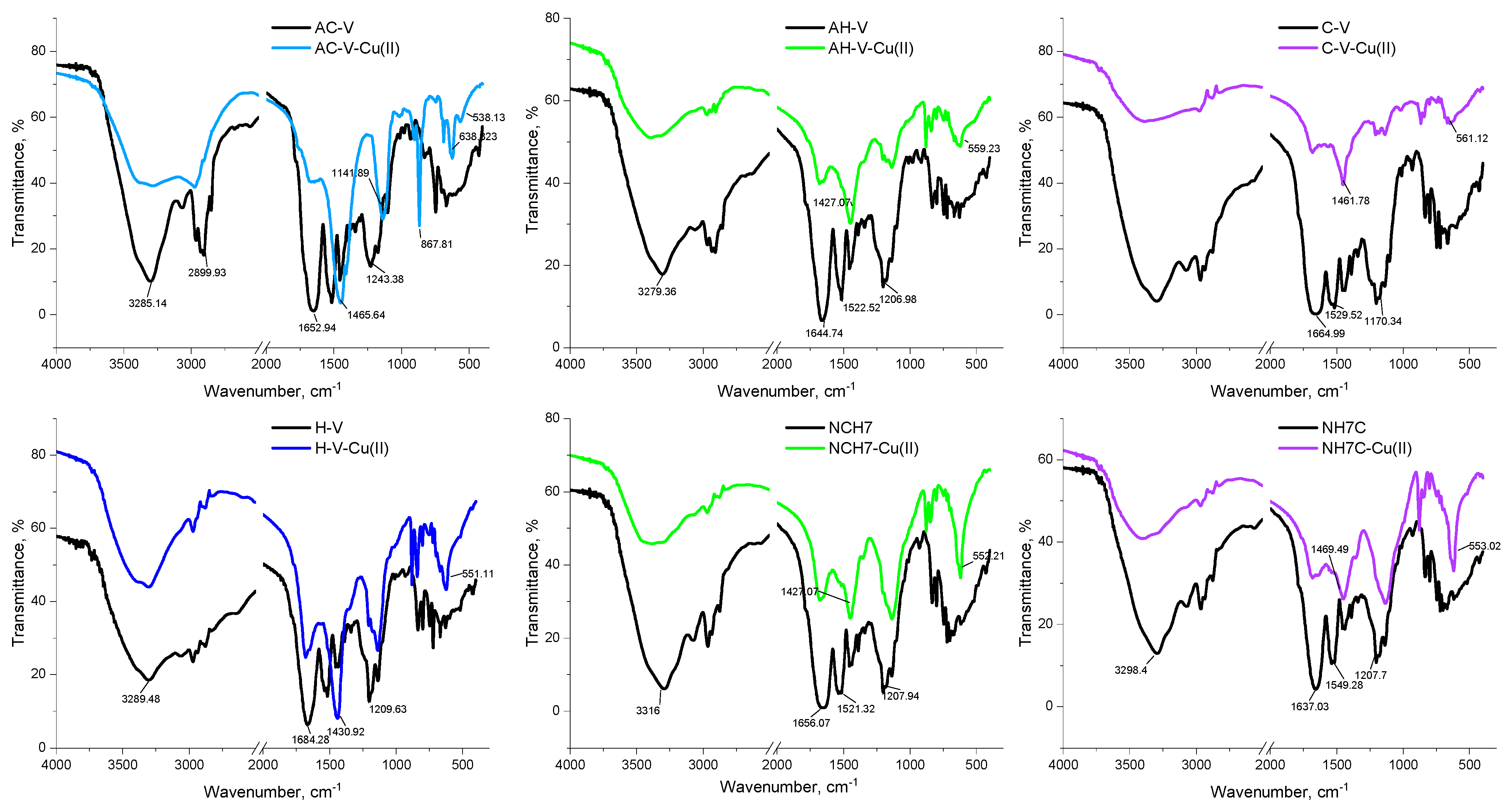



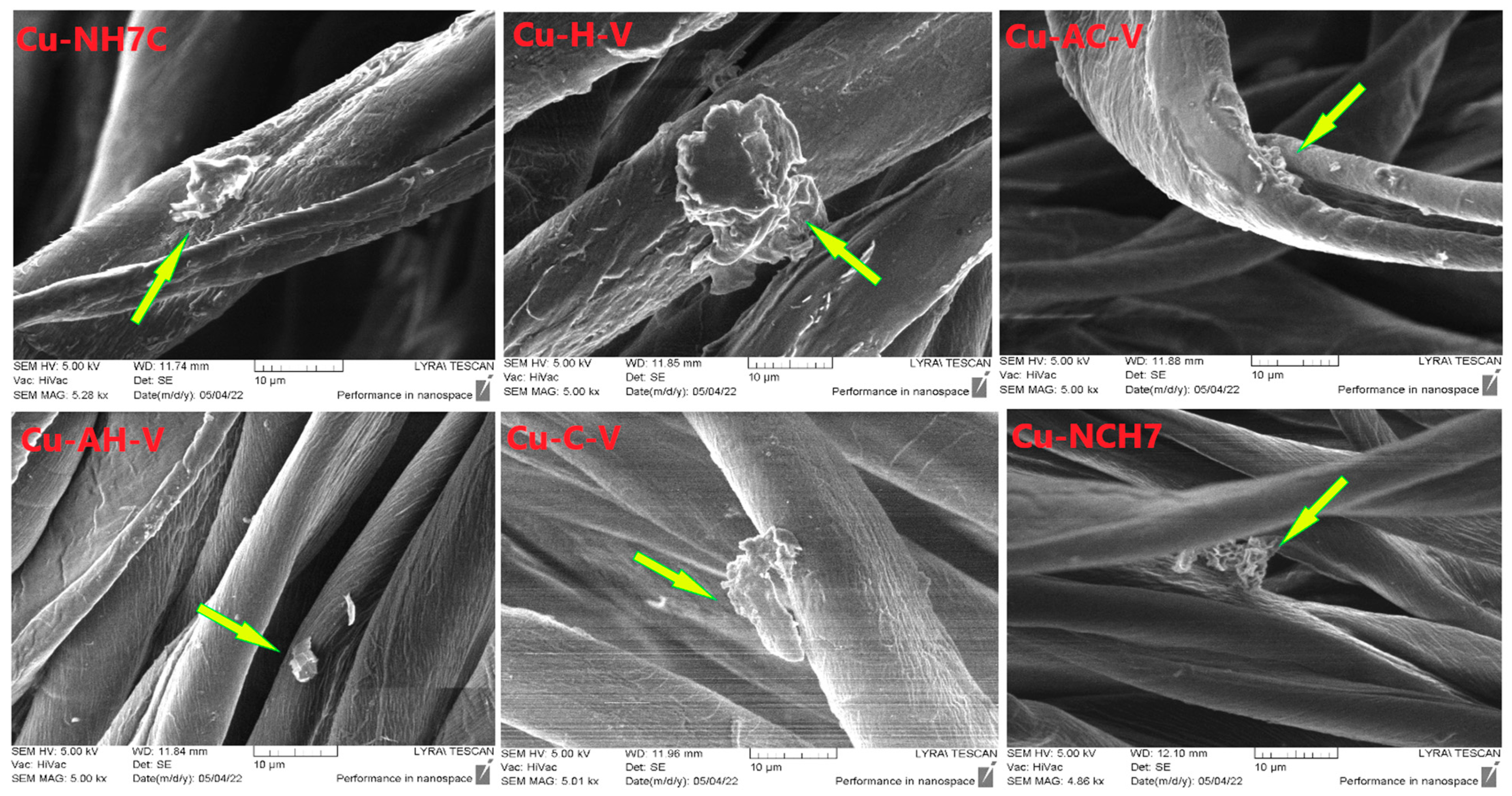
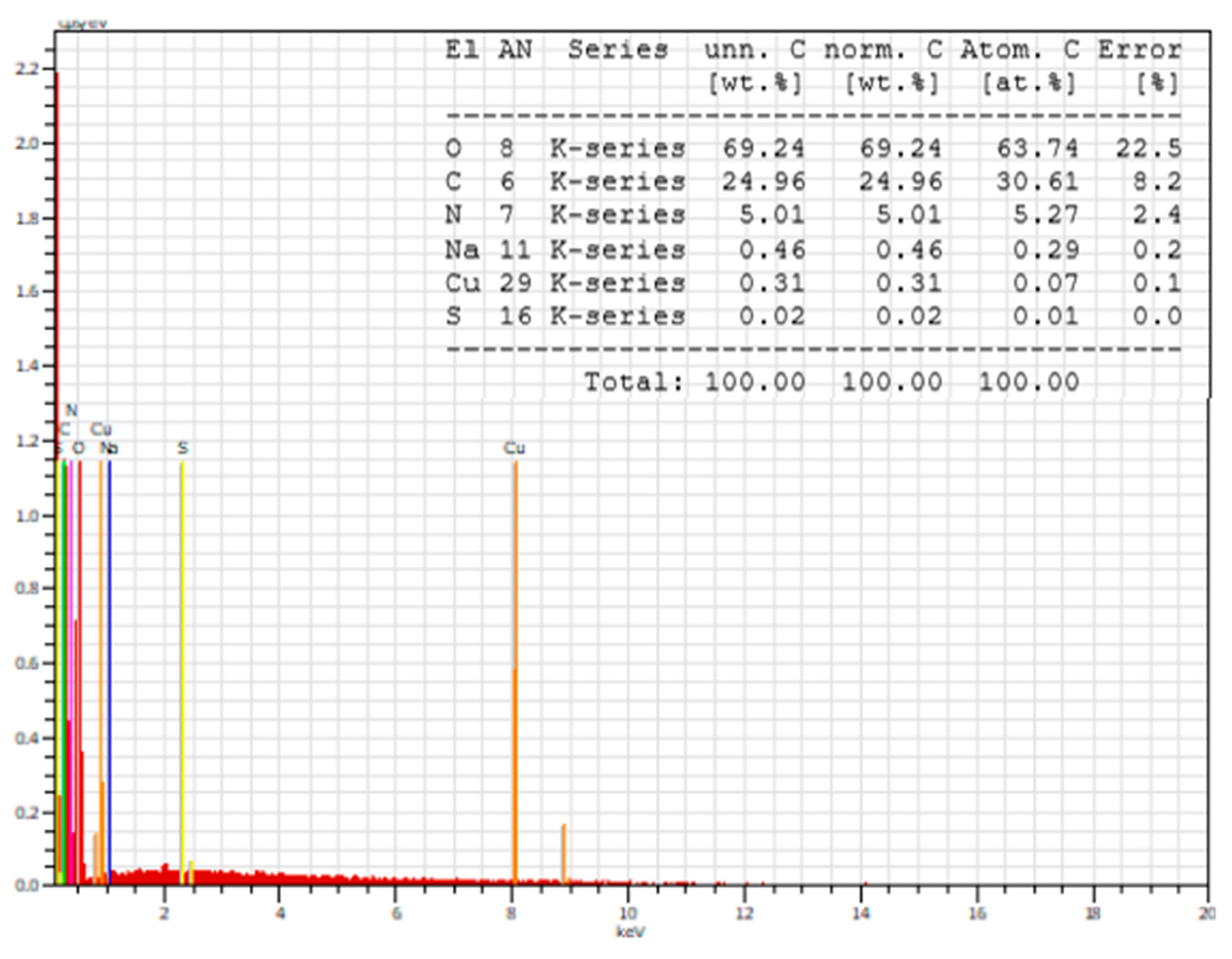

| Compounds | λabs [nm] | ε, [L/(mol.cm)] at 272 nm | ε, [L/(mol.cm)] at 537 nm | λem [nm] | Stokes Shift [cm−1] | Percentage Increase (%) in Stokes Shift Relative to That of the Peptides Given in [23] |
|---|---|---|---|---|---|---|
| Cu-L1 (C-V) | 272 | 5.40 × 106 | 226 | 357 | 8753 | 54.65 |
| Cu-L2 (AC-V) | 270 | 6.90 × 106 | 302 | 358 | 9104 | 54.65 |
| Cu-L3 (H-V) | 274 | 4.75 × 106 | 328 | 359 | 8641 | 48.78 |
| Cu-L4 (AH-V) | 273 | 6.25 × 106 | 181 | 360 | 8852 | 56.67 |
| Cu-L5 (NH7C) | 275 | 7.65 × 106 | 282 | 359 | 8508 | 48.51 |
| Cu-L6 (NCH7) | 275 | 8.01 × 106 | 451 | 359 | 8508 | 48.51 |
| Species | CV (L1) | AC-V (L2) | H-V (L3) | AH-V (L4) | NH7C (L5) | NCH7 (L6) |
|---|---|---|---|---|---|---|
| pK a | ||||||
| pKa(1–12) | 5.18 | 5.43 | 4.75 | 4.84 | 5.07 | 4.78 |
| a b | ||||||
| Cu(II) | 0.22 | 0.10 | 0.13 | 0.18 | 0.21 | 0.11 |
| lgβ c | ||||||
| Cu(II) | 4.92 | 5.72 | 5.31 | 5.18 | 5.09 | 5.65 |
| Organometallic Compound | Frozen Solution pH = 7.21 | Frozen Solution pH = 10.98 | Solid State |
|---|---|---|---|
| Cu(II)-AHV | g‖—not determined g⊥~2.063 AII—not determined | gz = 2.180 gx = 2.090 gy = 2.056 AII = 20.1 mT | g‖ = 2.345 g⊥ = 2.074 AII = 13.3 mT |
| Cu(II)-HV | After dissolving the complex g‖ = 2.24 g⊥ = 2.065 AII = 16.0 mT | gz = 2.170 gx = 2.075 gy = 2.036 AII = 20.6 mT | g‖ = 2.345 g⊥ = 2.070 AII = 13.3 mT |
| Complex forming in solution g‖ = 2.24 g⊥ = 2.065 AII = 16.0 mT | - | - | |
| Cu(II)-CV | After dissolving the complex g‖ = 2.242 g⊥~2.065 AII = 16.0 Mt Complex forming in solution g‖—not determined g⊥~2.065 AII—not determined | gz = 2.170 gx = 2.072 gy = 2.033 AII = 20.6 mT | gz = 2.331 gx = 2.14 gy = 2.035 AII = 14.4 mT gz = 2.171 gx = 2.082 gy = 2.034 AII = 20.8 mT |
| Cu(II)-ACV | g‖—not determined g⊥~2.067 AII- not determined | gz = 2.176 gx = 2.072 gy = 2.042 AII = 20.6 mT | g‖ = 2.32 g⊥ = 2.06 AII = 13.0 mT |
| Cu(II)-NH7C | g‖—not determined g⊥~2.069 AII—not determined | gz = 2.181 gx = 2.065 gy = 2.042 AII = 20.0 mT | g‖ = 2.35 g⊥ = 2.065 AII = 13.0 mT |
| Cu(II)-NCH7 | g‖—not determined g⊥~2.067 AII—not determined | gz = 2.190 gx = 2.07 gy = 2.038 AII = 20.0 mT |
| Virus | Δlog 30 min | Δlog 60 min | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cu-CV | Cu-HV | Cu-ACV | Cu-AHV | Cu-NH7C | Cu-NCH7 | Cu-CV | Cu-HV | Cu-ACV | Cu-AHV | Cu-NH7C | Cu-NCH7 | |
| HRSV-2 | 0.1 | 0.1 | 0.1 | 0 | 0 | 0.1 | 1.0 | 0.8 | 1.1 | 0.8 | 1.2 | 1.0 |
| HAdV-5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Compound | Cytotoxicity |
|---|---|
| CC50 (µM/mL) in HEp-2 Cells Peptide [23]/Cu-Peptide | |
| C-V/Cu-CV | 10.7/38 |
| H-V/Cu-HV | 12.2/49.7 |
| AC-V/Cu-ACV | 110/36.6 |
| AH-V/Cu-AHV | 106/9.0 |
| NH7C/Cu-NH7C | 170/13.1 |
| NCH7/Cu-NCH7 | 139/26.6 |
| Ribavirin | 2058 |
| Virus | Δlog 30 min | Δlog 60 min | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cu-CV | Cu-HV | Cu-ACV | Cu-AHV | Cu-NH7C | Cu-NCH7 | Cu-CV | Cu-HV | Cu-ACV | Cu-AHV | Cu-NH7C | Cu-NCH7 | |
| HRSV-2 | 0.1 | 0 | 0 | 0 | 0 | 0 | 0.7 | 0.7 | 0.7 | 0.5 | 0.5 | 0.8 |
| HAdV-5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgieva, S.; Todorov, P.; Staneva, D.; Grozdanov, P.; Nikolova, I.; Grabchev, I. Metal–Peptide Complexes with Antimicrobial Potential for Cotton Fiber Protection. J. Funct. Biomater. 2023, 14, 106. https://doi.org/10.3390/jfb14020106
Georgieva S, Todorov P, Staneva D, Grozdanov P, Nikolova I, Grabchev I. Metal–Peptide Complexes with Antimicrobial Potential for Cotton Fiber Protection. Journal of Functional Biomaterials. 2023; 14(2):106. https://doi.org/10.3390/jfb14020106
Chicago/Turabian StyleGeorgieva, Stela, Petar Todorov, Desislava Staneva, Petar Grozdanov, Ivanka Nikolova, and Ivo Grabchev. 2023. "Metal–Peptide Complexes with Antimicrobial Potential for Cotton Fiber Protection" Journal of Functional Biomaterials 14, no. 2: 106. https://doi.org/10.3390/jfb14020106
APA StyleGeorgieva, S., Todorov, P., Staneva, D., Grozdanov, P., Nikolova, I., & Grabchev, I. (2023). Metal–Peptide Complexes with Antimicrobial Potential for Cotton Fiber Protection. Journal of Functional Biomaterials, 14(2), 106. https://doi.org/10.3390/jfb14020106











